Two Complexes of Spindle Checkpoint Proteins Containing Cdc20 and Mad2 Assemble during Mitosis Independently of the Kinetochore in Saccharomyces cerevisiae (original) (raw)
Abstract
Favored models of spindle checkpoint signaling propose that two inhibitory complexes (Mad2-Cdc20 and Mad2-Mad3-Bub3-Cdc20) must be assembled at kinetochores in order to inhibit mitosis. We have directly tested this model in the budding yeast Saccharomyces cerevisiae. The proteins Mad2, Mad3, Bub3, Cdc20, and Cdc27 in yeast were quantified, and there are sufficient amounts to form stoichiometric inhibitors of Cdc20 and the anaphase-promoting complex. Mad2 is present in two separate complexes in cells arrested in mitosis with nocodazole. There is a small amount of Mad2-Mad3-Bub3-Cdc20 and a much larger amount of a complex that contains Mad2-Cdc20. We use conditional mutants to show that both Mad2 and Mad3 are essential for establishment and maintenance of the spindle checkpoint. Both spindle checkpoint complexes containing Mad2 form in mitosis, not in response to checkpoint activation. The kinetochore is not required to form either complex. We propose that the conversion of Mad1-Mad2 to Cdc20-Mad2, a key step in generating inhibitory checkpoint complexes, is limited to mitosis by the availability of Cdc20 and is kinetochore independent.
The spindle checkpoint is a conserved regulatory system that assures the correct segregation of chromosomes to daughter cells during mitosis (1, 4, 24, 36, 41). The checkpoint inhibits anaphase onset until all chromosomes are attached to the mitotic spindle with bipolar orientation and a functional checkpoint is crucial to prevent aneuploidy (1). Analysis of both meiotic cells and mitotic cells has shown that a single unattached chromosome is sufficient to inhibit anaphase onset (26). Direct demonstration for a role of the kinetochore in the spindle checkpoint has come from laser ablation experiments in higher cells and genetic experiments with Saccharomyces cerevisiae that abrogate the spindle checkpoint in the absence of kinetochore function (15, 28, 33). Detailed mapping experiments in yeast have localized spindle checkpoint function to the Ndc80 complex, which is an evolutionarily conserved complex of four proteins within the kinetochore (29, 40). The kinetochore is absolutely required for the spindle checkpoint but the nature of the requirement is mysterious and is a critical unanswered question.
The MAD1-3, BUB1-3, and MPS1 genes that are required for the spindle checkpoint were originally defined in genetic screens using budding yeast, and homologs have been found in virtually all eukaryotic organisms (19, 25, 39). Elegant genetic experiments in budding yeast showed that Cdc20 is the direct target of spindle checkpoint inhibition (21). Cdc20 is a specificity factor for an E3 ubiquitin ligase called the anaphase-promoting complex or cyclosome (APC/C) and regulates the ubiquitylation and subsequent proteolysis of a mitotic inhibitor called securin or Pds1 in budding yeast (36, 41). Securin/Pds1 destruction is a key event for initiating entry into anaphase, and securin/Pds1 proteolysis is inhibited by the spindle checkpoint (1, 4, 41). Interestingly, most of the spindle checkpoint proteins localize to kinetochores that are detached from the mitotic spindle, either in prometaphase or after treatment with microtubule poisons, and the amount of staining diminishes as kinetochores capture microtubules and establish bipolar orientation (8). Recent experiments have demonstrated that the spindle checkpoint proteins dynamically associate with unattached kinetochores with very short half-lives (18, 23, 34).
In vitro assays for APC/C activity have been developed, and soluble inhibitors have been identified (12, 13, 37, 38). Candidate approaches have shown that Mad2 and BubR1 (Mad3 in budding yeast) can inhibit APC/C activity and Mad2 can act synergistically with BubR1 to form a more effective inhibitor (12, 38). Biochemical purification of a mitotic checkpoint complex (MCC) from HeLa cells identified a potent inhibitor of APC/C activity in vitro that is made up of BubR1, hBub3, hMad2, and p55CDC/Cdc20 (37). Recently it has been suggested that BubR1 is a more potent inhibitor of Cdc20-APC/C when tested in combination with Mad2 and Bub3 (41). Therefore, both approaches identified MCC as a potent stoichiometric inhibitor of the APC/C.
Yeast forms MCC (Bub3, Mad2, Mad3, and Cdc20) in nocodazole-treated cells, suggesting that it assembles when kinetochores are detached from microtubules and in response to checkpoint activation (16). Similarly, Mad2 and Cdc20 form two complexes in response to nocodazole treatment, one being MCC and the other a separate complex containing Mad2-Cdc20 (41). MCC is not present in mitotic Xenopus egg extracts but is induced to form when sperm and nocodazole are added, suggesting that the assembly is catalyzed by the presence of unattached kinetochores. These observations have led to the dominant model for the role of the kinetochore in generating the spindle checkpoint signal (1, 41). Unattached kinetochores are believed to be the site of assembly of inhibitory complexes, primarily MCC and Mad2-Cdc20, and the dynamic nature of kinetochore associations with checkpoint proteins are believed to reflect this assembly process (18, 23, 34).
We have addressed several aspects of this popular model by first quantifying the amount of the individual checkpoint proteins in yeast. We show that Mad2, Mad3, and Bub3 proteins are in excess of both Cdc20 and Cdc27 (a component of the APC/C) and there are sufficient quantities to act as stoichiometric inhibitors. However, we show that there are very small quantities of the MCC in nocodazole-treated cells and most Cdc20 interacts with Mad2 in a separate complex. Despite the small amounts of MCC, it is critical for maintenance of a spindle checkpoint. We show that both complexes containing Mad2 and Cdc20 assemble when cells enter mitosis and assembly does not depend on activating the spindle checkpoint. Finally, we show that neither MCC nor Mad2-Cdc20 assembly requires kinetochores.
MATERIALS AND METHODS
Strains and media.
The strains used for this work are listed in Table 1. Standard yeast medium was used throughout (3). Cells were grown in at 30°C unless otherwise stated. The final concentration of glucose, raffinose, and galactose used in the medium was 2%. Alpha factor (1 mM) was diluted 15,000-fold to arrest bar1 mutants. Hydroxyurea (Sigma) was used at 200 mM, and nocodazole or methyl [5-(2-thienyl-carbonyl)-1H-benzimidazol-2-yl]-carbamate (Sigma) was used at 15 μg/ml. To arrest cells for the degron experiment, nocodazole and 25 μg/ml of carbendazim (Sigma) were used, and to keep the cells arrested at 37°C, nocodazole and 50 μg/ml carbendazim were used.
TABLE 1.
Strains
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| BY4741 | MATahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 | 2 |
| BY4742 | _MAT_α his3_Δ_1 leu2_Δ_0 lys2_Δ_0 ura3_Δ_0 | 2 |
| 2818 | MATahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 BUB3-13MYC::KANMX4 | This study |
| 2819 | MATahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 CDC20-13MYC::KANMX4 | This study |
| 2845-2-2 | MATahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 MAD3-13MYC::KANMX4 | This study |
| 2832-8-2 | MATaade2-1 bar1::loxP can1 cyh2 his3 leu2-3,112 trp1 ura3-52 PDS1-13MYC::HIS3 GA10-UBR1:HIS3 mad3-dt:URA3 | This study |
| 2832-10-2 | MATaade2-1 bar1::loxP can1 cyh2 his3 leu2-3,112 trp1 ura3-52 PDS1-13MYC::HIS3 GA10-UBR1:HIS3 | This study |
| 2850 | MATaade2-1 bar1::loxP can1 cyh2 his3 leu2-3,112 trp1 ura3-52 PDS1-13MYC::HIS3 GA10-UBR1:HIS3 mad2-dt:URA3 | This study |
| 2873 | _MAT_α his3_Δ_1 leu2_Δ_0 lys2_Δ_0 ura3_Δ_0 MAD3-PrA::URA3 | This study |
| 2876-2-1 | MATahis3_Δ_1 leu2_Δ_0 ura3_Δ_0 MAD3-PrA::URA3 CDC20-13MYC::KANMX4 | This study |
| 2898-11-3 | MATabar1::loxP leu2_Δ_0 his3_Δ_1 ura3_Δ_0 MAD3-PrA::URA3 CDC20-13MYC::KANMX4 P_GAL1-PDS1-MDB_::LEU2 | This study |
| 3403-48-2 | MATal bar1::loxP leu2_Δ_0 his3_Δ_1 ndc10-1 ura3_Δ_0 MAD3-PrA::URA3 CDC20-13MYC::KANMX4 P_GAL1-PDS1-MDB_::LEU2 PDS1-HA::URA3 | This study |
| 3404 | MATa1 bar1::loxP leu2_Δ_0 his3_Δ_1 ura3_Δ_0 MAD3-PrA::URA3 CDC20-13MYC::KANMX4 P_GAL1-PDS1-MDB_::LEU2 PDS1-HA::URA3 | This study |
| 8056 | MATaade2-1 bar1::loxP can1 cyh2 his3_Δ_1 leu2-3,112 trp1-289 ura3-52 MAD2-13MYC::TRP1 | This study |
| 9545 | MATaade2-1 bar1::loxP can1 cyh2 his3_Δ_1 leu2,3-112 trp1-289 ura3-52 CDC27-13MYC::TRP1 | This study |
Preparation of yeast extract.
