A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs (original) (raw)
Abstract
A widely expressed protein containing UBA (ubiquitin-associated) and UBX (ubiquitin-like) domains was identified as a substrate of SAPKs (stress-activated protein kinases). Termed SAKS1 (SAPK substrate-1), it was phosphorylated efficiently at Ser200 in vitro by SAPK3/p38γ, SAPK4/p38δ and JNK (c-Jun N-terminal kinase), but weakly by SAPK2a/p38α, SAPK2b/p38β2 or ERK (extracellular-signal-regulated kinase) 2. Ser200, situated immediately N-terminal to the UBX domain, became phosphorylated in HEK-293 (human embryonic kidney) cells in response to stressors. Phosphorylation was not prevented by SB 203580 (an inhibitor of SAPK2a/p38α and SAPK2b/p38β2) and/or PD 184352 (which inhibits the activation of ERK1 and ERK2), and was similar in fibroblasts lacking both SAPK3/p38γ and SAPK4/p38δ or JNK1 and JNK2. SAKS1 bound ubiquitin tetramers and VCP (valosin-containing protein) in vitro via the UBA and UBX domains respectively. The amount of VCP in cell extracts that bound to immobilized GST (glutathione S-transferase)–SAKS1 was enhanced by elevating the level of polyubiquitinated proteins, while SAKS1 and VCP in extracts were coimmunoprecipitated with an antibody raised against S5a, a component of the 19 S proteasomal subunit that binds polyubiquitinated proteins. PNGase (peptide N-glycanase) formed a 1:1 complex with VCP and, for this reason, also bound to immobilized GST–SAKS1. We suggest that SAKS1 may be an adaptor that directs VCP to polyubiquitinated proteins, and PNGase to misfolded glycoproteins, facilitating their destruction by the proteasome.
Keywords: peptide N-glycanase (PNGase), p97, stress-activated protein kinase (SAPK), ubiquitin-associated domain (UBA domain), ubiquitin-like domain (UBX domain), valosin-containing protein (VCP)
Abbreviations: DTT, dithiothreitol; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; HEK-293, human embryonic kidney; JNK, c-Jun N-terminal kinase; KESTREL, kinasesubstrate tracking and elucidation; MAP kinase, mitogen-activated protein kinase; MGP, misfolded glycoprotein; ORF, open reading frame; PEG, poly(ethylene glycol); PNGase, peptide N-glycanase; SAKS1, stress-activated protein kinase substrate-1; SAPK, stress-activated protein kinase; UBA, ubiquitin-associated; UBX, ubiquitin-like; VCP, valosin-containing protein
INTRODUCTION
The human genome encodes approx. 30000 proteins and 500 protein kinases. Since approx. 30% of mammalian proteins are thought to contain covalently bound phosphate, this implies that, on ‘average’, a protein kinase would be expected to phosphorylate approx. 20 substrates in vivo. In fact, the average number of substrates is likely to be far greater, since many proteins are phosphorylated at multiple sites by several protein kinases. However, in only a few cases have a significant number of the physiological substrates of a protein kinase been identified, because really powerful methodologies to tackle this problem are still lacking. Approaches that have been used with some success include the screening of expression libraries for protein kinase substrates [1], yeast two-hybrid analysis [2,3], the screening of protein sequence databases with known consensus sequences for particular protein kinases [4,5] and the use of ATP derivatives that can only be used as substrates by mutated protein kinases [6,7].
We have developed a simple approach to identify putative substrates of protein kinases that we have termed KESTREL for kinase substrate tracking and elucidation [8]. In this method, cell extracts that have been fractionated by a single step of ion-exchange chromatography are phosphorylated for a few minutes in the absence or presence of the protein kinase of interest and [γ32P]ATP of very high specific radioactivity. In order to enhance the probability of detecting physiologically relevant substrates, closely related protein kinases are studied in parallel to identify substrates that are phosphorylated relatively selectively. Wherever possible, MnATP is used instead of MgATP to reduce ‘background’ phosphorylation. Substrates that are phosphorylated rapidly, stoichiometrically and at similar rates to authentic physiological substrates are purified and identified by tryptic mass fingerprinting. The phosphorylation site(s) is identified and phospho-specific antibodies are raised that are sufficiently sensitive to investigate whether the endogenous protein is phosphorylated at that site(s) in vivo, in response to signals that activate the protein kinase [8]. Relatively specific inhibitors of the protein kinase and/or cells that do not express the protein kinase are then used to examine whether the protein is a physiological substrate for a particular protein kinase. Using this method, we identified EF2 (elongation factor 2) kinase as a likely substrate of SAPK (stress-activated protein kinase) 4 (also called p38δ), and showed that it was inactivated in response to cellular stresses and pro-inflammatory cytokines as a result of the phosphorylation of Ser359, the residue that was targeted specifically by SAPK4/p38δ in vitro [8].
SAPK4/p38δ is one member of a subfamily of MAP kinases (mitogen-activated protein kinases) that become activated when cells are exposed to a cell-damaging agent, such as osmotic shock, chemical insult or UV radiation, or stimulated by pro-inflammatory cytokines. They include the isoforms of SAPK1/JNK (c-Jun N-terminal kinase), SAPK2a/p38α and SAPK2b/p38β2 (collectively referred to as SAPK2/p38), SAPK3/p38γ and SAPK4/p38δ [9]. The SAPK2/p38 isoforms are potently inhibited by the anti-inflammatory drug SB 203580 [10], but the other SAPKs are inhibited far less strongly or not at all [11].
In the present paper, we have used KESTREL to identify a new physiological substrate for SAPKs, termed SAKS1 (SAPK substrate-1). This protein contains a UBA (ubiquitin-associated) domain that binds to polyubiquitin in vitro (and presumably to polyubiquitinated proteins in vivo) and a UBX (ubiquitin-like) domain that binds to VCP (valosin-containing protein), a member of the AAA family of ATPases. We also show that VCP interacts with PNGase (peptide N-glycanase) in cell extracts, an enzyme that removes high-mannose-containing oligosaccharides from MGPs (misfolded glycoproteins). Since proteins that are covalently linked to Lys48 of ubquitin are targeted to the proteasome for degradation, we suggest that SAKS1 is a scaffolding protein that targets VCP (and hence PNGase) to polyubiquitinated proteins. This may promote the VCP-dependent unfolding of such proteins and (in the case of MGPs) their deglycosylation by PNGase, thereby facilitating their proteolytic destruction.
MATERIALS AND METHODS
Materials
[γ32P]ATP and materials for protein purification were obtained from Amersham Biosciences (Little Chalfont, Bucks., U.K.), unlabelled ATP and DTT (dithiothreitol) were from Roche Molecular Biochemicals (Lewes, U.K.), pre-cast polyacrylamide gels, running buffer and transfer buffer were from Invitrogen (Paisley, U.K.), Immobilon P membranes were from Millipore (Bedford, U.K.) and cell culture media were from Biowhittaker (Wokingham, U.K.). SB 203580, MG-132 and proteasome inhibitor-1 were obtained from Calbiochem (Nottingham, U.K), microcystin LR and myelin basic protein were from Gibco-BRL (Paisley, U.K.). Tetra-ubiquitin was purchased from Affiniti Research Products (Exeter, U.K.), sodium arsenite was obtained from Fisher Scientific (Loughborough, U.K.), and Complete™ proteinase inhibitor cocktail tablets were from Roche. Peptides were synthesized by Dr G. Bloomberg (University of Bristol, Bristol, U.K.). PD 184352 was made by custom synthesis. Other chemicals were purchased from Merck (Poole, U.K.) or Sigma-Aldrich (Poole, U.K.).
