Retroviral Splicing Suppressor Sequesters a 3′ Splice Site in a 50S Aberrant Splicing Complex (original) (raw)
Abstract
Retroviral replication requires both spliced and unspliced mRNAs. Splicing suppression of avian retroviral RNA depends in part upon a _cis_-acting element within the gag gene called the negative regulator of splicing (NRS). The NRS, linked to a downstream intron and exon (NRS-Ad3′), was not capable of splicing in vitro. However, a double-point mutation in the NRS pseudo-5′ splice site sequence converted it into a functional 5′ splice site. The wild-type (WT) NRS-Ad3′ transcript assembled an ∼50S spliceosome-like complex in vitro; its sedimentation rate was similar to that of a functional spliceosome formed on the mutant NRS-Ad3′ RNA. The five major spliceosomal snRNPs were observed in both complexes by affinity selection. In addition, U11 snRNP was present only in the WT NRS-Ad3′ complex. Addition of heparin to these complexes destabilized the WT NRS-Ad3′ complex; it was incapable of forming a B complex on a native gel. Furthermore, the U5 snRNP protein, hPrp8, did not cross-link to the NRS pseudo-5′ splice site, suggesting that the tri-snRNP complex was not properly associated with it. We propose that this aberrant, stalled spliceosome, containing U1, U2, and U11 snRNPs and a loosely associated tri-snRNP, sequesters the 3′ splice site and prevents its interaction with the authentic 5′ splice site upstream of the NRS.
The generation of proteomic diversity occurs in part by alternative splicing, the process by which a pre-mRNA can be spliced using different combinations of 5′ and 3′ splice sites (42). The regulation of alternative splicing is also important for retroviral replication (39). A simple retrovirus, Rous sarcoma virus (RSV), uses a single 5′ splice site and two 3′ splice sites to generate two singly spliced mRNA species, which are translated into the Env and Src proteins (7). In addition, a large fraction of retroviral primary transcripts are not spliced. Instead, a pool of unspliced pre-mRNA is maintained and used both as mRNA for the Gag and Pol proteins and as packaged genomic RNA.
RSV encodes no known accessory proteins; its RNA splicing is regulated by _cis_-acting RNA sequences within the RSV genome. The env branchpoint is suboptimal; mutations that increase env splicing impair retroviral replication (14, 22). The splicing of the RSV src gene is negatively regulated by two _cis_-acting elements. The first, known as the suppressor of src splicing, is found upstream of the distal src 3′ splice site (1), and the second, known as the negative regulator of splicing (NRS), is downstream of the viral 5′ splice site within the gag gene (2, 35, 37).
The NRS suppresses splicing from an upstream 5′ splice site both in vivo and in vitro (16, 34, 35, 37). It has been shown to bind several splicing factors, including SF2/ASF and U1 and U11 snRNPs (16, 19, 32, 33). The terminal regions of the NRS are both essential for splicing suppression (35). SF2/ASF and hnRNPH bind to the purine-rich 5′ portion of the NRS, which functions as a splicing enhancer (12, 31). U1 snRNP binds to a pseudo-5′ splice site near the 3′ end of the NRS, which contains a 5/8 match to a U2-dependent 5′ splice site consensus sequence (19, 33). Three nonconsensus U's are present at positions −2, +3, and +4 relative to the pseudo-splice site (Fig. 1A). Mutation of any one of these U's to a consensus A converts the NRS into a functional 5′ splice site in vivo (38). Surprisingly, mutation of the two U's at +3 and +4 to nonconsensus C's abolishes splicing suppression (38). U11 snRNP also binds to the NRS RNA in vitro at a perfect 5′ splice site consensus sequence for a U12-dependent intron that begins at +5 relative to the U2-dependent pseudo-5′ splice site (16) (Fig. 1A). U11 snRNP recognizes a minor class of 5′ splice sites (46). However, the role of U11 snRNP in NRS-mediated splicing suppression remains unclear (19, 33).
FIG. 1.
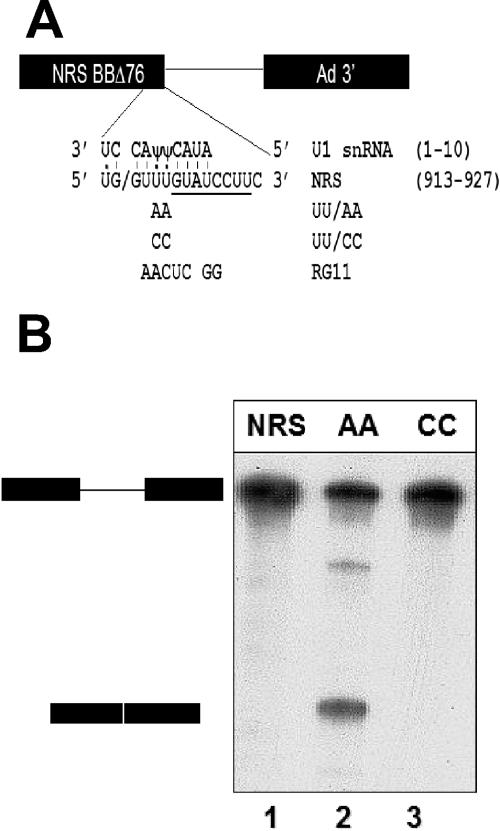
Mutated NRS sequence (UU/AA) functions as a 5′ splice site in vitro. A) The NRS sequence was fused to the adenovirus major late intron and 3′ splice site (13) to generate a splicing substrate. The 5′ splice site-like sequence of the WT NRS (RSV nt 913 to 927) is shown base paired with the 5′ end of U1 snRNA. The slash indicates the potential splice junction. In addition to the five matches to the consensus U2-dependent 5′ splice site (positions −1, +1, +2, +5, and +6), U:U and U:Ψ interactions at −2, +3, and +4 are shown. Note that the U1 base pairing extends beyond the consensus sequence to +7 and +8. In addition, the NRS contains a perfect consensus U12-dependent 5′ splice site, extending from +5 to +12 relative to the splice junction (underlined). The mutations utilized in this study are shown below the wild-type sequence. UU/AA makes the NRS a closer match to the consensus 5′ splice site. UU/CC and RG11 are alternative nonconsensus sequences. B) NRS-Ad3′ RNAs were incubated in an in vitro splicing reaction for 180 min and electrophoresed on a 6% acrylamide-8 M urea gel. Positions of the pre-mRNA substrate and the spliced product are shown on the left.