Cells were suspended in 0.1 N NaOH and incubated at the room temperature for 5 min to make yeast extracts for Western blots. Following NaOH treatment the cells were washed twice with water, resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and boiled for 5 min. The supernatant was collected after centrifugation and the concentration of protein was estimated by the Bradford assay. Equal amounts of protein were loaded on each lane.
Whole-cell extracts were prepared by breaking cells with glass beads in 50 mM HEPES (pH 7.5), 25 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 50 mM NaF, 1 mM dithiothreitol, 10% glycerol supplemented with a cocktail of protease inhibitors (Complete, Roche). The lysates were centrifuged at 10,000 rpm in an SS34 rotor for 10 min to remove the glass beads and cell debris. Clarified extracts were prepared by centrifugation at 45,000 rpm for 45 min.
Immunoprecipitations, gel filtration, and immunoblotting.
Immunoprecipitations were done from whole-cell extracts containing 2 mg of protein and incubated with antibody coupled agarose beads at 4°C for 1 h. The beads were washed four times with 10 volumes of buffer, boiled in SDS-PAGE sample buffer for 5 min, and spun down and the supernatants were loaded on the gel. Cdc20-13Myc was isolated using protein G-Sepharose (Amersham) preincubated with 9E10 antibodies. Mad3-prA was immunoprecipitated with immunoglobulin G-Sepharose (Amersham).
For gel filtration chromatography, whole-cell extracts containing 2 mg of protein was loaded onto a Superose 6 column (Amersham Pharmacia Biotech) equilibrated with 40 mM Tris-HCl, pH 7.9, 350 mM NaCl, 0.1% Tween, and 10% glycerol. Thirty 500-μl fractions were collected after the first 6 ml had passed, and alternate fractions were analyzed by Western blots. The gel filtration column was calibrated with gel filtration standards from Bio-Rad.
For immunoblotting, the protein samples were run on SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and probed with antibodies. Equal amounts of whole-cell extracts and immunodepleted extracts were loaded on the gel. Immunoprecipitations, equivalent to 10-fold more protein than that was loaded as whole-cell extract, were loaded for immunoblotting. Myc-tagged proteins were detected with mouse monoclonal antibody 9E10. Mouse monoclonal antibody 12CA5 was used to detect hemagglutinin (HA)-tagged proteins. Goat polyclonal antibodies raised against peptides mapping at the carboxy terminus of Mad2 (Santa Cruz Biotechnology) were used for the detection of Cdc20 and Mad2. Mad3-PrA was detected with anti-protein A (PrA) antibody raised in a rabbit (Sigma). Detection was done with Supersignal solution (Pierce).
Flow cytometry.
Approximately 106 cells were harvested and fixed with 70% ethanol. The fixed cells were treated with 50 mg/ml pepsin (Sigma) for 30 min at 37°C, washed and treated with 50 mg/ml RNase A at 37°C for 2 h. Following RNase treatment the cells were stained with Sytox green (1 μM) and analyzed using a Becton Dickinson FACScan flow cytometer.
Construction of epitope-tagged strains.
To make a yeast strain containing a PrA tag (calmodulin binding domain, tobacco etch virus protease cleavage site, and protein A) at the C-terminal end of the endogenous MAD3, the tag and the marker gene URA3 were PCR amplified from the plasmid pBS1479 using the primers MAD3TAP5 (5′-TGACAAGTCGAGTTCGTCTTTCATATCGTACCCACCACAGCGTTCCATGGAAAAGAGAAG-3′) and MAD3TAP3 (5′-GTCGGCCGTCGATGTGTTTACGATTGGCCAGTATACTTACTCATACGACTCACTATAGGG-3′). Wild-type yeast strain BY4742 was transformed with this cassette and the transformants were selected on SC-uracil plates. Integration of the cassette at the right locus was verified by PCR using primers flanking the site of integration and expression of the fusion protein was confirmed by Western blotting with anti-PrA antibodies.
Yeast strains containing 13-Myc tags at the C-terminal end of endogenous BUB3, MAD3, and CDC20 were made by PCR amplification of the 13Myc-TADH1-KANMX6 cassette from the plasmid pFA6a-13Myc-kanMX6 (27) followed by transformation of wild-type yeast strain (BY4741) with the PCR product and selection of the transformants on G418 plates (500 μg/ml). The primers used for the amplifications are BUB3F2 (5′-AAACGCAAGTTCAATATACATAATATTTGACTATGAGAACCGGATCCCCGGGTTAATTAA-3′), BUB3R1 (5′-GGAATGTTCTATCATACTACACGAATCTTCACGAAGATAGAATTCGAGCTCGTTTAAAC-3′), MAD3F2 (5′-CAAGTCGAGTTCGTCTTTCATATCGTACCCACCACAGCGTCGGATCCCCGGGTTAATTA-3′), MAD3R1 (5′-TTGGCCAGTATACTTACTCATTCATGGGATTAGTTTTATTGAATTCGAGCTCGTTTAAAC-3′), CDC20F2 (5′-TACAAGGAGGCCCTCTAGTACCAGCCAATATTTGATCAGGCGGATCCCCGGGTTAATTAA-3′), and CDC20R1 (5′-ATTATATGCCTTGACATGAACTTTTATTTTTTTTATTTTAGAATTCGAGCTCGTTTAAAC-3′).
To construct strains carrying 13-Myc-tagged Mad2 and Cdc27, the 13-Myc TADH1 -TRP1 cassette was amplified from the plasmid pFA6a-13Myc-TRP1 and transformed into wild-type yeast strain 2629-1-2. The transformants were selected on complete plates minus tryptophan. The primers used for amplifications are CYTAG1 (5′-CAACGATCATAAAGTTGGTGCGCAGGTCAGCTATAAATATCGGATCCCCGGGTTAATTAA-3′) and CYTAG2 (5′-TGGACTTCCGTCTTTTTTTTTTTTTTTGACTTGAATTCTAGAATTCGAGCTCGTTTAAAC-3′) for Mad2 and TG27F (5′-AGTTATCATCGATGAATTACAAAAATGTCATATGCAAGAACGGATCCCCGGGTTAATTAA-3′) and TG27R (5′-TTAACATGTTTCAATTTGTTATGTTTTGGCCAACCTGCGTGAATTCGAGCTCGTTTAAAC-3′) for Cdc27. For all fusion constructs integrations at the appropriate genomic locus was confirmed by PCR and expression of the fusion proteins of expected molecular weight by Western blotting with anti-Myc antibodies.