Antibodies
The peptide PGPVPSpSPSQEPP (where pS is phosphoserine), corresponding to residues 195–207 of SAKS1, was coupled separately to BSA and keyhole-limpet haemocyanin, these were then mixed and antibodies were raised in a rabbit at Diagnostics Scotland (Penicuik, U.K.). The antisera were affinity-purified on a phosphopeptide-antigen–Sepharose column, then passed through another column to which the unphosphorylated form of the peptide had been bound. The flowthrough fractions were collected and used at 1.0 μg/ml for immunoblotting. An antibody that immunoprecipitates SAKS1 and recognizes phosphorylated and non-phosphorylated SAKS1 protein equally well was generated by injecting sheep with GST (glutathione S-transferase)-tagged SAKS1. The antibody was affinity-purified on GST–Sepharose and then on GST–SAKS1–Sepharose.
Antibodies that recognize SAPK2a/p38 phosphorylated at Thr180 and Tyr18, and SAPK1/JNK phosphorylated at Thr183 and Tyr185, were purchased from New England Biolabs (Hitchin, U.K.), the anti-VCP antibody was from BD Biosciences (Oxford, U.K.) and the anti-His6 antibody was from Novagen (Nottingham, U.K.). The anti-FLAG monoclonal antibody M2 was obtained from Sigma-Aldrich, and anti-S5a (also called Rpn10) and anti-S4 (also called Rpt2) were from Affiniti Research Products. Rabbit anti-sheep IgG, goat anti-rabbit IgG, and rabbit anti-mouse IgG horseradish-peroxidase-conjugated antibodies were from Perbio Science UK (Tattenhall, Cheshire, U.K.) and an anti-ubiquitin antibody was from Dako (Ely, U.K.).
Purification of a 40 kDa substrate for SAPK3/p38γ from rabbit skeletal muscle
Rabbit muscle extracts (140 ml, 15 mg/ml), prepared as described previously [12], were passed through a 0.2-μm-pore-size filter, applied to a 200 ml column of SP-Sepharose at ambient temperature and washed with buffer A [30 mM Mops/NaOH, pH 7.0, 0.1 mM EGTA, 5% (v/v) glycerol, 0.1% (v/v) 2-mercaptoethanol and 0.03% (w/v) Brij 35] until no further protein was detected in the eluate. Bound proteins were eluted with buffer A containing 1 M NaCl, and the solution was fractionated from 0 to 6, 6 to 12, 12 to 18, 18 to 24 and 24 to 30% (w/v) PEG [poly(ethylene glycol)]-6000. The 18–24% PEG-6000 pellet was collected by centrifugation at 10000 g for 30 min at 4 °C, resuspended in buffer A and chromatographed on a 20 ml Source 15S column. The column was developed with a 162 ml hyperbolic salt gradient to 1 M NaCl at a flow rate of 2 ml/min. Fractions of 1.7 ml were collected, and an aliquot of each was incubated for 5 min at 30 °C with 10 m-units of activated SAPK3/p38γ and 20 nM Mn[γ-32P]ATP (4×106 c.p.m./nmol) in a total volume of 0.03 ml. Reactions were stopped by denaturation in SDS, subjected to SDS/PAGE, transferred to Immobilon P membrane and autoradiographed. Fractions containing the 43 kDa substrate (eluting at 0.05–0.07 M NaCl) were pooled and exchanged into buffer B [30 mM Mes/NaCl, pH 6.0, 0.1 mM EGTA, 5% (v/v) glycerol, 0.1% (v/v) 2-mercaptoethanol and 0.03% (w/v) Brij 35] using a Vivascience spin column (Millipore). The material was then chromatographed on a 1 ml column of heparin–Sepharose (HiTrap) equilibrated in the same buffer. The column was developed with a 30 ml gradient to 1 M NaCl in buffer B at a flow rate of 1 ml/min. The peak fraction containing the 43 kDa substrate (eluting at 0.3 M NaCl) was diluted in buffer C (30 mM Tris/HCl, pH 7.5, 1 mM DTT), concentrated to 0.1 ml by passage through a Centricon 10 membrane using a Vivascience spin column and analysed as described in the Results section.
DNA constructs and protein expression
Protein kinases were expressed in Escherichia coli strain BL21 and purified as described previously [13]. Human cDNA encoding full-length SAKS1 and a variety of subfragments were cloned and inserted into pGEX6P1 vectors for expression in E. coli and mammalian cells. Full details are provided in the Supplementary section (available at http://www.BiochemJ.org/bj/384/bj3840391add.htm). A vector expressing His6–VCP (human) was kindly provided by Dr Colin Gordon, MRC Human Genetics Unit, Edinburgh.
GST–SAKS1, GST-fusions of fragments of SAKS1, GST–FLAG–PNGase and His6–VCP were transformed into E. coli BL21 and expression was induced with IPTG (isopropyl β-D-thiogalactoside) for 16 h at 26 °C. Recombinant proteins were purified on glutathione–Sepharose (GST-fusion proteins) or Ni2+-nitrilotriacetate agarose beads (His6–VCP), dialysed into 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA, 50% (v/v) glycerol and 0.1% (v/v) 2-mercaptoethanol, and stored at −20 °C.
Separation of phosphorylated from unphosphorylated SAKS1
GST–SAKS1 (2 mg/ml) was phosphorylated to 0.3 mol of phosphate per mol of protein by incubation for 60 min at 30 °C with 2 units/ml SAPK3/p38γ and MgATP in a reaction volume of 0.25 ml [13]. After dilution to 0.1 mg/ml in the manufacturer's ‘phosphoprotein lysis buffer’, the partially phosphorylated GST– SAKS1 was applied to a phosphoprotein-purification column (Qiagen, Crawley, U.K.). The flowthrough fractions did not contain phosphorylated GST–SAKS1, as judged by immunoblotting with the phosphospecific antibody and these fractions were discarded. The phospho-GST–SAKS1, which was retained on the column, was eluted with ‘phosphoprotein elution buffer’ and used for the binding experiments mentioned in the Results section.
Fibroblasts from mice deficient in particular protein kinases
Primary fibroblasts were derived from E13.5 embryos from wild-type mice and knockout mice that do not express either SAPK3/p38γ or SAPK4/p38δ, and were cultured as described [14]. Information on the generation and characterization of these mice is available on request from Ana Cuenda, MRC Protein Phosphorylation Unit, Dundee. Immortalized embryonic fibroblasts that do not express JNK1 or JNK2 [15] were maintained in the same culture medium as described in [14].
Cell culture, cell transfection and cell lysis
Mammalian HEK-293 (human embryonic kidney) fibroblasts were cultured at 37 °C in DMEM (Dulbecco's modified Eagle's medium), supplemented with 10% foetal bovine serum, 50 units/ml of penicillin G and 50 μg/ml of streptomycin (Life Technologies). Cells were exposed to 0.5 M sorbitol, 0.5 mM sodium arsenite or UV-C radiation (200 J/m2), followed by incubation at 37 °C for 30 min under 5% CO2, as indicated in the Figure legends. Transfection of HEK-293 cells was carried out using the calcium phosphate method [16] with 2 μg of FLAG–PNGase plasmid DNA or 4 μg of His6–VCP DNA.