When the NRS is inserted into the intron of a heterologous adenovirus splicing substrate, it inhibits its splicing in vitro (16). In the absence of ATP, the NRS interacts with a downstream adenovirus 3′ splice site (Ad3′) to form a commitment complex containing U1 snRNP and ASF/SF2 (8, 15). These data led to the hypothesis that the NRS functions as a decoy 5′ splice site that interacts with a downstream 3′ splice site to prevent its splicing. Here, we investigated whether the NRS can form a larger RNP complex, perhaps even a spliceosome, in the presence of ATP.
RNA splicing in eukaryotic cells is carried out by a large, dynamic RNA-protein complex called the spliceosome, which appears to be assembled on the pre-mRNA through a series of intermediates (reviewed in references 4, 36, and 43). The first of these, termed the E or commitment complex, is characterized by ATP-independent binding of U1 snRNP at the 5′ splice site. Secondly, U2 snRNP binds to the branchpoint sequence to form the ATP-dependent pre-spliceosome or A complex. The U4/U6.U5 tri-snRNP complex is then recruited to form the B complex. Recent work has identified a spliceosomal intermediate called BΔU1, in which U1 snRNP has been dissociated but the tri-snRNP has yet to undergo the rearrangements required to form the catalytically active C or B* complex, containing U2, U5, and U6 snRNPs (27). Despite extensive characterization of the intermediates that occur during assembly, the possibility exists that the spliceosome may be recruited to the pre-mRNA as a preformed unit (28, 45). In this model of spliceosome assembly, the tri-snRNP is present but loosely associated during the early stages. In either the stepwise or penta-snRNP model of assembly, multiple, mutually exclusive RNA-RNA interactions occur.
To further investigate the mechanism of NRS-mediated splicing suppression, we have studied the interaction between the NRS and a downstream 3′ splice site sequence (Ad3′) in vitro. We have compared a splicing substrate containing the NRS pseudo-5′ splice site with one containing an authentic 5′ splice site, derived from the NRS by UU/AA mutations at positions +3 and +4 (Fig. 1A). Both of these RNAs assembled ATP-dependent, ∼50S RNP complexes, which contained all five major spliceosomal snRNPs. In contrast to the functional spliceosome, the NRS-Ad3′ complex was disrupted by heparin. In addition, the U5 snRNP 220-kDa protein, hPrp8, failed to cross-link to the NRS RNA as it did to the functional 5′ splice site, suggesting that the U5 snRNP was positioned differently than it was in an authentic spliceosome. We propose that the inability of the NRS pseudo-5′ splice site to bind hPrp8 properly prevents it from becoming a functional 5′ splice site. However, the NRS can still generate an aberrant spliceosomal complex that sequesters the 3′ splice site, thus suppressing splicing.
MATERIALS AND METHODS
In vitro transcription.
The templates for in vitro transcription of the NRS-Ad3′ RNAs were generated by PCR from the NRS fragment BBΔ76 linked to the adenovirus major late intron and 3′ exon (16). Mutations were introduced by PCR, and the templates were sequenced to confirm the mutations. The 300-nucleotide (nt) NRS-Ad3′ RNAs were transcribed with T7 polymerase, a gift from Yun-Xing Wang, as previously reported (16).
Splicing reactions.
Approximately 10 fmol of uniformly 32P-labeled RNA was incubated in a 20-μl splicing reaction for 3 h at 30°C. Each reaction mixture contained 60% HeLa cell nuclear extract in buffer D (10) with 2.6 mM polyvinyl alcohol, 4.2 mM ATP, 4.2 mM MgCl2, 5.2 mM creatine phosphate, and 0.4 U of RNasin. The reaction was digested with proteinase K, extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a 6% acrylamide-8 M urea gel.
Velocity sedimentation.
Total volumes of 11 ml of 10% to 30% sucrose gradients were made in buffer D containing 4.2 mM MgCl2. For the gradients shown in Fig. 2, ∼10 fmol of 32P-labeled RNA was incubated in a 20-μl splicing reaction mixture, layered on top of the gradient, and centrifuged in a Beckman SW41Ti rotor at 35,000 rpm for either 4.5 h (Fig. 2A) or 8 h (Fig. 2B). In the latter case, heparin was added to a final concentration of 0.8 mg/ml immediately after the reaction.
FIG. 2.