To tag the MAD3 gene with a degron tag, the HindIII site (181 nucleotides downstream of the first ATG) in the MAD3 gene was eliminated by fusion PCR. A TCA codon (SER) was changed to TCT because the codons have similar usage (Saccharomyces Database). Two PCRs were done from genomic DNA to amplify DNA fragments overlapping the HindIII site of the genome using primer sets MAD3DEG1 (5′-GGGCAAGCTTCCGGGGGGATGAAAGCGTACGCAAAGAAACG-3′), MAD3DEG2 (5′-CAGTCTTTGTTCGAAGCTAGATTTAACTTG-3′), MAD3DEG3 (5′-CCAAGTTAAATCTAGCTTCGAACAAAGACT-3′), and MAD3DEG4 (5′-GCGCGTCGACGTTTAAGCTCACCCAAATTGG-3′). The fragments were purified and fusion PCR was done with MAD3DEG1 and MAD3DEG4. The fusion PCR product was cloned into the vector TA (Invitrogen). A fragment containing the PCR product was excised from the TA vector with SalI and HindIII and subcloned into the HindIII and XhoI sites of the degron vector pPW66R (10). The degron fusion was integrated at the MAD3 locus by digesting the plasmid with BglII. Correct integration was confirmed by PCR with a pair of primers to amplify a fragment from the HA in the plasmid to a site within the gene that is downstream of the integration site. The degron tag contains an HA epitope. Expression of the fusion protein was confirmed by a Western blot using anti-HA antibodies.
A fusion PCR strategy was used to integrate the degron tag at the 5′ end of the endogenous MAD2 gene (30). The 335 nucleotides upstream of the first ATG of MAD2 gene was amplified with primer sets MAD2TAPN1 (5′-TTCCTTGGTCCACTGTTTC-3′) and MAD2TAPN2 (5′-GTGCTATTGGGCCCCTTTG 3′) and the 5′ end of the Kluyveromyces lactis URA3 gene was amplified from the plasmid PWJ1077 (30) using the primer set MAD2TAPN11 (5′-CAAAGGGGCCCAATAGCACTGCTCGAGGCAAGTAATGTGTG-3′) and MAD2TAPN5 (5′-CTTGACGTTCGTTCGACTGATGAGC-3′). Fusion PCR was done to fuse these two PCR products with MAD2TAPN1 and MAD2TAPN5 (fusion PCR 1). The degron tag was amplified from the plasmid pPW66R using MAD2DEG1 (5′-GTGACTGGGAAAACCCTGGCG CATTACCGACATTTGGGCGC-3′) and MAD2DEG2 (5′-GTGATATTGATTGTGACATAGCTTGCCCTCCTAAAAATGC-3′) and the 5′ end of the MAD2 gene (274 nucleotides) was amplified with MAD2TAPN9 (5′-ATGTCACAATCAATATCACTAAAGG-3′) and MAD2TAPN10 (5′-CCACCACCTCTCCCTCATCC-3′).
Fusion PCR was used to combine the degron tag at the 5′ end of the MAD2 gene with the primer set MAD2DEG1 and MAD2TAPN10 (fusion PCR 2). The 3′ end of the K. lactis URA3 gene was amplified from the plasmid PWJ1077 (30) using the primer set MAD2TAPN4 (5′-GAGCAATGAACCCAATAACGAAATC-3′) and MAD2TAPN6 (5′-CGCCAGGGTTTTCCCAGTCAC-3′). It was fused to the N terminal end of the Degron-MAD2 cassette by a fusion PCR with MAD2TAPN4 and MAD2TAPN10 using the fusion PCR 2 and the PCR amplified 3′ end of the K. lactis URA3 gene as template (fusion PCR 3). Fusion PCR 1 and fusion PCR 3 were used to transform yeast cells and the transformants were selected on SC-uracil plates. PCRs using combinations of primers flanking the integration site and that for the degron tag confirmed the integration of the _URA3_-degron-MAD2 cassette in the MAD2 locus. Western blotting with anti-HA antibody was done to confirm the expression of fusion protein of right size.
Expression and purification of recombinant Mad2-13Myc and Mad3-13Myc from Escherichia coli.
MAD2 gene carrying a C-terminal Myc tag was PCR amplified from a yeast strain carrying Mad2-13Myc using the primer setMAD2PET5 (5′-CTAGCTAGCATGTCACAATCAATATCACTAAAGGG-3′) and 13MYC-CTER (5′-ACGCGTCGACAGAAGTGGCGCGAATTCAC-3′). The PCR product was purified, digested with _Nhe_1 and _Sal_1 and cloned into the _Nhe_1 and _Sal_1 site of the expression vector pET28a. Sequencing was done to verify the in frame fusion of the PCR product and the presence of 13-Myc tags in the cloned fragment. Similarly MAD3-13 MYC was PCR amplified from a yeast strain carrying 13-Myc-tagged Mad3 using the primer set MAD3PET5 (5′-CTAGCTAGCATGAAAGCGTACGCAAAG-3′) and 13MYC-CTER (5′-ACGCGTCGACAGAAGTGGCGCGAATTCAC-3′), cloned in pET28a and sequenced. The recombinant His-tagged fusion proteins were expressed in E. coli strain BL21(DE3, pLysS) (Novagen), purified using Ni2plus-nitrilotriacetic acid -agarose (QIAGEN). Twofold serial dilution of known concentration of bovine serum albumin and purified protein was run on a gel. The concentration of the purified protein was determined by Bradford assay using bovine serum albumin as the standard.
Degron experiment.
The cells were grown overnight in YM-1 plus raffinose plus galactose medium containing 0.1 mM copper sulfate at 23°C. To activate the checkpoint 15 μg/ml nocodazole and 25 μg/ml carbendazim was added to cycling cells. After 80% of the cells arrested as large budded cells the proteolysis of the degron-tagged proteins was induced by a shift to growth in prewarmed (37°C) complete medium containing raffinose and galactose, and the cells were allowed to grow at 37°C. Nocodazole (15 μg/ml) and carbendazim (50 μg/ml) was added to the medium to keep the spindle checkpoint signal activated. Samples were collected for Western blotting and cell morphology study for every 45 min starting from 0 min after shifting the cells to 37°C.
RESULTS
Quantification and cofractionation of the components of MCC and APC/C.
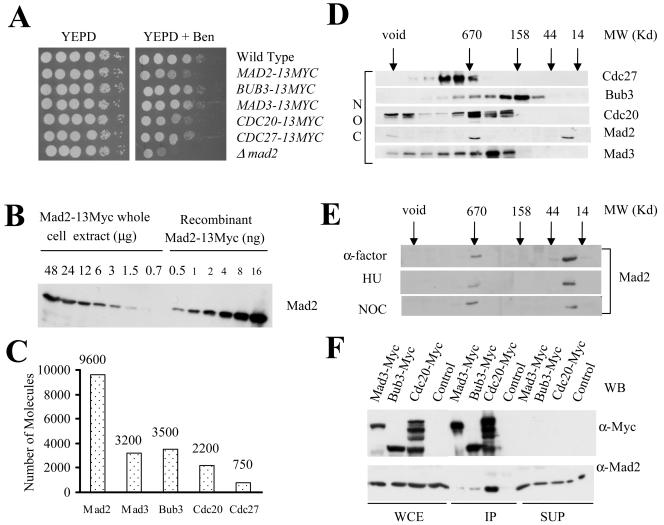
If Mad2 or MCC is a stoichiometric inhibitor of APC/C, then the intracellular levels of Mad2 must be in excess of both Cdc20 and the APC/C. To measure the levels of MCC and APC/C proteins, the endogenous copies of Mad2, Mad3, Bub3, Cdc20, and Cdc27 were tagged at the C terminus with 13Myc. The 13Myc peptide did not interfere with the function of any of the proteins. CDC20 and CDC27 are essential genes, and both of the Myc-tagged versions were viable and grew with normal kinetics (Fig. 1A). MAD2, MAD3, and BUB3 are nonessential genes, and mutants are sensitive to low concentrations of the drug benomyl (19, 25). Serial dilutions of cells were spotted onto benomyl-containing YPD plates to determine if the 13Myc affected the function of any of the checkpoint genes (Fig. 1A). Strains containing Mad2-13Myc, Mad3-13Myc, and Bub3-13Myc were similar to the wild type with respect to growth on YPD plates containing benomyl. The strain containing Mad2-13Myc was slightly benomyl sensitive but was much more resistant than the mad2 mutant (Fig. 1A). Furthermore, the Mad2-13Myc cells were able to arrest in mitosis and retained high viability when grown in the presence of nocodazole (not shown). Therefore, the strains were not compromised for spindle checkpoint function. We also determined the benomyl sensitivity of Cdc20-13Myc and Cdc27-13Myc and in both cases the benomyl sensitivity was similar to that of the wild type (Fig. 1A) suggesting that neither protein is compromised for the spindle checkpoint. Therefore, the introduction of the Myc tag at the C-terminal end of each gene did not eliminate the function of any of the five proteins tested.