All the cells used in the present study were lysed in 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 50 mM sodium β-glycerophosphate, 0.27 M sucrose, 1 μM mycrocystin-LR, 0.1 mM PMSF, 1% (v/v) Triton X-100, 0.1% (v/v) 2-mercaptoethanol and Complete™ proteinase inhibitor cocktail. Lysates were centrifuged at 13000 g for 15 min at 4 °C, the supernatants (termed cell extract) were removed, quick-frozen in liquid nitrogen and stored at −80 °C until use. Where required, cells were pre-incubated for 1 h with 10 μM SB 203580 and/or 2 μM PD 184352 before stimulation.
Immunoprecipitation of proteins from cell extracts
Cell extracts (0.5 mg of protein) were immunoprecipitated with 1 μg of anti-SAKS1 or anti-S5a antibody coupled to Protein-G–Sepharose. For immunoprecipitation of SAPK3/p38γ, immunoprecipitations were carried out using 4 μg of anti-GST–SAPK3 in the presence of 0.5% (w/v) SDS. After incubation for 1 h at 4 °C, the captured proteins were centrifuged at 13000 g, the supernatants were discarded and the beads washed twice in lysis buffer containing 0.5 M NaCl, then twice in lysis buffer alone. Samples were denatured in SDS subjected to SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted using the ECL® (enhanced chemiluminescence) detection system (Amersham Biosciences).
Protein kinase assays
Protein kinase assays were carried out as described previously [8,13]. One unit of activity was that amount which catalysed the phosphorylation of 1 nmol of phosphate into myelin basic protein in 1 min.
RESULTS
Identification of a substrate of SAPK3/P38γ
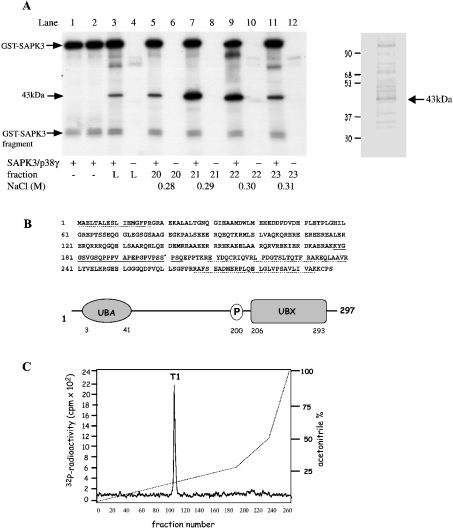
This project started as an attempt to identify putative substrates for SAPK3/p38γ, a MAP kinase family member of unknown function present at high concentrations in skeletal muscle [17,18]. This led to the detection of a 43 kDa substrate for SAPK3/p38γ after chromatography of the 18–24% PEG fraction on Source 15S (results not shown). The fractions containing the substrate were pooled and subjected to chromatography on HiTrap heparin–Sepharose from which it eluted at 0.3 M NaCl (Figure 1A). The peak fraction was subjected to SDS/PAGE, which revealed that the 32P-labelled protein co-migrated with a major protein staining band (Figure 1A, right-hand panel). This protein was eluted from the gel, digested with trypsin and the masses of the resultant peptides used to interrogate the NCBI and Swiss-Prot data- bases (see Supplementary Table 1 at http://www.BiochemJ.org/bj/384/bj3840391add.htm). This provisionally identified the protein as the product of an ORF (open reading frame) (GenBank® accession number BC000902), which encoded a widely expressed protein of unknown function that contains a UBA domain at its N-terminus and a UBX domain [19] at its C-terminus (Figure 1B). The identity of the protein, termed SAKS1, was confirmed by Edman sequencing of two of these peptides, RPLQELGLVP and LPDGTSLTQTFR (Figure 1B).
Figure 1. Characterization of the 43 kDa substrate of SAPK3/p38γ as SAKS1.
(A) Skeletal muscle extracts were chromatographed on SP-Sepharose, fractionated from 18 to 24% PEG and chromatographed on Source 15 S. The fractions containing the 43 kDa substrate of SAPK3/p38γ were pooled and applied to heparin–Sepharose (see the Materials and methods section). Aliquots of each fraction (1 μl) were phosphorylated for 5 min at 30 °C in a 0.03 ml incubation with 2 mM MnCl2 and 20 nM [γ-32P]ATP (4×106 c.p.m./nmol) in the presence or absence of SAPK3/p38γ. Reactions were terminated in SDS, subjected to SDS/PAGE, transferred on to PVDF membranes and autoradiographed. Lanes 1 and 2 show autophosphorylation of GST–SAPK3/p38γ, and lanes 3 and 4 show the pooled fractions from Source 15 S that were applied to heparin–Sepharose (L, load). Lanes 5–12 are fractions from the heparin–Sepharose column containing the 43 kDa substrate, which were incubated with MnATP in the presence (+) or absence (−) of SAPK3/p38γ. The positions of autophosphorylated GST–SAPK3/p38γ, a small proteolytic fragment of GST–SAPK3/p38γ, and the 43 kDa substrate are marked. An aliquot of fraction 21 was also subjected to SDS/PAGE and stained with SYPRO orange (right-hand panel). (B) Amino acid sequence and domain structure of the 43 kDa protein from (A), termed SAKS1. Peptides identified by MS (broken lines) or Edman sequencing (solid line) and the phosphorylation site Ser200 (*) are indicated. (C) Identification of the phosphorylation site on SAKS1. SAKS1 from heparin–Sepharose was phosphorylated for 60 min at 30 °C with SAPK3/p38γ and 0.1 mM [γ-32P]ATP (106 c.p.m./nmol) and subjected to SDS/PAGE as in (A). The phosphorylated band was excised, digested with trypsin, and the peptides were separated by chromatography on a Vydac C18 column equilibrated in 0.1% (v/v) TFA (trifluoroacetic acid). The column was developed with an acetonitrile gradient in 0.1% TFA (broken line), and fractions of 0.4 ml were collected. The site of phosphorylation in the single 32P-labelled peptide T1 (solid line) was identified as described in the text.
SAKS1 was maximally phosphorylated, digested with trypsin and chromatographed on a Vydac C18 column to reveal a single peak of 32P-radioactivity T1 (Figure 1C). MS showed that T1 contained a mixture of two peptides corresponding to residues 178–209 and 179–209 of human SAKS1 plus one phosphate group (results not shown). The two peptides arise from incomplete cleavage of the Lys-Lys bond between residues 177 and 178. Solid-phase sequencing identified the site of phosphorylation as Ser200 (results not shown). This is as expected, since SAPK3/p38γ, like other MAP kinase family members, normally phosphorylates serine and threonine residues that are followed by proline, and Ser200 lies in the only Ser-Pro sequence in the peptide.
SAKS1 was phosphorylated by SAPK4/p38δ and SAPK1/JNK at slightly lower rates then SAPK3/p38γ, but only phosphorylated weakly by SAPK2a/p38, SAPK2b/p38β2 or ERK (extracellular-signal-regulated kinase) 2, when the activity of each protein kinase was matched to myelin basic protein. SAPK4/p38δ and SAPK1/JNK1 phosphorylated the same site in SAKS1 (results not shown).