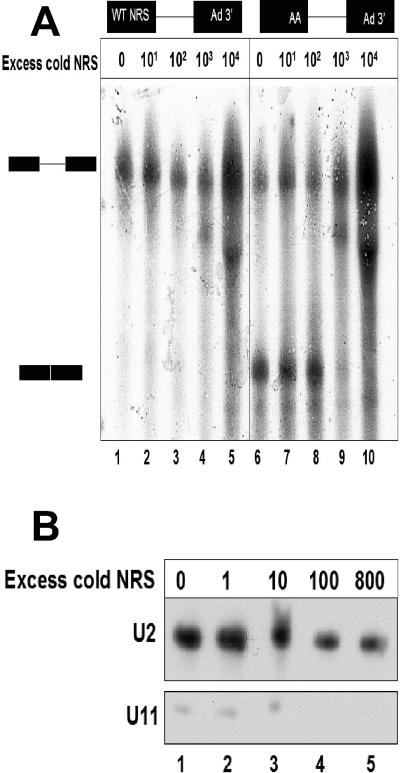
NRS-Ad3′ RNA assembles an ∼50S complex in the absence of heparin but fails to assemble a 35S complex in the presence of heparin. A) NRS-Ad3′, UU/AA-Ad 3′, and RG11-Ad3′ RNA substrates, plus the Ad3′ intron and exon portion alone, were incubated in a splicing reaction for 30 min at 30°C and layered onto an 11-ml sucrose gradient in the absence of heparin. The gradients were centrifuged at 35,000 rpm for 4.5 h at 20°C in a Beckman SW41 Ti rotor. Fraction 1 is the heaviest fraction. The Escherichia coli 50 S ribosomal subunit was used as a marker. Only fractions 12 to 24 are shown for clarity. B) The NRS-Ad3′, RG11-Ad3′, and UU/AA-Ad 3′ RNAs were incubated in splicing reactions as described above, and heparin was added after the incubation and before layering onto the gradient. Gradients were sedimented as described above for 8 h. Chicken rRNAs were used as markers.
Native gel electrophoresis.
Native gel analysis was carried out as previously described (9).
Affinity selection.
In this report two different types of affinity selection were performed. For each affinity selection performed (see Fig. 4), a 100-μl splicing reaction was carried out with a reaction mixture containing 1.2 pmol of biotinylated RNA. After velocity sedimentation without heparin, as shown in Fig. 2A, the 50S peak fractions were collected (1 ml total) and mixed with 25 μl of a 1:1 slurry of streptavidin-agarose beads (Pierce). Incubation occurred overnight at 4°C in buffer D plus 0.1% NP-40 and 4 mM MgCl2. RNA was harvested from the beads, after four washes in Buffer D containing 100 mM KCl, by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation and subjected to Northern analysis as described previously (16). The affinity selection performed (see Fig. 5B) used RNA covalently bound to agarose beads as previously published (6).
FIG. 4.
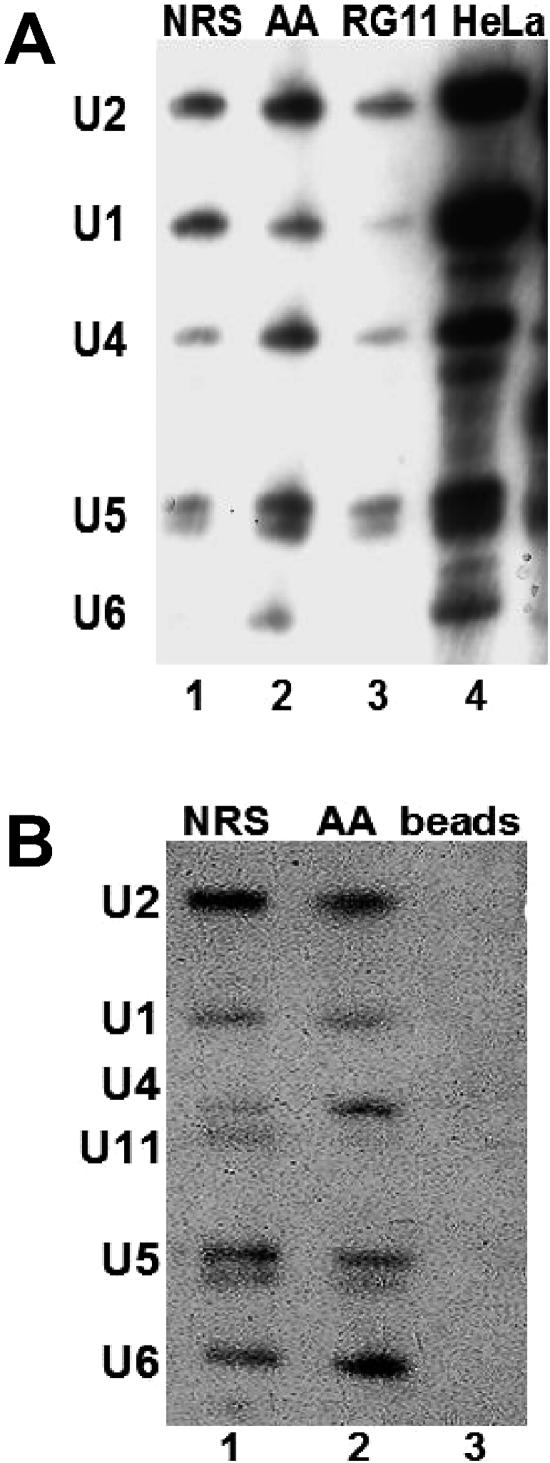
The ∼50S splicing complex contains all five major spliceosomal snRNAs. A) Biotinylated NRS-Ad3′ RNAs were incubated in splicing reactions and layered onto a sucrose gradient in the absence of heparin. The ∼50S peak fractions were affinity selected using streptavidin-agarose beads. The affinity-selected RNA was harvested and electrophoresed, and a Northern analysis was performed, using probes for the major spliceosomal snRNAs. The HeLa control lane contains total RNA extracted from HeLa nuclear extract. B) Affinity selection was carried out as described for panel A. The Northern blot was probed to detect the five major spliceosomal snRNAs and then reprobed with a probe complementary to U11 snRNA. The lane marked “Beads” exhibits a control affinity selection carried out in the absence of biotinylated RNA.