FIG. 1.
(A) Benomyl sensitivity of the yeast cells expressing 13-Myc-tagged Bub3 (2818), Mad2 (8056), Mad3 (2845-2-2), Cdc20 (2819) or Cdc27 (9545) protein; 10-fold serial dilutions of cells were spotted on YPD and YPD plus benomyl (15 μg/ml). The plates were incubated at 23°C for 4 days before being photographed. (B) Quantification of Mad2 in nocodazole-treated yeast cells. Anti-Myc immunoblot of 2-fold serial dilution of yeast whole-cell extract made from cells expressing Mad2-13Myc and recombinant Mad2-13Myc is shown. The numbers represent the amount of whole-cell extract (in μg) and the purified protein (in ng) loaded/lane. (C) The estimated number of molecules of proteins present per yeast cell arrested in M phase. (D) Gel filtration profiles of the checkpoint proteins (Bub3, Mad2, and Mad3), Cdc20, and APC (Cdc27) present in nocodazole-treated yeast cells. Whole-cell extracts were made from nocodazole-treated cells expressing Myc-tagged Bub3, Mad3, Cdc20, and Cdc27 proteins. The extracts were separated on Superose 6 column. Alternate fractions were probed with anti-Myc antibodies or anti-Mad2 antibodies. The elution profiles of the molecular weight standards in the same column are indicated by arrows. (E) Spindle checkpoint induces association of Mad2p with other proteins to form a high-molecular-weight protein complex. Wild-type cells were arrested in G1, S, and M phase with α-factor, hydroxyurea, and nocodazole, respectively. Clarified extracts were fractionated on a Superose 6 column. Alternate fractions were probed with anti-Mad2 antibodies. The arrows indicate the elution profiles of the gel filtration standards. (F) More Mad2 coimmunoprecipitated with Cdc20 compared to Bub3 and Mad3. Cycling cultures of cells expressing Mad3-13 Myc (2845-2-2), Bub3-13Myc (2818), or Cdc20-13Myc (2819) were arrested in M phase with nocodazole. Myc-tagged proteins were immunoprecipitated from the whole-cell extracts using anti-Myc antibody-coupled beads. The whole-cell extracts, immunoprecipitates, and supernatants were probed with anti-Myc and anti-Mad2 antibodies. An untagged strain (BY4741) was used as a control. Equal amounts of whole-cell extracts and supernatants were loaded; 5-fold more protein was loaded in the IP lane.
To determine the relative stoichiometry of the Myc-tagged proteins, we constructed 13Myc-tagged Mad2 and Mad3 in bacterial expression vectors and purified them to homogeneity from E. coli. The concentration of endogenous Mad2 present in yeast whole-cell extracts was estimated by comparing the intensities of the bands after serial twofold dilutions to known amounts of recombinant Mad2-13Myc protein. (Fig. 1B). Molecular weight is the major variable in quantification experiments using Western blots and probably reflects differential transfer for proteins of different molecular weights (P.T.S and A.P., unpublished). Purified recombinant Mad3-13Myc is similar in size to Bub3-13Myc, Cdc20-13Myc, and Cdc27-13Myc and is identical to Mad3-13Myc and was used as standard for quantification of these proteins using serial twofold dilutions as described above (data not shown). The quantification data for all of the proteins are summarized in Fig. 1C and shows that Mad2 is the most abundant of these five proteins. We estimate that the relative amounts of the other components of MCC (Cdc20, Bub3, Mad3) is 2- to 3-fold less and Cdc27, a component of the APC/C, is approximately 10% of that of Mad2. Therefore, the spindle checkpoint proteins are equivalent or in excess of both Cdc20 and the APC/C protein Cdc27, suggesting that the formation of a stoichiometric inhibitor is possible.
We determined the proportion of checkpoint proteins that were assembled into complexes by size exclusion chromatography. Clarified extracts from nocodazole-treated cells expressing C-terminal 13Myc-tagged Mad3, Bub3, Cdc20, or Cdc27 were fractionated over a Superose 6 column, and alternate fractions were immunoblotted with anti-Myc antibody. The fractionation profiles, Fig. 1D, demonstrate that Bub3, Mad3, Cdc20, and Cdc27 comigrated as a large multiprotein complex (670 kDa) when extracts were prepared from cells grown in the presence of nocodazole to activate the spindle checkpoint. The fractions from the Mad3-13Myc gel filtration column were probed with anti-Mad2 antibodies prepared against a carboxy-terminal peptide. Mad2 was present in two different peaks, as previously demonstrated (7, 14). In one of the peaks, Mad2 cofractionated with the Bub3, Mad3, Cdc20, and Cdc27 complex, and the other peak corresponded to the monomer or dimer form of Mad2. However, only a small fraction of Cdc20-13Myc, Mad3-13Myc, Bub3-13Myc, and Cdc27-13Myc comigrated with Mad2 in the 670-kDa fraction. This suggests that a small proportion of these proteins form MCC. We did not detect monomer forms of Mad3, Bub3, Cdc20, or Cdc27.
Mad2, Mad3, and Bub3 are constitutively expressed in wild-type yeast cells and Mad3 is continuously associated with Bub3 regardless of cell cycle stage (7, 14, 16). Increased amount of Mad2 has been shown to interact with Mad3 and Bub3 in nocodazole-treated cells (16). The gel filtration profiles of Mad2 in the alpha-factor, hydroxyurea, or nocodazole treated cells is shown in Fig. 1E. We quantified the Mad2 in both regions and found that approximately 80% of the Mad2 was present in the monomer and 20% in the high-molecular-weight complex in α-factor or hydroxyurea-treated cells. In contrast, approximately 50% of Mad2 was in a high-molecular-weight complex in nocodazole-treated cells. Taken together, our data suggest that a Mad2 assembles into one or more larger complexes in response to nocodazole treatment. However, the data suggest that only a small amount of MCC is formed.
Mad2 and Cdc20 exist as a separate subcomplex independent of MCC.
We used immunoprecipitation to directly measure the association of Mad2 in MCC. If Bub3, Mad3, Cdc20, and Mad2 were a part of a single multiprotein complex (MCC), the amount of Mad2 that coimmunoprecipitate with Bub3-13Myc, Mad3-13Myc, or Cdc20-13Myc should be the same. Whole-cell extracts from cells expressing Mad3-13 Myc, Bub3-13Myc, or Cdc20-13Myc were prepared and Myc-containing proteins were purified by immunoprecipitation. The pellets and the supernatants were probed with anti-Myc and anti-Mad2 antibodies (Fig. 1F). A greater amount of Mad2 coimmunoprecipitated with Cdc20-13Myc than with either Bub3-13Myc or Mad3-13Myc. This was not caused by differential precipitation of the three complexes, as all of the Myc-tagged proteins were quantitatively depleted. These data are consistent with previous observations that there are two separate complexes of Mad2 and Cdc20 and suggest that MCC is much less abundant than Mad2-Cdc20 (1).
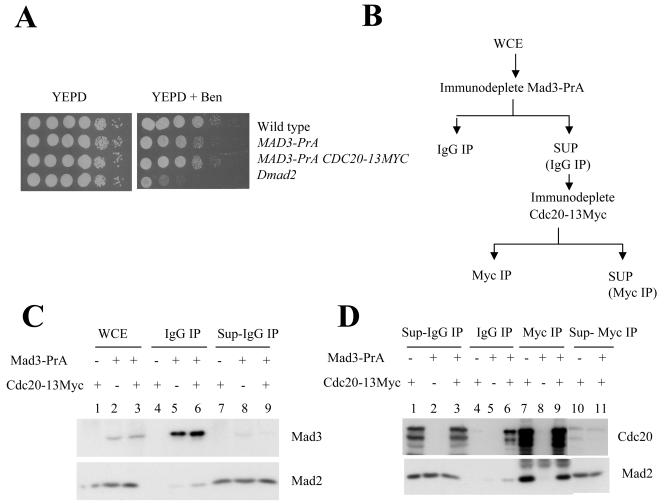
We performed sequential immunoprecipitations to separate the two Mad2-containing complexes in spindle checkpoint-activated cells. The design of the experiment is shown in Fig. 2B. Two strains with either a C-terminal protein A (PrA) tag on the endogenous Mad3 (Mad3-PrA) or expressing both Mad3-PrA and Cdc20-13Myc were constructed. Neither strain was benomyl sensitive (Fig. 2A), and the strains were checkpoint proficient. Extracts made from the strains expressing single tags were used as controls for this and subsequent experiments. Mad3-PrA was depleted from extracts prepared from nocodazole-treated cells using agarose beads coupled to human IgG, and Cdc20-13Myc was subsequently isolated from the supernatants using agarose beads coupled to anti-Myc.