An immunoprecipitating antibody raised against the full-length SAKS1 protein was used in immunoblotting experiments to show that SAKS1 is widely distributed in mammalian cells (see Supplementary Figure 1A at http://www.BiochemJ.org/bj/384/bj3840391add.htm). A phospho-specific antibody was also raised that only recognized SAKS1 phosphorylated at Ser200 (see Supplementary Figure 1B at http://www.BiochemJ.org/bj/384/bj3840391add.htm). Both antibodies were then used to examine whether SAKS1 was phosphorylated at Ser200 in HEK-293 cells where it is highly expressed (see Supplementary Figure 1A at http://www.BiochemJ.org/bj/384/bj3840391add.htm).
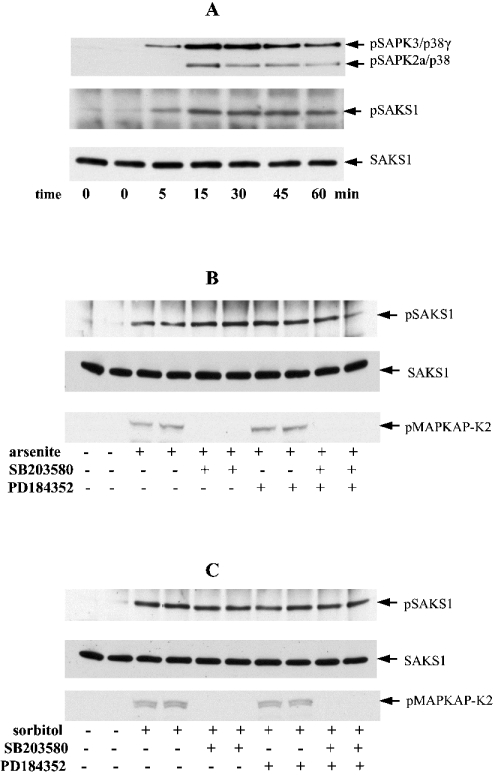
Osmotic shock (0.5 M sorbitol) induced the activation of SAPK3/p38γ and other SAPKs (Figure 2A) after 5 min, which became maximal after 15 min, and was sustained for at least 1 h. The phosphorylation of SAKS1 at Ser200 followed similar kinetics (Figure 2A). In order to obtain information about the protein kinase(s) responsible for phosphorylating SAKS1, we incubated HEK-293 cells with SB 203580 before exposure to osmotic shock (0.5 M sorbitol) and a chemical insult (sodium arsenite) [11,16]. These experiments revealed that the phosphorylation of SAKS1 induced by sodium arsenite (Figure 2B), osmotic shock (Figure 2C) or UV-C radiation (results not shown) was unaffected by SB 203580, consistent with the involvement of another SAPK(s) [21]. In contrast, SB 203580 prevented the phosphorylation of MAPKAP-K2 (MAP-kinase-activated protein kinase-2), a known substrate of SAPK2a/p38 [10], in the same cells (Figures 2B and 2C). PD 184352, which inhibits activation of the MAP kinases ERK1/ERK2 [22] had no effect on arsenite- or osmotic-shock-induced phosphorylation of SAKS1 in the presence or absence of SB 203580 (Figures 2B and 2C). Consistent with this finding, EGF (epidermal growth factor), a strong activator of ERK1/ERK2 and only a weak activator of SAPKs, did not induce any phosphorylation of SAKS1 (results not shown).
Figure 2. Activation of SAPK3/p38γ and SAPK2a/p38, and phosphorylation of SAKS1 at Ser200, induced by stress in HEK-293 cells.
(A) Cells were exposed to an osmotic shock (0.5 M sorbitol) for the times indicated. SAKS1 and SAPK3/p38γ were immunoprecipitated from the cell extracts (see the Materials and methods section) with antibodies coupled to protein-G–Sepharose. After washing the beads, bound proteins were denatured in SDS and subjected to SDS/PAGE on 10% gels. After transfer on to nitrocellulose, the membranes were immunoblotted with an antibody that recognizes the phosphorylated forms of both SAPK3/p38γ (pSAPK3/p38γ) and SAPK2/p38 (pSAPK2a/p38), an antibody that recognizes the phosphorylated form of SAKS1 (pSAKS1) and an antibody that recognizes phosphorylated and unphosphorylated SAKS1 equally well (SAKS1). pSAPK2/p38 is present because the anti-SAPK3/p38γ antibody immunoprecipitates some of this enzyme. (B, C) HEK-293 cells were incubated for 1 h without (−) or with (+) 10 μM SB 203580 and/or 2 μM PD 184352, then exposed for 60 min to a chemical stress, 0.5 mM sodium arsenite (B), or for 15 min to 0.5 M sorbitol (C). The cell lysates were then immunoblotted as in (A). A further aliquot of cell lysate (50 μg of protein) was immunoblotted (without prior immunoprecipitation) using an antibody that recognizes MAPKAP-K2 (MAP-kinase-activated protein kinase 2) phosphorylated at Thr334 (pMAPKAP-K2).
Phosphorylation of SAKS1 in fibroblasts deficient in different SAPKs
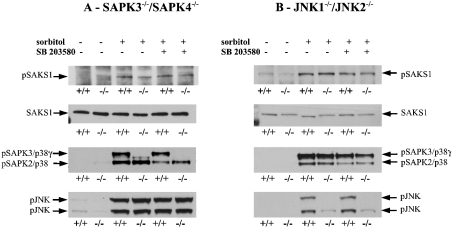
In order to investigate further which protein kinases might mediate the phosphorylation of SAKS1 at Ser200, we carried out experiments using primary fibroblasts from mice that do not express either SAPK3/p38γ or SAPK4/p38δ (J. S. C. Arthur and A. Cuenda, unpublished work), and in immortalized fibroblasts from mice that do not express JNK1 and JNK2 [15]. These studies revealed that osmotic shock induced a similar phosphorylation of SAKS1 at Ser200 in the wild-type or the ‘double-knockout’ fibroblasts (Figures 3A and 3B). The osmotic-shock-induced phosphorylation of Ser200 was unaffected by SB 203580 in both double-knockout fibroblasts, indicating that SAPK2a/p38 or SAPK2b/p38β2 cannot compensate for the lack of SAPK3/p38γ and SAPK4/p38δ or the lack of JNK1 and JNK2.
Figure 3. Osmotic-shock-induced phosphorylation of SAKS1 is not attenuated in fibroblasts that do not express either SAPK3/p38γ and SAPK4/p38δ or JNK1 and JNK2.
(A) Primary mouse fibroblasts were isolated from day-13.5 embryos of wild-type (+/+) mice and mice that do not express SAPK3/p38γ and SAPK4/p38δ (−/−). The fibroblasts were cultured [14] and incubated for 1 h with or without 10 μM SB 203580, then exposed for 30 min to 0.5 M sorbitol. Following cell lysis, SAKS1 was immunoprecipitated from 2 mg of cell-lysate protein, subjected to SDS/PAGE and immunoblotted for the phosphorylation of SAKS1 at Ser200 and with antibodies that recognize the phosphorylated forms of SAPK3/p38γ, SAPK2/p38 and JNK. Antibodies used were against the phosphorylated form of SAKS1 (pSAKS1), both the phosphorylated and unphosphorylated SAKS1 equally (SAKS1), the phosphorylated forms of both SAPK3/p38γ (pSAPK3/p38γ) and SAPK2/p38 (pSAPK2/p38), and the phosphorylated JNK. (B) As in (A), except that immortalized fibroblasts from wild-type mice (+/+) and mice that do not express JNK1 and JNK2 (−/−) were used.