FIG. 5.
Incubation of excess NRS RNA with a splicing reaction failed to activate the WT NRS as a 5′ splice site. A) The WT NRS-Ad3′ RNA was incubated in splicing reactions with various amounts of a cold NRS sequence, ranging from 0 to 10,000-fold molar excess (lanes 1 to 5). The UU/AA mutant NRS-Ad3′ RNA was incubated under conditions identical to those of a positive control for splicing (lanes 6 to 10). B) Affinity depletion of U11 snRNA from splicing reactions. A biotinylated NRS fragment was titrated into a splicing reaction for 30 min. The NRS RNA was affinity selected using streptavidin-agarose beads, and the selected pellet was discarded. The remaining RNA from the HeLa nuclear extract was isolated, and a Northern analysis was performed with probes for U11 and U2 snRNAs.
Site-specific labeling.
The generation of RNA with a single site-specific, radioactive phosphate was performed essentially as previously described (30). Between 500 and 1,000 pmol of gel-purified NRS-Ad3′ RNA transcripts was annealed with an equimolar amount of the following chimeric oligonucleotide: 5′-CCACTCCCCACATAAGGAG-3′ (Oligos, Etc.). The first 4 nt were 2′ deoxynucleotides, and the remaining 16 were 2′-O-methyl ribonucleotides. The RNA was digested with RNase H at a site between positions +1 and +2 of the 5′ splice site-like sequence, and the fragments were gel purified. The 3′ half was phosphatased with calf intestinal phosphate and kinased with T4 polynucleotide kinase (both from New England Biolabs) and [γ-32P]ATP (Perkin-Elmer) and then ligated with an equimolar amount of the 5′ half, using a bridging DNA oligonucleotide (Operon), 5′-GGGAAGGATACAAACCACTCCCCACATAAGG-3′, and a rapid ligation kit (Invitrogen). The products were analyzed on a 6% acrylamide-8 M urea gel and gel purified. UV irradiation, RNase digestion, and electrophoresis on a sodium dodecyl sulfate (SDS)-6% polyacrylamide gel were performed as described previously (49).
Immunoprecipitation.
After digestion with 2 mg/ml RNase A (Calbiochem) for 30 min at 37°C, the residual site-specifically labeled RNA and cross-linked protein were immunoprecipitated. Rabbit antibody (1 μl) raised against the C-terminal fragment of human Prp8 (26) was incubated with the cross-linked material for 1 h at 4°C. Pansorbin (Calbiochem) was added and incubated for an additional 15 min at 4°C. The immunoprecipitate was washed three times in buffer D and centrifuged through a 6% polyacrylamide gel containing SDS.
RESULTS
Mutagenesis converts the NRS into a 5′ splice site.
To study the mechanism of splicing suppression by the NRS, we first characterized the interaction between the NRS and a downstream 3′ splice site in splicing reactions carried out in vitro. For the sake of comparison, we needed a positive control that would assemble authentic splicing complexes. Previous work demonstrated that mutation of any one of the three nonconsensus U's in the NRS 5′ splice site-like sequence to consensus A's generates a functional 5′ splice site in vivo (38). Here, we used the UU917/918AA (UU/AA) double-point mutant, which makes the NRS sequence a 7/8 match to the 5′ splice site consensus sequence with two A's instead of two nonconsensus U's at positions +3 and +4 relative to the splice junction (Fig. 1A).
To test the ability of the UU/AA NRS mutant to function as a 5′ splice site in vitro, it was fused to an adenovirus intron and exon sequence called Ad 3′ (Fig. 1A) and incubated in HeLa cell nuclear extract under splicing conditions. Gel electrophoresis to separate the spliced and unspliced RNAs revealed that the UU/AA-Ad3′ pre-mRNA was efficiently spliced (Fig. 1B, lane 2), whereas the NRS-Ad 3′ substrate generated no detectable spliced products (Fig. 1B, lane 1). Thus, mutation of two nucleotides in the NRS 5′ splice site-like sequence converted it into a functional 5′ splice site; similar results have been observed in vivo in chicken embryo fibroblasts (38). The UU/AA mutant will be used as a positive control in our subsequent studies because it differs from the wild-type (WT) NRS sequence at only 2 out of 151 nt and yet it functions as a 5′ splice site instead of as a splicing suppressor.
We also tested the effect of an additional NRS mutation, UU917/918CC, on splicing in vitro. This UU/CC mutant changed the nonconsensus bases at +3 and +4 from U to C (Fig. 1A). Since C is also nonconsensus at these positions, the UU/CC NRS mutant sequence can potentially form the same Watson-Crick base pairs with U1 snRNA as the wild-type NRS sequence. Nevertheless, this UU/CC mutation greatly reduces binding of U1 snRNP to the NRS in vitro (5) and functions as neither a splicing suppressor nor a splice site in vivo (38). Accordingly, we observed no splicing of the UU/CC mutant NRS-Ad3′ RNA in vitro (Fig. 1B, lane 3). In addition, we tested a more extensive NRS mutant, called RG11 (Fig. 1A), which fails to bind both U1 and U11 snRNPs (16, 33). The RG11-Ad3′ splicing substrate also failed to splice in HeLa nuclear extract (data not shown).
Thus, we found that binding of U1 snRNP to the NRS was necessary but not sufficient for splicing in vitro, since the wild-type NRS sequence binds U1 snRNP nearly as well as the UU/AA mutant (5); however, only the latter construct is capable of splicing.