FIG. 2.
Cdc20-Mad2 forms a complex independent of Mad3. (A) The benomyl sensitivity of the strains expressing Mad3-PrA (2873) and Mad3-PrA Cdc20-13Myc (2876-2-1) is shown. Serial tenfold dilutions of cells were spotted on YPD and YPD plus benomyl (15 μg/ml) plates. The plates were incubated at 23°C for 4 days and photographed. (B) The flow chart of the sequential immunoprecipitation from yeast whole-cell extract is shown. Mad3-PrA was immunodepleted from the extracts using IgG beads, and Cdc20-13Myc was immunoprecipitated from the resulting supernatants using anti-Myc antibody beads. (C) Mad3-PrA was immunodepleted from whole-cell extract made from nocodazole-treated cells expressing Mad3-PrA, Cdc20-13Myc (2876-2-1), and Mad3-PrA (2873). Extract made from nocodazole-treated cells expressing Cdc20-13Myc (2819) was used as the control. The whole-cell extracts (lanes 1 to 3), IgG immunoprecipitates (lanes 4 to 6), and supernatants of IgG IP (lanes 7 to 9) were probed with anti-PrA and anti-Mad2 antibodies; 50 μg whole-cell extracts and the supernatants were loaded per lane and the amount of protein loaded in the IP lane was 10-fold more than that loaded in the whole-cell extract lane. (D) Cdc20-13Myc was immunoprecipitated from the Mad3-PrA depleted extracts using anti-Myc antibody beads. The supernatants of the Mad3-PrA depleted extracts (lanes 1 to 3), anti-Myc immunoprecipitates (lanes 7 to 9), and the supernatants of anti-Myc immunoprecipitations (lanes 10 to 11) were probed with anti-Myc and anti-Mad2 antibodies. The IgG immunoprecipitates were probed with anti-Myc antibodies (lanes 4 to 6) to detect coimmunoprecipitation of Cdc20-13Myc with Mad3-PrA.
The Western blot analysis of the whole-cell extracts, supernatants, and IgG immunoprecipitates probed with anti-PrA and anti-Mad2 antibodies showed that greater than 95% of the Mad3-PrA was removed from the extract with IgG beads (Fig. 2C) and a small fraction of Mad2 copurified with Mad3-PrA (Fig. 2C, lanes 5 and 6), consistent with our previous observation. A small amount of Cdc20 copurified with Mad3-PrA (Fig. 2D, lanes 4 to 6) demonstrating that this complex contains Mad2, Mad3-PrA, and Cdc20-13Myc and therefore is MCC (37).
Cdc20-13Myc was purified from the supernatant by anti-Myc immunoprecipitation using beads, and immunoblots were probed with anti-Myc and anti-Μad2 antibodies (Fig. 2D). Mad2 coimmunoprecipitated with Cdc20-13Myc from Mad3-PrA-depleted extracts (Fig. 2D, lane 9), showing that Mad2 and Cdc20 form a separate complex that does not contain Mad3. The amount of Mad2 that coprecipitated with Cdc20-13Myc from an extract lacking Mad3-PrA was similar to the amount of Mad2 that coprecipitated with Cdc20-13Myc from Mad3-PrA-depleted extract (Fig. 2D, lanes 7 and 9). This confirms that a small fraction of Cdc20-13Myc and Mad2 coprecipitated with Mad3-PrA using IgG and shows that little Cdc20 is associated with MCC compared to the amount in the Mad2-Cdc20 complex.
MCC is essential for the maintenance of the spindle checkpoint.
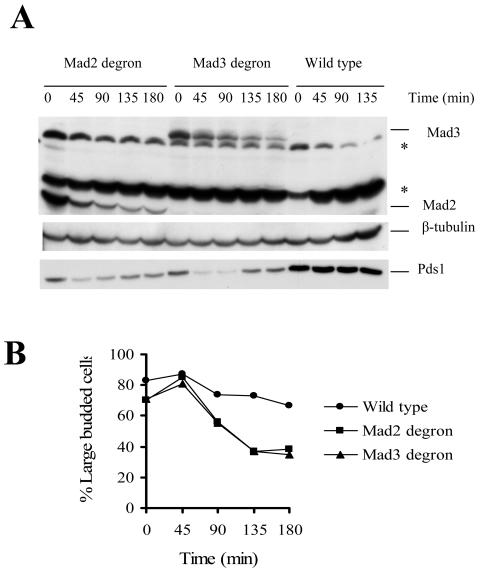
The spindle checkpoint is activated when kinetochores are unoccupied and maintained until bipolar attachment of the kinetochore to the spindle is achieved. One possible rationale for the existence of two different complexes of spindle checkpoint proteins is that one complex has a role in establishing the arrest and the other is required to maintain it. We used temperature-inducible degron fusions to construct conditional mad2 td and mad3 td mutants in which we also epitope tagged the mitotic inhibitor Pds1 with 13Myc (10). Pds1 stability is a measure of cells passing from metaphase to anaphase, as the protein levels are high when cells are in metaphase, as in spindle checkpoint-activated cells, and drop precipitously as cells enter anaphase (9).
We grew wild-type, mad2 td and mad3 td cells under permissive conditions for the degron and added nocodazole and carbendazim to the medium until greater than 80% of the cells were arrested as large budded cells, indicative of mitosis. We induced the degradation of Mad2 and Mad3 and assayed for the ability of the cells to stay arrested by monitoring Pds1-13Myc stability. The degron-tagged proteins contained an HA epitope so that we could follow the stability of Mad2 and Mad3. Pds1 levels began to decline as soon as the Mad2 or Mad3 levels dropped (Fig. 3A). The Pds1 levels rise again because cells enter the next cell cycle. This was confirmed by the cytology of the cells that showed that the cells could not maintain the mitotic arrest as the levels of either protein was reduced (Fig. 3B). There was no significant change in Pds1 level of the wild type over time. We conclude that despite the small amount of MCC in the cell it is an important mitotic inhibitor and that MCC is required for maintenance as well as establishment of the spindle checkpoint.
FIG. 3.
MCC is required for the maintenance of the spindle checkpoint function. Cells expressing degron-tagged Mad2 (2850), degron-tagged Mad3 (2832-8-2), and no degron tag (wild type) (2832-10-2) were grown overnight at 23°C in YM-1 containing raffinose, galactose, and CuSO4 (100 μM). The cycling cells were arrested in M phase with carbendazim (25 μg/ml) and nocodazole (15 μg/ml). The cells were washed, resuspended in prewarmed (37°C) complete medium containing galactose, raffinose, carbendazim, and nocodazole. Aliquots of cells were collected at the indicated time points, and protein samples were prepared. Time zero represents the time at which the cells were shifted to 37°C. (A) Anti-HA immunoblot showing the levels of the degron-tagged Mad3-HA and Mad2-HA fusion proteins. Anti-Myc antibody was used to detect Pds1-13 Myc. Immunoblotting with α-tubulin antibody was done as a loading control. Asterisks indicate nonspecific bands. (B) The percentage of large budded cells in the degron-tagged Mad2, degron-tagged Mad3, and wild-type cells at different time points as indicated above.
MCC formation is cell cycle regulated and independent of the spindle checkpoint.
A key experiment to support the model that an unattached kinetochore serves as a platform of assembly of the mitotic inhibitors is that MCC formation in budding yeast occurs in nocodazole-treated cells but is at very low levels in cells arrested in S phase by hydroxyurea and absent in α-factor-treated cells arrested in G1 (16). This was interpreted to mean that MCC formation was induced by the spindle checkpoint (16). However, there is an alternative explanation. Nocodazole-treated cells are in mitosis, and the hydroxyurea and α-factor-treated cells are not. MCC assembly may reflect cell cycle dependence and not spindle checkpoint activation.