As expected, the active phosphorylated form of SAPK3/p38γ was not detected in cells that do not express SAPK3/p38γ and SAPK4/p38δ (Figure 3A), while the active phosphorylated forms of JNK were not detected in the JNK1/JNK2 double-knockout cells (Figure 3B). The lack of expression of SAPK3/p38γ and SAPK4/p38δ did not affect the extent of activation of JNK (Figure 3A), while the lack of expression of JNK1 and JNK2 did not affect the activation of SAPK3/p38γ and SAPK2/p38 (Figure 3B).
SAKS1 interacts with polyubiquitin and VCP
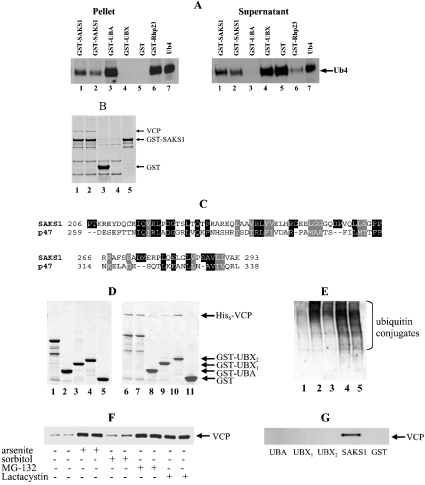
The UBA domain binds to polyubiquitin or mono-ubiquitin, and we therefore investigated whether this was also the case for the UBA domain of SAKS1. SAKS1 or Rhp23 (a protein known to interact with polyubiquitin [23]) bound ubiquitin tetramers (Figure 4A). The isolated UBA domain of SAKS1 bound the ubiquitin tetramers even more strongly, while the isolated UBX domain of SAKS1 or GST did not bind detectably to ubiquitin tetramers (Figure 4A). In contrast, SAKS1 or the isolated UBA domain only bound ubiquitin monomers very weakly (results not shown).
Figure 4. SAKS1 binds ubiquitin tetramers and VCP.
(A) Full-length GST–SAKS1 (lanes 1 and 2), a fragment containing the UBA domain, GST–UBA (3–41) (lane 3), a fragment containing the UBX domain, GST–UBX1 (206–293) (lane 4), GST (lane 5) or GST–Rhp23 (lane 6) each at 0.2 μM were coupled individually to 20 μl of glutathione–Sepharose beads, then incubated with 0.05 μg of tetra-ubiquitin (Ub4) in 0.05 ml 50 mM Tris/HCl, pH 7.5, 50 mM NaCl, 1% (w/v) Triton X-100, 10% (v/v) glycerol and 0.1 mg/ml BSA. After mixing for 30 min at 4 °C and centrifugation, the supernatant was retained and the beads washed three times. The supernatant and bound material were denatured in SDS, electrophoresed on a 4–12% polyacrylamide gradient gel, transferred on to nitrocellulose membranes and immunoblotted for the presence of ubiquitin. Lane 7, 0.05 μg of Ub4 control. The GST–Rhp23 was included as a protein known to bind polyubiquitin [23]. (B) GST–SAKS1 (lanes 1 and 2), GST (lane 3), each at 10 μg, or buffer (lane 4) were coupled to glutathione–Sepharose beads and incubated for 1 h at 4 °C with 4 mg of HEK-293 cell-lysate protein. The suspensions were centrifuged, the supernatants were discarded, and the beads were washed twice in lysis buffer containing 0.25 M NaCl and twice in lysis buffer alone. Samples were denatured in SDS, electrophoresed on a 4–12% polyacrylamide gel and stained with colloidal Coomassie Blue (Invitrogen). Lane 5 is purified GST–SAKS1 and the 97 kDa protein (VCP) binding specifically to GST–SAKS1 is indicated. (C) The UBX domains of SAKS1 and p47 are compared with identities highlighted in black and conservative replacements in grey. (D) A 10 μg volume of bacterially expressed full-length GST–SAKS1 (lane 1), GST–UBA (3–41) (lane 2), GST–UBX1 (206–293) (lane 3), GST–UBX2 (180–297) (lane 4) or GST (lane 6) was denatured in SDS, electrophoresed on a 4–12% polyacrylamide gradient gel and stained with colloidal Coomassie Blue. A volume of 10 μg of bacterially expressed full-length GST–SAKS1 (lanes 6 and 7), GST–UBA (3–41) (lane 8) GST–UBX1 (206–293) (lane 9), GST–UBX2 (180–297) (lane 10) or GST (lane 11) were coupled to 20 μl of glutathione–Sepharose in 0.5 ml of 50 mM Tris/HCl, pH 7.5, 0.15 M KCl, 1 mM DTT, 1 mM MgCl2, 1 mM ATP and 0.03% (w/v) Brij 35. After washing the beads, 10 μg of His6–VCP in 0.5 ml of buffer was added and the suspension was mixed end-over-end for 30 min at 21 °C. After centrifugation for 1 min at 13000 g, the supernatant was removed, the beads were washed, and attached proteins were subjected to SDS/PAGE as in (B). Protein staining bands migrating more rapidly than GST–SAKS1 (lanes 1 and 2) are proteolytic fragments truncated at the C-terminus. (E) HEK-293 cells were left unstimulated (lane 1), exposed for 1 h to 0.5 mM sodium arsenite (lane 2), incubated for 30 min with 0.5 M sorbitol (lane 3) or incubated for 3 h with 20 μM MG-132 (lane 4) or 20 μM lactacystin (lane 5). Following cell lysis, 15 μg of lysate protein was subjected to SDS/PAGE and immunoblotted with an anti-ubiquitin antibody. (F) Cells were left unstimulated or exposed to sodium arsenite, sorbitol, MG-132 or lactacystin as in (E). Following cell lysis, 3 mg of lysate protein was incubated with 2 μg of GST–SAKS1 bound to glutathione–Sepharose, and the VCP bound to glutathione–Sepharose was analysed by immunoblotting. (G) Cells were incubated with sodium arsenite as in (B) and the VCP associated with full-length GST–SAKS1, GST–UBA, GST–UBX1 (206–293), GST–UBX2 (180–297) and GST were immunoblotted as in (F).
To investigate if any other proteins interact with SAKS1, we studied whether proteins present in HEK-293 cell extracts were capable of binding to added GST–SAKS1. These experiments revealed a 97 kDa protein that bound specifically to SAKS1 (Figure 4B), which was identified by tryptic digestion and MS as VCP (also called p97) (see Supplementary Table 2 at http://www.BiochemJ.org/bj/384/bj3840391add.htm).