NRS-Ad 3′ RNA assembles an ∼50S complex that is unstable in the presence of heparin.
To determine the reason for the splicing defect in the NRS, we first compared the RNP complex that assembled on the NRS-Ad3′ substrate with that of the UU/AA NRS-Ad3′ functional pre-mRNA. As negative controls, we used the RG11-Ad3′ nonfunctional RNA and the Ad3′ intron/exon transcript by itself. After incubation of the radiolabeled pre-mRNAs in HeLa nuclear extract with splicing buffer and ATP, the complexes formed were analyzed by velocity sedimentation through sucrose gradients in the absence of heparin. We observed that both the wild-type NRS-Ad3′ and the UU/AA NRS-Ad3′ RNAs assembled a major ∼50S RNP complex (Fig. 2A). This was consistent with previous studies in which an ∼50S spliceosome assembled onto splicing substrates after similar incubation conditions (13, 17). In contrast, the Ad3′ sequence alone, containing the adenovirus major late intron, 3′ splice site, and downstream exon sequence but no 5′ splice site, did not assemble a 50S complex (Fig. 2A). The RG11-Ad3′ substrate repeatedly assembled a complex with slightly lower mobility than the ∼50S splicing complex (Fig. 2A). Although both the NRS-Ad3′ and UU/AA NRS-Ad3′ RNA substrates assembled complexes with very similar sedimentation rates in sucrose gradients, the striking functional differences between these two NRS sequences made it seem unlikely that the RNP complexes that formed on them were identical.
To test the stability of these ∼50S complexes, we next treated the splicing reactions with heparin prior to velocity sedimentation analysis. Heparin is a polyanion that disrupts loose protein-RNA interactions (21). The addition of heparin to a spliceosome assembly reaction typically results in the formation of 35S, 25S, and 15S complexes, corresponding to B/C, A, and E/H complexes, respectively (18). The UU/AA NRS-Ad3′ pre-mRNA was capable of forming all three splicing complexes, including the 35S B/C spliceosomal complex (Fig. 2B); this is consistent with our previous observation of its ability to splice (Fig. 1B, lane 2). In contrast, the NRS-Ad3′ and the RG11-Ad3′ transcripts formed only 25S and 15S complexes, which may include presplicing complexes (Fig. 2B). In summary, while the ∼50S complex that formed on the NRS-Ad3′ RNA appeared to be identical to that which formed on an authentic splicing substrate (UU/AA NRS-Ad3′), the two complexes differed in their stability in the presence of heparin.
The NRS-Ad3′ RNA does not assemble a B complex.
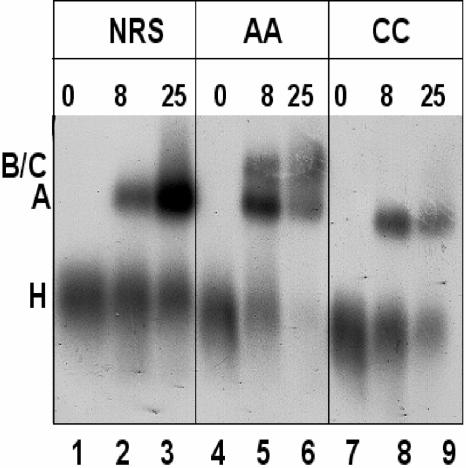
Velocity sedimentation in the presence of heparin suggested that the NRS-Ad3′ RNA could assemble a 25S A complex but was unable to form a 35S B/C complex. To confirm this result, we performed native gel electrophoresis on the RNP complexes. The wild-type NRS, UU/AA, and UU/CC NRS-Ad3′ RNAs were incubated in HeLa nuclear extract for various lengths of time, treated with heparin, and electrophoresed on a native agarose gel (Fig. 3). The assembly of normal spliceosomal intermediates, H, A, and B/C, on the UU/AA NRS-Ad 3′ RNA, is exhibited in Fig. 3 (lanes 4 to 6). At zero time, the labeled RNA was associated with hnRNPs in an H complex. After 8 min of incubation, most of the RNA was present in a presplicing (A) complex, and some of it was in a B complex containing the tri-snRNP. After 25 min of incubation, there appeared to be less total RNA associated with the RNP complexes, probably due to it being spliced, and it was distributed through A and B/C complexes. In contrast, both the wild-type NRS and the UU/CC mutant were unable to form normal B or C complexes (Fig. 3, lanes 1 to 3 and 7 to 9) and yet both were capable of generating A complexes. Interestingly, the WT NRS A complex appears to be more abundant than either the AA or CC complex after 25 min of incubation, and it was frequently associated with a more slowly migrating smear (Fig. 3; compare lane 3 with lanes 6 and 9).
FIG. 3.
The NRS-Ad3′ RNA generates a stable presplicing complex. Wild-type NRS, UU/AA, and UU/CC NRS-Ad3′ sequences were incubated in a splicing reaction for 0, 8, or 25 min at 30°C, heparin was added, and reactions were electrophoresed through a nondenaturing agarose gel. Positions of H, A, and B/C splicing complexes are indicated on the left.
When splicing reactions were carried out in the absence of ATP, only the H complexes were detected by this method (data not shown). As expected, formation of both the A and the B/C complexes required ATP. Since the Ad3′ RNA alone and the RG11-Ad3′ RNA were also capable of forming A complexes (data not shown), we concluded that the intact NRS sequence was not necessary to form an A complex in this assay. However, the NRS sequence was required to form the ∼50S RNP complex observed on gradients in the absence of heparin (Fig. 2A).