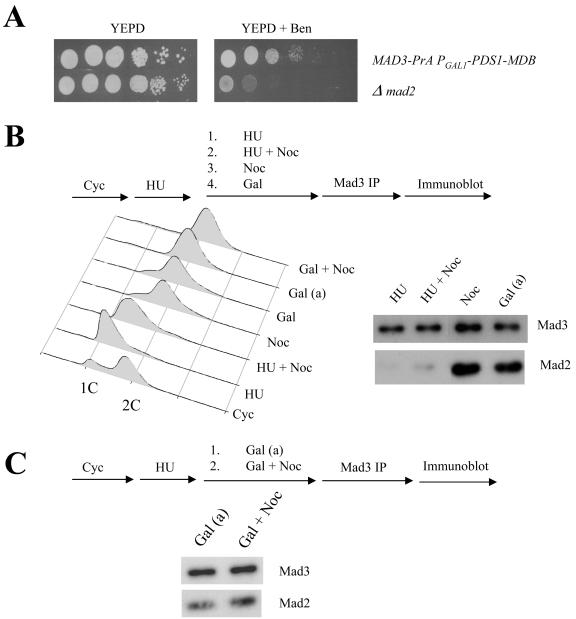
We directly tested whether MCC formation responds to unattached kinetochores or assembles in mitosis even when kinetochore attachments are not perturbed. We followed MCC production in a strain expressing Mad3-PrA by the association of Mad2 with Mad3-PrA and we included a PGAL1 -PDS1-MDB, which is a dominant galactose-inducible PDS1 mutant that has a mutation in the destruction box so that the mutated Pds1 does not turn over (9). Excess expression of the mutant Pds1 causes cells to arrest in mitosis and should not affect kinetochore-microtubule attachments. The strain is benomyl resistant, showing that the spindle checkpoint is intact (Fig. 4A). We grew cells in YM-1, arrested them in hydroyurea and purified Mad3-PrA using IgG beads and showed they contained a small amount of associated Mad2 (Fig. 4B). We repeated the experiment but added nocodazole to the cells previously arrested with hydroxyurea. Nocodazole did not stimulate the formation of additional MCC in cells previously arrested in hydroxyurea (Fig. 4B). In contrast, when we induced unattached kinetochores and arrested cells in mitosis by growing them in the presence of nocodazole, we identified significantly more MCC (Fig. 4B) as previously described (16). However, we found equivalent amounts of Mad2 associated with Mad3-PrA in cells grown in the presence of galactose to induce PDS1-MDB (Fig. 4B). Adding nocodazole to cells previously arrested in mitosis with PDS1-MDB did not stimulate formation of additional MCC (Fig. 4C). We conclude that MCC formation in yeast is cell cycle dependent and does not depend on the presence of unattached kinetochores.
FIG. 4.
Mad3 associates with Mad2 independently of the spindle checkpoint. **(**A) Benomyl sensitivity of strain 2898-11-3 (Mad3-PrA P_GAL1_ -PDS1-MDB) was checked by spotting 10-fold serial dilutions of the cells on YPD and YPD plus benomyl (15 μg/ml). The photographs were taken after 4 days of incubation of the plates at 23°C. **(**B) A flow diagram of the experiment is shown. Wild-type cells (2898-11-3) containing P_GAL1_ -PDS1-MDB and Mad3-PrA were grown overnight in YM-1 and raffinose at 30°C. The cells were arrested in S phase with hydroxyurea (200 mM) for 3 h. Following arrest, the culture was divided into several aliquots. An aliquot of the cells were collected (HU). To another aliquot of cells, nocodazole (15 μg/ml) was added. The cells were allowed to grow and collected after 1.5 h (hydroxyurea plus Noc). To arrest cells in M phase by the activation of the spindle checkpoint, an aliquot of hydroxyurea-treated cells were washed three times and released into YM-1 with raffinose and nocodazole. The cells were allowed to grow for 1.5 h and collected (Noc). Another aliquot of cells were treated with hydroxyurea for 2.5 h, galactose was added, and the culture was allowed to grow for 30 min to induce the expression of P_GAL1_ -PDS1-MDB. The cells were washed three times, resuspended in YM-1 and raffinose and galactose, and harvested after 1.5 h (Gal). Cells were prepared for flow cytometry, and protein extracts were made from the samples collected. Flow cytometry profiles of the cells collected in different growth conditions is shown. 1C and 2C represent the DNA content of the cells. Mad3-PrA was immunoprecipitated from the whole-cell extracts made from the cells, and the immunoprecipitates were probed with anti-PrA and anti-Mad2 antibodies. (C) A similar experiment except after induction with galactose the culture was divided into two. One set was allowed to grow for 1.5 h in the presence of raffinose and galactose, and samples were collected (Gal [a]). Nocodazole was added to the other half in addition to raffinose and galactose. The cells were allowed to grow and collected after 1.5 h (Gal plus Noc). Flow cytometry and immunoprecipitations were done as described for B. The flow cytometry result is shown in B.
One possible caveat is that PDS1-MDB may activate the spindle checkpoint, and therefore the abundant association of MCC would be indicative of the indirect effect of Pds1-MDB on spindle function. We tested this directly as described below (Fig. 5C) and found that PDS1-MDB does not activate the spindle checkpoint.
FIG. 5.
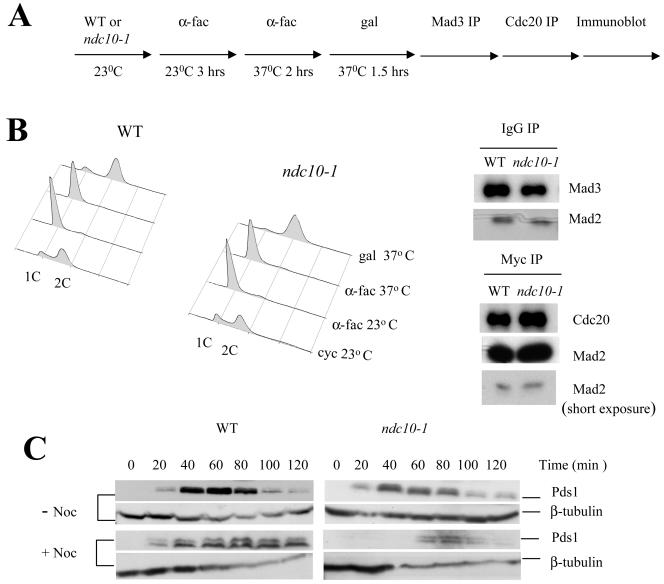
Mad2 associates with Mad3 and Cdc20 in the M phase independently of the functional kinetochore. (A) Flow diagram of the experiment is shown. Wild-type (3404) and ndc10-1 (3403-47-2) cells expressing P_GAL1_-PDS1-MDB, Mad3-PrA, Cdc20-13Myc, and Pds1-HA were grown overnight at 23°C in YM-1 and raffinose. The cells were arrested in G1 phase with α-factor. Following G1 arrest at 23°C the cells were filtered, resuspended in prewarmed (37°C) YM-1 plus raffinose containing α-factor, and allowed to grow at 37°C. After 1.5 h galactose was added, and cells were grown for 30 min to induce the expression of P_GAL1_ -PDS1-MDB. The cells were released from α-factor arrest and allowed to grow in YM-1, raffinose, and galactose at 37°C for 90 min, and samples were collected for immunoprecipitations. (B) Flow cytometry analysis of the cells collected at the indicated time points. 1C and 2C represent the DNA content of the cells. Μad3-PrA was immunoprecipitated with IgG beads from the whole-cell extracts made from the wild-type and ndc10-1 cells. The immunoprecipitates were probed with anti-PrA and anti-Mad2 antibodies. Cdc20-13Myc was immunoprecipitated from the Mad3-PrA-depleted extracts using anti-Myc beads, and the immunoprecipitates were probed with anti-Myc and anti-Mad2 antibodies. (C) Wild-type and ndc10-1 cells were released from the α-factor arrest into galactose at 37°C as described for A in the absence (−Noc) or presence (+Noc) of nocodazole (15 μg/ml). Samples were collected every 20 min for 120 min; 0 min represents the time at which the cells were released from the α-factor arrest. Whole-cell extracts were made from the collected cells and probed with anti-HA antibodies to detect the HA-tagged endogenous Pds1. An α-tubulin immunoblot was done as a loading control.