VCP is reported to interact with another UBX-domain-containing protein, termed p47 [24]. Since the isolated UBX domain of p47 (see Figure 4C) is capable of interacting with VCP in vitro [25], we examined whether this was also the case for SAKS1. VCP bound to full-length SAKS1 and equally well to UBX2 (residues 180–297), a fragment that contains the UBX domain, but not the UBA domain (Figure 4D). VCP also bound to UBX1 (residues 206–293), the minimal UBX domain (residues 206–293), although more weakly (Figure 4D). These results indicate that VCP binds to the UBX domain of SAKS1, and suggest that the conformation of the UBX domain may be stabilized by amino acid sequences that surround it. VCP did not bind to the isolated UBA domain (Figure 4D).
To study the effect of phosphorylation on the interaction of SAKS1 with VCP and ubiquitin, we phosphorylated SAKS1 in vitro with SAPK3/p38γ and separated phosphorylated from unphosphorylated protein as described in the Materials and methods section. We then examined the binding of phospho-SAKS1 and dephospho-SAKS1 to their binding partners as in Figure 4(D). These experiments showed that phosphorylation of SAKS1 did not impair its ability to bind to ubiquitin tetramers. However, in several independent experiments, phosphorylated SAKS1 was found to bind only approx. 50% of the VCP bound by unphosphorylated SAKS1 (results not shown).
Binding of VCP to SAKS1 is enhanced by MG-132 and sodium arsenite, but not by osmotic shock
VCP is believed to facilitate the degradation of polyubiquitinated proteins by the proteasome [26]. In this process, VCP is thought to bind to the adaptor protein p47, which itself interacts with the Ufd1–Np14 complex that interacts with polyubiquitinated proteins [24]. It was therefore of interest to investigate whether SAKS1 might play a role analogous to p47. We incubated HEK-293 cells with the proteasome inhibitor MG-132, in order to elevate the intracellular level of polyubiquitinated proteins (Figure 4E). This caused a striking increase in the amount of endogenous VCP in the extracts that bound to added exogenous GST–SAKS1 (Figure 4F). Similar results were obtained with the structurally unrelated proteasome inhibitors, termed lactacystin (Figure 4F) and PI-1 (results not shown). Sodium arsenite has also been reported to inhibit the proteasome [27] and, like MG-132, incubating HEK-293 cells with this compound elevated the level of polyubquitinated proteins (Figure 4E).
The enhanced binding of VCP to added GST–SAKS1 in lysates from arsenite-treated cells was only observed with the full-length GST–SAKS1, and not with the isolated UBA or UBX domains (Figure 4G), even with GST–UBX2 that bound to VCP in vitro (Figure 4D). These observations are considered further in the Discussion.
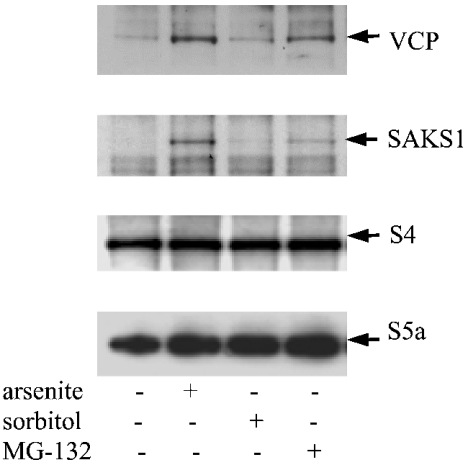
Endogenous SAKS1 and VCP in cell extracts co-immunoprecipitate with the S5a subunit of the 19 S proteasome
Polyubiquitinated proteins are known to interact with the S5a subunit of the 19 S proteasome [28]. If SAKS1 binds to polyubiquitinated proteins and VCP in cells, it would be predicted that SAKS1 and VCP should also be associated with the proteasome. Indeed, some of the VCP in cells has previously been reported to co-sediment with the proteasome during density-gradient centrifugation [29]. We therefore immunoprecipitated the endogenous S5a protein from HEK-293 cell extracts, and looked for the presence of SAKS1 and VCP by immunoblotting. These experiments showed that the endogenous SAKS1 and VCP in HEK-293 cell extracts were both immunoprecipitated by the anti-S5a antibody (Figure 5). The amount of SAKS1 and VCP in the S5a immunoprecipitates was enhanced by pre-treatment of the cells with MG-132 (Figure 5), lactacystin and PI-1 (results not shown).
Figure 5. SAKS1, VCP and S4 are present in anti-S5a immunoprecipitates.
HEK-293 cells were incubated without (−) or with (+) sodium arsenite, sorbitol or MG-132, as in Figure 4(E), and lysed. The S5a protein was immunoprecipitated from 3 mg of cell-lysate protein, and the immunoprecipitates were analysed for the presence of VCP, SAKS1, S4 and S5a by immunoblotting.
S4, another component of the 19 S proteasomal subunit, was also co-immunoprecipitated with the anti-S5a antibody (Figure 5), demonstrating that this antibody had immunprecipitated the intact proteasomal complex.
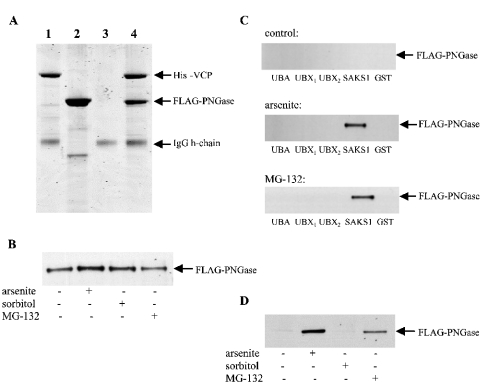
PNGase also interacts with SAKS1
While our work was in progress, it was reported that PNGase interacted with SAKS1 in yeast two-hybrid analysis [30], but whether the proteins were capable of interacting in vitro was not investigated. Surprisingly, we could not detect any interaction between bacterially expressed GST–FLAG–PNGase and untagged SAKS1 in vitro (results not shown). In contrast, we found that it was VCP and PNGase that formed a 1:1 complex in vitro. This could be demonstrated by mixing His6–VCP with a molar excess of FLAG–PNGase, followed by immunoprecipitation with an anti-His6 antibody in vitro (Figure 6A) or after co-transfection of FLAG–PNGase and His6–VCP in cells in a manner that did not depend on prior incubation of the cells with proteasome inhibitors (Figure 6B). This suggests that PNGase ‘interacts’ with SAKS1 in yeast two-hybrid analysis, because both proteins interact with VCP. Consistent with this interpretation, the transfected FLAG–PNGase in HEK-293 cell extracts (Figure 6C), like the endogenous VCP (Figure 4G), bound to added full-length GST–SAKS1, provided that the cells had first been incubated with MG-132 or sodium arsenite. The transfected PNGase, like the endogenous VCP, also co-immunoprecipitated with endogenous SAKS1, and this interaction was greatly enhanced in extracts from cells exposed to sodium arsenite or MG-132 (Figure 6D). As for VCP (Figure 4F), the ability of arsenite to enhance the amount of PNGase in cell lysates that bound to added GST–SAKS1 was only observed with full-length GST–SAKS1, and not with the isolated UBA or UBX domains (Figure 6C).
Figure 6. PGNase forms a complex with VCP and therefore interacts indirectly with SAKS1.