NRS-Ad3′ ∼50S complex contains all five spliceosomal snRNPs plus U11 snRNP.
To investigate the composition of the ∼50S complexes that assembled on the NRS-Ad3′, UU/AA-Ad 3′, and RG11-Ad3′ transcripts, we performed affinity selections using biotinylated RNA substrates. Complexes assembled in HeLa nuclear extract with ATP for 30 min were centrifuged on sucrose gradients in the absence of heparin, as shown in Fig. 2A. ∼50S peaks were collected, and the biotinylated RNA substrates, and associated proteins and RNPs, were affinity selected with streptavidin agarose. After phenol extraction, selected snRNAs were identified by Northern analysis. This analysis showed that all five spliceosomal snRNAs were present in the complexes assembled with both the NRS-Ad3′ and UU/AA NRS-Ad3′ RNAs (Fig. 4A, lanes 1 and 2). The RG11-Ad3 substrate failed to bind U1 snRNA but was associated with the other spliceosomal snRNPs (Fig. 4A, lane 3). The absence of U1 snRNP may account for its decreased mobility in sucrose gradients (Fig. 2A).
The affinity-selected NRS-Ad3′ and UU/AA NRS-Ad3′ RNAs were also probed for U11 snRNA. U11 snRNA was present in the NRS-Ad3′ ∼50S complex but was barely detectable in the UU/AA ∼50S complex (Fig. 4B, lanes 1 and 2). In contrast, splicing reactions that had been incubated for only 8 min prior to affinity selection, and sedimented at ∼30S in the absence of heparin, exhibited U1, U2, and U11 snRNAs in both NRS-Ad3′ and UU/AA-Ad3′ complexes (data not shown). However, after 30 min of incubation, significantly more U11 snRNA was present in the wild-type NRS complex than in the functional splicing complex (Fig. 4B, lanes 1 and 2). This is interesting in light of the fact that the U12-dependent 5′ splice site sequences (+5 to +12 relative to the U2-dependent splice junction) are identical in the wild-type and UU/AA mutant NRS sequences (underlined in Fig. 1A). Perhaps, the arrangement of snRNPs is different in the NRS-Ad3′ 50S complex, allowing the U11 snRNP to gain access to the U12-dependent splice site.
The presence of all five major spliceosomal snRNPs in the ∼50S complex suggests that both the wild-type NRS and the UU/AA NRS-Ad3′ RNAs may form a penta-snRNP complex (28, 45) in the absence of heparin. However, the mutant is capable of splicing and the WT NRS is not. Another interpretation of the data in Fig. 4 (lane 2) is that a heterogeneous mixture of different complexes, including A, BΔU1, and B*, was formed with the functional splicing substrate, and that together, these complexes contained all five of the spliceosomal snRNPs. In some but not all affinity selection experiments, the ratio of U1/U2 and U4/U2 snRNAs was higher for the NRS-Ad3′ substrate than for functional splicing substrates. This could indicate that the U1 and U4 snRNPs were not being efficiently displaced to generate the B*/C catalytic spliceosome in the NRS-Ad3′ complex.
Does the NRS-U11 snRNP interaction inhibit splicing of NRS-Ad 3′?
We considered three possible mechanisms for NRS-mediated splicing suppression. (i) The NRS may recruit a _trans_-acting suppressor that is not normally present in a spliceosome and which prevents splicing. (ii) There may be some critical component of a normal spliceosome that is missing from the NRS splicing complex. (iii) The NRS RNP complex may have all the components of a normal spliceosome, but its configuration may not be compatible with splicing.
Because the affinity selection experiment shown in Fig. 4B suggested that U11 snRNP was preferentially bound to the WT NRS, we tested the hypothesis that this interaction was responsible for the inability of the WT NRS-Ad3′ RNA to splice. To this end, we titrated an excess of NRS RNA into an NRS-Ad3′ splicing reaction in an attempt to sequester U11 snRNP away from the splicing substrate and to correct the splicing defect. However, addition of up to a 10,000-fold excess of cold, WT NRS RNA failed to rescue splicing of the WT NRS-Ad3′ RNA (Fig. 5A, lanes 1 to 5).
The UU/AA NRS-Ad3′ splicing substrate was used as a positive control; it was also titrated with NRS RNA in an identical series of splicing reactions (Fig. 5A, lanes 6 to 10). This substrate was capable of splicing when up to a 100-fold excess of cold NRS RNA was added, but splicing was inhibited upon addition of a 1,000-fold or greater excess of RNA (Fig. 5A, lanes 9 and 10). We also examined a narrower range of intervals between 100- and 1,000-fold excess RNA (data not shown); however, at no point was the NRS-Ad3′ RNA capable of splicing.
We monitored the efficiency of the NRS fragment in depleting U11 snRNP from the HeLa nuclear extract, using biotinylated NRS RNA (ranging up to an 800-fold excess of the splicing substrate shown in Fig. 5A) and streptavidin agarose beads. After affinity selection, the remaining unselected RNA was used in a Northern analysis with probes complementary to U2 and U11 snRNAs (Fig. 5B). After selection with 100-fold excess NRS RNA, there was no detectable U11 snRNA remaining in the extract (Fig. 5B, lane 4). In contrast, the levels of U2 snRNA were not affected by incubation with the NRS RNA. These results suggest that binding of U11 snRNP to the WT NRS RNA is not solely responsible for the NRS-Ad3′ splicing defect.
The U5 snRNP protein hPrp8 does not cross-link to the NRS RNA.