Mad2 associates with Mad3 and Cdc20 in mitosis in the absence of functional kinetochores.
The previous experiments do not rule out a role for the kinetochore in assembling either MCC or the Mad2-Cdc20 complex. Therefore, we tested the requirement of the kinetochore in assembling MCC and Mad2-Cdc20 by using an ndc10-1 mutant so kinetochores can be destroyed in a temperature-dependent fashion (5, 11). The design of the experiment is shown in Fig. 5A. Cells containing ndc10-1, PGAL1 -PDS1-MDB, Mad3-PrA, and Cdc20-13Myc were grown under permissive conditions for ndc10-1 and arrested in G1 using α-factor. The temperature was raised to 37°C for two hours to inactivate Ndc10. Cells were released from α-factor in the presence of galactose to induce expression of the ectopic PDS1-MDB, and cells were collected 90 min later, at a time when wild-type cells normally enter anaphase (Fig. 5C).
The efficiency of the synchronization steps was confirmed by flow cytometry and is shown in Fig. 5B. We detected MCC as Mad2 associated with Mad3-PrA, and we identified the independent Mad2-Cdc20 complex by sequential immunoprecipitation (Mad2 associated with Cdc20-13Myc in the supernatant after removing Mad3-PrA) as described in Fig. 2B. Similar amounts of MCC were present in both wild-type and ndc10-1 cells, and Mad2-Cdc20 was also present in both wild-type and ndc10-1 cells (Fig. 5B). This confirms that both Mad2-containing complexes form in mitosis and suggests that the kinetochore is not required for the formation of either checkpoint complex.
It is essential to confirm that kinetochores are completely inactive in the ndc10-1 mutant. We used an exquisitely sensitive assay that takes advantage of the observation that a single kinetochore in capable of inducing a checkpoint signal (26). We tagged the endogenous wild-type Pds1 protein with a 3HA epitope at the PDS1 locus. The Pds1 protein has an intact destruction box and is subject to spindle checkpoint control. During the experiment described above, we released cells from α-factor and grew them in the presence or absence of nocodazole. We assayed the stability of the endogenous Pds1-3HA over time. If even a single kinetochore remained intact in the ndc10-1 mutant, then the Pds1-3HA would remain stable.
In the absence of nocodazole, Pds1-3HA turned over in both wild-type and ndc10-1 cells between 80 and 90 min after release from α-factor. The instability of the endogenous Pds1-3HA in wild-type cells expressing PGAL1 -PDS1-MDB shows that the spindle checkpoint was not activated by ectopic expression of the mutant Pds1. In the presence of nocodazole, endogenous Pds1-3HA was stabilized in wild-type cells because the checkpoint was active, but Pds1-3HA turned over in nocodazole-treated ndc10-1 cells, showing that every kinetochore had been inactivated (Fig. 5C). We conclude that both complexes of Mad2-containing spindle checkpoint proteins, mitotic MCC and Mad2-Cdc20, form in mitosis in the complete absence of kinetochore function, and the role of the kinetochore in spindle checkpoint signaling is independent of complex assembly.
DISCUSSION
MCC is a minor complex in vivo.
We have quantified the spindle checkpoint proteins of the MCC in yeast and shown that they are present in sufficient quantities to act as stoichiometric inhibitors of the APC/C. The two protein complexes that contain Mad2, MCC and Mad2-Cdc20, are present in different quantities in yeast cells, and the minor component is MCC, the most potent in vitro spindle checkpoint inhibitor identified to date. We used conditional mutants to establish a checkpoint arrest and then remove Mad2 and Mad3. These experiments show that that both Mad2 and Mad3 are important for maintaining a spindle checkpoint arrest. MCC and Mad2-Cdc20 formation is not induced by the spindle checkpoint rather the complexes forms in cells in mitosis regardless of the state of the spindle. Finally, the formation of both MCC and Mad2-Cdc20 is completely independent of the kinetochore.
The target of the spindle checkpoint is Cdc20 and mutants that prevent Mad2 from interacting with Cdc20 abrogate the signal (21). There are two complexes that form in vivo, Mad2-Cdc20 and MCC, and therefore they are excellent candidates for mitotic inhibitors (12, 37, 38). Cells arrested in mitosis contain both as separable complexes, but the relationship between the two is not clear. One possible explanation is that the two complexes are fundamentally different. It has been proposed that the Mad2-Cdc20 complex sequesters Cdc20 from the APC/C and thereby serves as an inhibitor (1). MCC is thought to act stoichiometrically by binding to the APC/C and inhibiting it directly (37). We found that yeast cells arrested in mitosis with nocodazole contain very small quantities of MCC. Therefore, it is not likely that MCC is a stoichiometric inhibitor of APC/C and the dominant mitotic inhibitor given the small quantity. Despite the small quantity, it is a critical inhibitor because cells are unable to maintain a spindle checkpoint arrest in the absence of Mad3. We interpret this to mean that MCC is required for the maintenance of the spindle checkpoint, although we cannot rule out that there is another pool of Mad3 that is also important for maintenance.
Why is there such a small amount of MCC if it is such a potent inhibitor of the APC/C? One possibility is that MCC is catalytic. Another possibility is that it is a stoichiometric inhibitor of a special pool of APC/C or perhaps only a small fraction of the APC/C is active in the cell.
Complexes are not induced to form in response to activating the spindle checkpoint.
There are discrepancies in the literature about whether the association of Mad2 and Cdc20 is induced by the spindle checkpoint. MCC forms in yeast cells grown in the presence of nocodazole, is present in low amounts in cells grown in hydroxyurea and is absent from cells arrested in G1 with α-factor (16). MCC is not present in mitotic extracts of Xenopus oocytes and is induced to form after adding sperm and nocodazole to activate the spindle checkpoint (6). In contrast, several earlier studies using mammalian cells showed that Mad2 associated with Cdc20 in response to nocodazole and can be interpreted to mean either formation of MCC or Mad2-Cdc20 (12, 37, 38). Sudakin et al. showed that MCC is present in interphase HeLa cells at a time when there are no mature kinetochores and the checkpoint is silenced and the MCC is active as an in vitro inhibitor of the APC/C (37). Our data clearly show that MCC and Mad2-Cdc20 form in mitosis and levels do not increase in response to checkpoint signals.
What regulates the formation of complexes in mitosis is unclear. Mitotic phosphorylation may play a role but a more likely candidate is the levels of Cdc20 protein. The mRNA expression of Cdc20 is periodic and peaks at the metaphase to anaphase transition in yeast (35). Cdc20 is a substrate of Cdh1, a specificity factor for the APC/C that is active throughout the G1 phase of the cell cycle (20). Therefore, there is no Cdc20 mRNA in G1 and the protein is degraded by Cdh1-APC/C. Mad2, Mad3, and Bub3 are constitutively expressed (7, 14, 16). The simplest interpretation of our data is that Cdc20 is limiting for both Mad2-Cdc20 and MCC. Mad2-Cdc20 and MCC cannot be formed in G1 and the small amount of MCC that is detected in hydroxyurea-arrested cells is expected since cells arrest before the peak in Cdc20 synthesis (35).
Mitosis per se is not sufficient for forming MCC in Xenopus laevis because cytostatic factor (CSF)-arrested Xenopus egg extracts do not contain MCC even though all of the subunits are present (6). We suspect that this reflects a significant difference in embryonic systems compared to somatic cells that may reflect the need to keep the spindle checkpoint inactive until the midblastula transition. Perhaps there is an active mechanism to keep the checkpoint inactive during the early embryonic cell divisions. For example, Emi1 is a protein that binds and inhibits Cdc20 from S phase until the early stages of mitosis, and Emi1 is a component of CSF (31, 32). Since both Emi1 and Mad2 bind to the amino terminus of Cdc20, it is likely that Emi1 binding to Cdc20 in CSF-arrested extracts inhibits the formation of MCC.
Kinetochore is not required for formation of the complexes.
A previous report suggested that MCC formation is kinetochore independent (14). Our data confirm the principle conclusions of the previous report but extend the conclusion by two important observations. First, our quantification shows that MCC is present in low quantities in cells and therefore accounts for a very small fraction of inhibited Cdc20. We measured the formation of the independent Mad2-Cdc20 complex, which is much more abundant, and we show that its formation is also kinetochore independent.