(A) Bacterially expressed His6–VCP (2.5 μg) was immunoprecipitated in 0.5 ml of the buffer used in Figure 4(D) using an anti-His6 antibody coupled to protein-G–Sepharose. The supernatant was discarded, the beads were washed once with 1 ml of buffer, and incubated without (lane 1) or with (lane 4) a several-fold molar excess of FLAG–PNGase in 0.5 ml of buffer. The suspensions were mixed end-over-end for 30 min at 21 °C, and the beads were collected and washed as before. After denaturation in SDS, the solubilized proteins were electrophoresed on 4–12% polyacrylamide gradient gels, stained with colloidal Coomassie Blue and destained. Lane 2, same as lane 1 except that His6–VCP was omitted; lane 3, same as lane 1 except that His6–VCP and anti-His6 antibody were both omitted. The position of the IgG heavy (h)-chain is indicated. (B) Vectors expressing FLAG–PNGase and His6–VCP were transfected into HEK-293 cells. The cells were then either left untreated (control) or incubated with sodium arsenite, or MG-132 as in Figures 4(E) and 4(F). The cells were lysed and 1 mg of lysate protein was incubated for 1 h with 0.5 μg of anti-His6 antibody bound to protein-G–Sepharose. The Sepharose beads were washed and immunoblotted with an anti-FLAG antibody as in (B). (C) Vectors expressing FLAG–PNGase were transfected into HEK-293 cells. The cells were then incubated with sodium arsenite, sorbitol or MG-132 as in Figures 4(E) and 4(F). The cells were lysed and 3 mg of lysate protein was incubated with 2 μg of GST, GST–SAKS1 or the GST–SAKS1 fragments used in Figure 4(G), each bound to glutathione–Sepharose. The PNGase bound to glutathione–Sepharose was then analysed by immunoblotting with an anti-FLAG antibody. (D) The endogenous SAKS1 in cell extracts was immunoprecipitated from 2 mg of cell-lysate protein and the amount of FLAG–PNGase present in the immunoprecipitates was analysed by immunoblotting.
DISCUSSION
The present study provides further evidence that KESTREL is a useful method for identifying physiologically relevant phosphorylation sites in proteins. Using this procedure, we identified SAKS1 as a protein that is phosphorylated efficiently at Ser200 by several SAPKs in vitro, but is phosphorylated only poorly by several others. SAKS1 became phosphorylated at Ser200 when cells were exposed to several stresses, and the time course of phosphorylation of Ser200 paralleled the activation of SAPKs (Figure 2A). These experiments establish that SAKS1 is a physiological substrate for one or more proline-directed SAPKs, but the identity of the relevant protein kinase(s) is still unclear. SAPK2a/p38 and SAPK2b/p38β2 are not required, because the phosphorylation of SAKS1 was resistant to SB 203580, a relatively specific inhibitor of these enzymes, while ERK1 and ERK2 could be excluded because the phosphorylation of SAKS1 was unaffected by PD 184352 (Figures 2B and 2C). However, none of the other SAPKs known to be resistant to SB 203580 and PD 184352 (SAPK3/p38γ, SAPK4/p38δ, JNK1 and JNK2) appear to be required either, based on experiments with fibroblasts that do not express pairs of these enzymes (Figure 3). The MAP kinase family member ERK5 becomes activated in response to some cellular stresses [31], but EGF, a much stronger activator of ERK5, failed to elicit any phosphorylation of SAKS1. In summary, our experiments point to the possible involvement of a novel SAPK, although it cannot be excluded that, in the fibroblasts deficient in both JNK1 and JNK2, there is compensation by SAPK3/p38γ and/or SAPK4/p38δ and vice versa.
Although we identified SAKS1 as a substrate for SAPKs in vitro and in cells, we have thus far been unable to identify the role of this modification. The phosphorylation of SAKS1 at Ser200 in vitro does not appear to impair its ability to bind to ubiquitin tetramers, and only decreases to a small extent the binding of VCP to SAKS1 in vitro under the conditions we have studied so far, nor does it appear to affect the ATPase activity of VCP (H. McNeill, unpublished work). It will clearly be critical to understand how the SAKS1-containing complexes really operate in cells before a role for its phosphorylation can be understood.
SAKS1 was only known previously as an ORF encoded within the human genome and, to our knowledge, the protein as such has never been studied before. A clue to its function came from the presence of an N-terminal UBA domain and a C-terminal UBX domain, which together account for 40% of the amino acid sequence (Figure 1B). We have shown that SAKS1 binds to ubiquitin tetramers via the UBA domain (Figure 4A), and much more efficiently than to ubiquitin monomers (results not shown) and appears to bind to VCP via the UBX domain (Figure 4D). VCP is an abundant and widely expressed protein, and is a member of the AAA family of ATPases. It is the homologue of CDC48 in yeast and disruption of this gene [32] or that encoding the Drosophila homologue TER94 [33] is lethal, indicating that it plays one or more roles essential for cell growth and survival.
VCP does not itself bind to polyubiquitinated proteins [23], but has been reported to co-sediment with polyubiquitinated proteins, such as phosphorylated IκB (inhibitory κB) [29]. Since SAKS1 interacts with VCP and polyubiquitin, it is tempting to speculate that SAKS1 functions as an adaptor to couple VCP to polyubiquitinated proteins and the proteasome (Figure 7A). Polyubiquitinated proteins are themselves known to interact with the S5a subunit of the 19 S component of the proteasome [28], and, consistent with the role for SAKS1 proposed above, VCP and SAKS1 were co-immunoprecipitated from cell extracts with anti-S5a antibodies (Figure 5). The role of VCP itself may be to promote the ATP-dependent unfolding of polyubiquitinated proteins, thereby stimulating their destruction by the proteasome, the ATPase activity of VCP providing the energy required for this process [34].
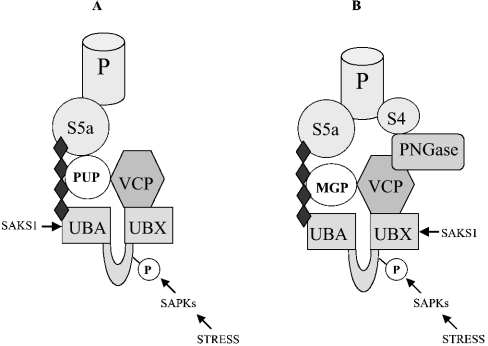
Figure 7. Schematic representation of the proposed role of SAKS1 in localizing VCP to ubiquitinated proteins and PNGase to MGPs.
The four black diamonds denote a polyubiquitin chain, which binds to the UBA domain of SAKS1. (A) VCP is directed to polyubiquitinated proteins (PUPs) via the adaptor protein SAKS1, allowing VCP to unfold PUPs in an ATP-requiring process. PUPs can then be degraded by the proteasome with which they interact via the S5a subunit of the 19 S proteasomal subunit (P). (B) PNGase is directed to polyubiquitinated MGPs via VCP and the adaptor protein SAKS1, allowing PNGase to deglycosylate MGPs, which can then be degraded by the proteasome. PNGase itself is reported to bind to the S4 component of the 19 S proteasome.