The NRS-Ad3′ substrate was unable to assemble a normal B splicing complex (Fig. 2B and 3), despite the fact that the tri-snRNP was present (Fig. 4). Furthermore, we were unable to restore splicing by titrating out a factor, such as U11 snRNP, that was critical for splicing suppression (Fig. 5). This suggested that some factor necessary for B complex formation, other than the five major spliceosomal snRNPs, was either missing or not properly oriented. To test the latter idea, we generated NRS-Ad3′ RNAs with a single radioactive phosphate between the highly conserved G and U at positions +1 and +2 relative to the splice junction. The RNAs were incubated in a splicing reaction and then irradiated with 254-nm UV light to transfer the radioactive phosphate to any nearby protein. After digestion with RNase A, the radiolabeled proteins were separated on a 6% polyacrylamide gel containing SDS (Fig. 6A).
FIG. 6.
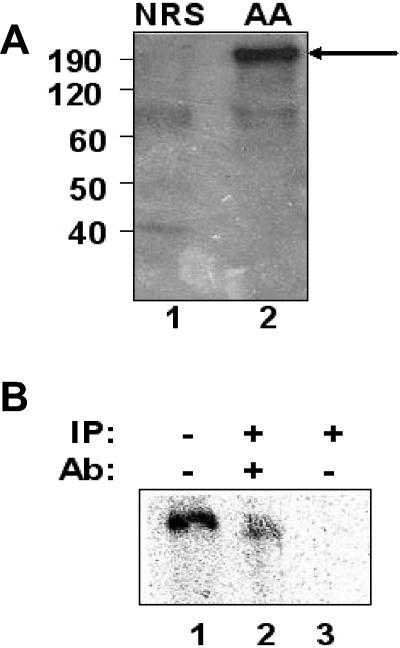
U5 hPrp8 is not properly positioned on the wild-type NRS-Ad3′ RNA. A) The wild-type NRS-Ad 3′ and UU/AA NRS-Ad3′ RNAs were labeled with a single radioactive phosphate between nucleotides G915 and U916, which correspond to positions +1 and +2 relative to the splice junction of the 5′ splice site consensus sequence. The RNAs were incubated in a splicing reaction, UV irradiated to cross-link any bound proteins, and digested with RNase A. The reaction was then denatured and analyzed by SDS-polyacrylamide gel electrophoresis. The sizes of the molecular mass markers are shown on the far left in kilodaltons. The UU/AA NRS-Ad3′ RNA demonstrated the presence of a cross-linked protein of approximately 200 kDa (lane 2, arrow) that was absent from the wild-type NRS-Ad 3′ reaction (lane 1). (B) The UU/AA-Ad3′ RNA was cross-linked as described above and either electrophoresed directly (lane 1) or immunoprecipitated (IP) with an antibody (Ab) against hPrp8 (lane 2) or with no antibody (lane 3).
We observed that a protein slightly larger than 200 kDa was cross-linked to the UU/AA NRS-Ad3′ 5′ splice site sequence but was absent from the wild-type NRS reaction (Fig. 6A). The most likely spliceosomal component of this size binding to the functional 5′ splice site is U5 220 kDa (hPrp8), which was shown previously to cross-link to this position of 5′ splice sites (29, 40, 50). To confirm the identity of the cross-linked species, we utilized an antibody, generated against the C-terminal fragment of hPrp8 (26), and observed that the cross-linked protein was precipitated only in the presence of this antibody (Fig. 6B, lane 2). Thus, hPrp8 is either not positioned properly or is unable to bind to the NRS pseudo-5′ splice site. Since hPrp8 is a component of the U5 snRNP, it is likely that this snRNP, and perhaps the entire tri-snRNP complex, is not properly positioned in this aberrant splicing complex. This may generate a stalled splicing complex which sequesters the 3′ splice site and suppresses its interaction with a productive 5′ splice site.
DISCUSSION
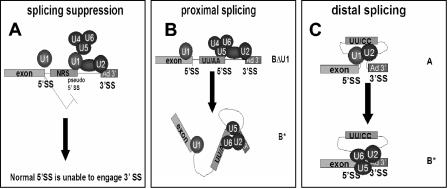
We have observed that the pseudo-5′ splice site of the NRS, when linked to a strong 3′ splice site, assembles an ∼50S spliceosome-like complex, which contains all five major spliceosomal snRNPs. In contrast to an authentic spliceosome, this splicing suppressor complex is unstable in the presence of heparin and migrates as a presplicing (A) complex on native gels. Further, we have found that hPrp8 does not cross-link to the NRS pseudo-5′ splice site, suggesting that the position of the U5 snRNP in the ∼50S complex is different from that in an authentic spliceosome. The inability of the NRS to bind hPrp8 may be responsible for its inability to splice, since hPrp8 is an important protein cofactor for the catalytic RNA core of the spliceosome (29, 40, 48, 50). We propose that the aberrant ∼50S RNP complex may be a stalled splicing intermediate, which sequesters the downstream 3′ splice site and suppresses its interaction with the authentic 5′ splice site upstream of the NRS sequence (Fig. 7A).
FIG. 7.
Model of NRS splicing suppression by sequestration of the 3′ splice site in an aberrant ∼50S complex. A) The wild-type NRS binds U1 snRNP and interacts with U2 snRNP bound at the downstream branch point sequence and with the tri-snRNP complex to form the ∼50S complex shown in Fig. 2A. Although the tri-snRNP is contained within this complex, it is not properly oriented for splicing. This complex sequesters the 3′ splice site and prevents an upstream 5′ splice site from interacting with it. U11 snRNP is also present in some of these complexes but is not shown on the model. B) Mutation of the nonconsensus UU to AA converts the NRS pseudo-5′ splice site into a functional 5′ splice site, allowing proximal splicing. Normal spliceosome assembly involves the interaction of both U1 snRNP and hPrp8 with the 5′ splice site. C) NRS mutants, such as UU/CC or RG11, which are unable to bind U1 snRNP cannot splice or suppress splicing. This allows the upstream, distal 5′ splice site to interact with the 3′ splice site. In this case, splicing occurs as if there were no NRS sequence present.