It was important to repeat the experiment to test the requirement of the kinetochore in the formation of MCC and Mad2-Cdc20 for three important reasons. The first is that the previous experiment assayed the formation of MCC in kinetochore mutants ndc10-1, spc24-1, and spc25-1 arrested with hydroxyurea (14). Hydroxyurea-treated cells contain very little Cdc20 and hence a very small amount of MCC and are arrested at the start of S phase, well before the onset of anaphase. The second is that the previous experiment used cell synchrony with the temperature-sensitive ndc10-1 mutant arrested in α-factor at the permissive temperature and then released to the cell cycle at the restrictive temperature. The experiment did not account for the time required to inactivate the Ndc10 protein. We have found that it takes two hours to completely eliminate the spindle checkpoint function in our ndc10-1 mutant strains (P. Tavormina, and D. Burke, unpublished observation). Thus, it was impossible to determine if the MCC detected in the previous experiment was formed before cells lost kinetochore function. The third reason is that it is also necessary to determine if the formation of Mad2-Cdc20 is kinetochore dependent and it was not tested previously. We found that there is no role for the kinetochore in the formation of either MCC or Mad2-Cdc20. Our data agree with those of Sudakin et al. (37), who isolated MCC from interphase cells, where there are no kinetochores.
Cdc20 and Mad1 both have similar regions that bind Mad2 (28). Moreover, peptides that correspond to the Mad2 binding domains of Mad 1 and Cdc20 induce a similar conformational change to Mad2 (28). Mad1 is required to localize Mad2 to kinetochores and it is believed that Mad1, in the kinetochore, catalyzes a change in Mad2 that promotes association with Cdc20 (17, 28, 37). Our data argue that this cannot be true. Both MCC and Mad2-Cdc20 can form in the complete absence of the kinetochore function although neither is capable of inhibiting APC/C activity. If Mad1 is catalyzing the association between Mad2 and Cdc20 then it is happening somewhere in the cell other than at the kinetochore. Mad1 and Mad2 have been localized to the nuclear pore through interactions with the Nup53 complex of proteins (22). It is possible that Mad2 is exchanged with Cdc20 during transport through the nuclear pore complex, however, this seems unlikely because nup53 mutants are checkpoint proficient.
Unattached kinetochores are clearly at the top of a signal transduction pathway that senses microtubule attachment and in its absence initiates spindle checkpoint signaling. If Mad2-Cdc20 complexes form in the absence of the kinetochore, what is the role of the kinetochore in the spindle checkpoint? More complex models that require the kinetochore as the site of assembly can be envisioned. Perhaps there is some other inhibitor that has not yet been identified, or perhaps there is a core of the MCC or of Mad2-Cdc20 that form in mitosis and there is an exchange of subunits at the kinetochore that convert it to an inhibitor. Alternatively, either or both complexes may get modified, for example by protein kinases, at the kinetochore. Sudakin et al. (37) suggested that the role of the kinetochore is to potentiate the APC/C so that it is responsive to MCC inhibition. We think this is unlikely to account for all of the inhibitory activity, given the small amount of MCC in yeast, but it is possible that one role of the kinetochore is to modulate the activity of the APC/C.
Checkpoint proteins are at kinetochores when the spindle checkpoint is active, and many associate dynamically, with very short half-lives (18, 23, 34). The simple model that the kinetochore is the site of Mad2-Cdc20 assembly is incorrect, but the dynamic nature of these important regulatory proteins is likely to be one of the keys to understanding the molecular details of the spindle checkpoint.
Acknowledgments
We thank Orna Cohen Fix and Doug Koshland for strains and reagents and the members of the Burke and Stukenberg laboratories for helpful discussions. We thank Andy Hoyt for critical comments on the manuscript.
The work was supported by a grant from the National Institutes of Health (GM40344).
REFERENCES
- 1.Bharadwaj, R., and H. Yu. 2004. The spindle checkpoint, aneuploidy, and cancer. Oncogene 23**:**2016-2027. [DOI] [PubMed] [Google Scholar]
- 2.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:**115-132. [DOI] [PubMed] [Google Scholar]
- 3.**Burke, D. J., D. Dawson, and T. Stearns.**2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Chan, G. K., and T. J. Yen. 2003. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog. Cell Cycle Res. 5**:**431-439. [PubMed] [Google Scholar]
- 5.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157**:**199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, R. H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158**:**487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, R. H., D. M. Brady, D. Smith, A. W. Murray, and K. G. Hardwick. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10**:**2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. H., J. C. Waters, E. D. Salmon, and A. W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274**:**242-246. [DOI] [PubMed] [Google Scholar]
- 9.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10**:**3081-3093. [DOI] [PubMed] [Google Scholar]
- 10.Dohmen, R. J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263**:**1273-1276. [DOI] [PubMed] [Google Scholar]
- 11.Enquist-Newman, M., I. M. Cheeseman, D. Van Goor, D. G. Drubin, P. B. Meluh, and G. Barnes. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12**:**2601-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13**:**755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, G., H. Yu, and M. W. Kirschner. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12**:**1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraschini, R., A. Beretta, L. Sironi, A. Musacchio, G. Lucchini, and S. Piatti. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20**:**6648-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner, R. D., A. Poddar, C. Yellman, P. A. Tavormina, M. C. Monteagudo, and D. J. Burke. 2001. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics 157**:**1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick, K. G., R. C. Johnston, D. L. Smith, and A. W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148**:**871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwick, K. G., R. Li, C. Mistrot, R. H. Chen, P. Dann, A. Rudner, and A. W. Murray. 1999. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics 152**:**509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell, B. J., B. Moree, E. M. Farrar, S. Stewart, G. Fang, and E. D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14**:**953-964. [DOI] [PubMed] [Google Scholar]
- 19.Hoyt, M. A., L. Totis, and B. T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66**:**507-517. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage, and D. Pellman. 2001. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154**:**85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang, L. H., L. F. Lau, D. L. Smith, C. A. Mistrot, K. G. Hardwick, E. S. Hwang, A. Amon, and A. W. Murray. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279**:**1041-1044. [DOI] [PubMed] [Google Scholar]
- 22.Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai, and R. W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159**:**807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallio, M. J., V. A. Beardmore, J. Weinstein, and G. J. Gorbsky. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158**:**841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37**:**251-282. [DOI] [PubMed] [Google Scholar]
- 25.Li, R., and A. W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell 66**:**519-531. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., and R. B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature 373**:**630-632. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. Mckenzie. III; D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Phillipsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 28.Luo, X., Z. Tang, J. Rizo, and H. Yu. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9**:**59-71. [DOI] [PubMed] [Google Scholar]
- 29.McCleland, M. L., R. D. Gardner, M. J. Kallio, J. R. Daum, G. J. Gorbsky, D. J. Burke, and P. T. Stukenberg. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17**:**101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid, R. J., M. Lisby, R. Rothstein. 2002. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350**:**258-277. [DOI] [PubMed] [Google Scholar]
- 31.Reimann, J. D., E. Freed, J. Y. Hsu, E. R. Kramer, J. M. Peters, and P. K. Jackson. 2001. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105**:**645-655. [DOI] [PubMed] [Google Scholar]
- 32.Reimann, J. D., and P. K. Jackson. 2002. Emi1 is required for cytostatic factor arrest in vertebrate eggs. Nature 416**:**850-854. [DOI] [PubMed] [Google Scholar]
- 33.Rieder, C. L., R. W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130**:**941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah, J. V., E. Botvinick, Z. Bonday, F. Furnari, M. Berns, and D. W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14**:**942-952. [DOI] [PubMed] [Google Scholar]
- 35.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9**:**3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stukenberg, P. T., and D. J. Burke. 2004. Analyzing the spindle checkpoint in yeast and frogs. Methods Mol. Biol. 280**:**83-98. [DOI] [PubMed] [Google Scholar]
- 37.Sudakin, V., G. K. Chan, and T. J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154**:**925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1**:**227-237. [DOI] [PubMed] [Google Scholar]
- 39.Weiss, E., and M. Winey. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132**:**111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152**:**349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14**:**706-714. [DOI] [PubMed] [Google Scholar]