We found that the isolated UBX2 domain of SAKS1, like the full-length protein, was able to bind to VCP in vitro (Figure 4D). However, the binding of endogenous VCP in cell extracts to exogenously added GST-fusion proteins could only be observed with full-length GST–SAKS1 (Figure 4G). This could be explained by a lower affinity of VCP for the isolated UBX2 domain compared with the full-length protein, because the concentration of VCP in the cell extracts is far lower than those used to study the binding of VCP in vitro. However, an intriguing alternative idea is that the binding of polyubiquitinated proteins to the UBA domain of SAKS1 may enhance the binding of VCP to the UBX domain. Consistent with this possibility, treating cells with proteasome inhibitors to elevate the level of polyubiquitinated proteins greatly enhanced the amount of VCP in cell extracts that bound to added full-length SAKS1 (Figure 4F).
SAKS1 was identified by others as a protein that interacted with PNGase in yeast two-hybrid analysis [30], but in the present paper, we give evidence that this interaction is likely to be indirect, and is explained by the ability of VCP to interact with both SAKS1 and PNGase (Figures 4B and 6A). PNGase removes high-mannose-containing oligosaccharides from MGPs [30], and our results suggest this may be facilitated by the formation of a complex between PNGase, VCP, SAKS1 and ubiquitinated MGPs, as illustrated schematically in Figure 7(B). PNGase has been reported to bind to the S4 component of the proteasome [30], so that the deglycosylation of MGPs by PNGase, followed by VCP-catalysed unfolding, may facilitate their destruction by the proteasome. In summary, our working hypothesis is that SAKS1 acts as scaffolding protein to enhance the unfolding and proteolytic destruction of a subset of proteins. While the present paper was undergoing revision, a similar conclusion was reached for UBX3, a UBX-domain-containing protein in Saccharomyces cerevisiae (the homologue of mammalian p47) that binds to cdc48 (cell-division control), the homologue of VCP in budding yeast [35].
Online data
Supplementary material
Acknowledgments
We thank Colin Gordon (MRC Human Genetics Unit, Edinburgh, U.K.) for the constructs expressing VCP and Rhp23, and Juan Jose Ventura and Roger Davis (University of Massachusetts, Worcester, MA, U.S.A.) for the immortalized embryonic fibroblasts from wild-type and JNK-knockout mice. We also thank Roger Davis and Graham Warren for helpful suggestions during this study. We thank our colleagues at Dundee for the following reagents: antibodies that recognize SAKS1 and SAPK3/p38γ (Jane Leitch), peptide sequencing and phosphorylation site analysis (Nick Morrice), protein kinases (the protein production team co-ordinated by Hilary McLauchlan and James Hastie) and the cloning of PNGase (Mark Peggie). The work was supported by the UK Medical Research Council, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. A. K. was the recipient of a Long-Term EMBO Fellowship and P. C. is a Royal Society Research Professor.
References
- 1.Fukunaga R., Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLaughlin M. M., Kumar S., McDonnell P. C., Van Horn S., Lee J. C., Livi G. P., Young P. R. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem. 1996;271:8488–8492. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 3.Balendran A., Casamayor A., Deak M., Paterson A., Gaffney P., Currie R., Downes C. P., Alessi D. R. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol. 1999;22:393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- 4.New L., Jiang Y., Zhao M., Liu K., Zhu W., Flood L. J., Kato Y., Parry G. C., Han J. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yaffe M. B., Lebarc G. G., Lai J., Obata T., Volinia S., Cantley L. C. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 6.Shah K., Liu Y., Deirmengian C., Shokat K. M. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Shah K., Yang F., Witucki L., Shokat K. M. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem. Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 8.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyriakis J. M., Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 10.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stress and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 11.Eyers P. A., Craxton M., Morrice N., Cohen P., Goedert M. Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem. Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 12.Cuenda A., Alonso G., Morrice N., Meier R., Cohen P., Nebreda A. R. Purification and cDNA cloning of SAPKK3, the major activator of RK/p38 in stress- and cytokine-stimulated monocytes and epithelial cells. EMBO J. 1996;15:4156–4164. [PMC free article] [PubMed] [Google Scholar]
- 13.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. C. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tournier C., Hess P., Yang D. D., Xu J., Turner T. K., Nimnual A., Bar-Sagi D., Jones S. N., Flavell R. A., Davis R. J. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 16.Cuenda A., Cohen P., Buée-Scherrer V., Goedert M. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPK33 (MKK6): comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertens S., Craxton M., Goedert M. SAP kinase-3, a new member of the family of mammalian stress-activated protein kinases. FEBS Lett. 1996;383:273–276. doi: 10.1016/0014-5793(96)00255-4. [DOI] [PubMed] [Google Scholar]
- 18.Lechner C., Zahalka M. A., Giot J.-F., Moller N. P. H., Ullrich A. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4355–4359. doi: 10.1073/pnas.93.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchberger A., Howard M. J., Proctor M., Bycroft M. The UBX domain: a widespread ubiquitin-like module. J. Mol. Biol. 2001;307:17–24. doi: 10.1006/jmbi.2000.4462. [DOI] [PubMed] [Google Scholar]
- Reference deleted.
- 21.Gum R. J., McLaughlin M. M., Kumar S., Wang Z., Bower M. J., Lee J. C., Adams J. L., Livi G. P., Goldsmith E. J., Young P. R. Acquisition of sensitivity of stress-activated protein kinases to the p38 inbitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 1998;273:15605–15610. doi: 10.1074/jbc.273.25.15605. [DOI] [PubMed] [Google Scholar]
- 22.Seebolt-Leopold J. S., Dudley D. T., Herrera R., Becelaere K. V., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumours in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson R. M., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 24.Meyer H. H., Wang Y., Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan X., Shaw A., Zhang X., Kondo H., Lally J., Freemont P. S., Matthews S. Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J. Mol. Biol. 2001;311:255–263. doi: 10.1006/jmbi.2001.4864. [DOI] [PubMed] [Google Scholar]
- 26.Dai R. M., Li C.-C. H. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 27.Klemperer N. S., Pickart C. M. Arsenite inhibits two steps in the ubiquitin-dependent proteolytic pathway. J. Biol. Chem. 1989;264:19245–19252. [PubMed] [Google Scholar]
- 28.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 29.Dai R., Chen E., Longo D. L., Gorbea C. M., Li C. H. Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 30.Park H., Suzuki T., Lennarz W. Identification of proteins that interact with mammalian peptide:N-glycanase and implicate this hydrolase in the proteasome-dependent pathway for protein degradation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11163–11168. doi: 10.1073/pnas.201393498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe J., Kusuhara M., Ulevich R. J., Berk B. C., Lee J. D. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 1996;271:16586–16590. doi: 10.1074/jbc.271.28.16586. [DOI] [PubMed] [Google Scholar]
- 32.Moir D., Stewart S. E., Osmond B. C., Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leon A., McKearin D. Identification of TER94, and AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol. Biol. Cell. 1999;10:3825–3834. doi: 10.1091/mbc.10.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golbik R., Lupus A. N., Koretke K. K., Baumeister W., Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. J. Biol. Chem. 1999;380:1049–1062. doi: 10.1515/BC.1999.131. [DOI] [PubMed] [Google Scholar]
- 35.Hartmann-Petersen R., Wallace M., Hofmann K., Koch G., Johnsen A. H., Hendil K. B., Gordon C. The UBX2 and UBX3 cofactors direct cdc48 to proteolytic and non-proteolytic ubiquitin-dependent processes. Curr. Biol. 2004;14:824–828. doi: 10.1016/j.cub.2004.04.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material