The differences between the NRS pseudo-5′ splice site and a normal 5′ splice site must originate with the three nonconsensus U's at −2, +3, and +4 (Fig. 1A), since mutation of any one of these to an A generates a functional 5′ splice site (38). We have searched a database of alternatively spliced introns (47) for the NRS pseudo-5′ splice site sequence: UG/GUUUGU. This sequence was utilized as a functional alternative 5′ splice site only three times in the 70,000 splice sites sequences that we examined. Nevertheless, the NRS is capable of binding U1 snRNP in vitro at a level similar to that of functional 5′ splice sites (5; our unpublished results), and U1 binding is essential for NRS-mediated splicing suppression (19, 33).
The unstable ∼50S NRS-Ad3′ complex may be a precursor to a B splicing complex. The tri-snRNP has previously been reported to be recruited to the spliceosome prior to B complex formation via interactions with the 5′ splice site (3, 29). Nilsen and coworkers have suggested that both U1 and U5 snRNPs are required to recognize a 5′ splice site and that pseudo-5′ splice sites are a result of U1 snRNP binding in the absence of hPrp8 binding (29). The NRS appears to be an example of such a pseudo-5′ splice site. We think that the adjacent U's at +3 and +4 in the NRS sequence, together with the nonconsensus U at position −2, may prevent proper positioning of hPrp8, possibly due to aberrant interactions with U1 snRNP. Maroney et al. (29) found that a similar 5′ splice site mutant (AG/GUCUGA) retained some cross-linking to hPrp8; their substrate differed from ours in that it contained a consensus A at position −2. Further, mutations in their 5′ splice site sequence at −1 and −2 abolished cross-linking to hPrp8 (29). We predict that an NRS mutant with an A at position −2 and UU at +3 and +4 would be able to bind hPrp8 properly, since it is able to splice in vivo (38).
One of our major conclusions is that the NRS-Ad3′ RNA cannot form a normal B splicing complex. However, the molecular details behind this remain unclear. In the traditional view of spliceosome assembly, two events must take place to form a B complex. U1 snRNP must dissociate from the 5′ splice site, and the U4/U6.U5 tri-snRNP must be recruited (4, 36, 44). The binding of U1 snRNP to the NRS has previously been demonstrated to be important for NRS activity; this binding appears to be dependent upon the base-pairing interaction between the NRS and U1 snRNA (19, 33) and probably also involves interactions with U1 proteins (11, 25). We think that the wild-type NRS forms non-Watson-Crick UU:ψψ base pairs with U1 snRNA at positions +3 and +4 (Fig. 1A). In contrast, the UU/CC RG11 mutants, which fail to bind U1 snRNP, are inactive in splicing suppression (5, 38). The NRS may also form a U:U base pair with U1 snRNA at position −2 (Fig. 1A). These nonconsensus base pairs may interfere with the displacement of U1 snRNP from the 5′ splice site and/or the proper binding of U5 and U6 snRNPs.
Interestingly, the consensus yeast 5′ splice site:U1 snRNA duplex, which features a conserved U:ψ interaction at the +4 position, has a slower off rate in nuclear extract than a duplex with complete Watson-Crick base pairing (23). A mammalian RNA helicase, p68, has been shown to facilitate the unwinding of U1 snRNA from a 5′ splice site (20, 24). It will be interesting to see whether the NRS is capable of recruiting this or some other helicase and whether it can unwind the NRS:U1 snRNA helix.
In our model, the NRS-Ad3′ RNA assembles an ∼50S complex that is very similar in composition to a normal spliceosome (Fig. 7A). Further, U11 snRNP is also associated with some of the complexes assembled with the NRS-Ad3′ RNA, but not with the UU/AA mutant after a 30 min incubation (Fig. 4B), even though both share a consensus U12-dependent 5′ splice site. We bound U11 snRNP in the extract with an excess of NRS RNA but did not rescue splicing of NRS-Ad3′ (Fig. 5). Thus, U11 snRNP binding to the NRS-Ad3′ RNA is not solely responsible for its inability to splice. We think the preferential binding of U11 snRNP to the NRS-Ad3′ RNA complex may be a reflection of the disordered state of this complex. Remarkably, U6 snRNA is complementary to 12 out of 13 nt in the NRS 5′ splice site-like sequence (+2 to +14). If U6 snRNA were base paired with this sequence, it would be expected to interfere with binding of U11 snRNP. Kinetic studies showed that U11 snRNP bound to the UU/AA-Ad3′ pre-mRNA, as well as to the NRS-Ad3′, after short incubation times, suggesting that it was dissociated by the tri-snRNP complex at later times. Similarly, our finding of an increased amount of U4 snRNA in some of our NRS-Ad3′ affinity selection experiments (data not shown) is consistent with a failure of unwinding of the U4/U6 snRNA interaction. Thus, it appears that the tri-snRNP does not bind properly to the NRS-Ad3′ RNA and that this may allow U11 snRNP to bind to a fraction of the complexes.
The major question regarding NRS function is how does it suppress the splicing of an upstream functional 5′ splice site? We hypothesize that the formation of a stalled, ∼50S complex bridges the NRS to the downstream 3′ splice site and prevents the upstream, functional 5′ splice site from engaging this 3′ splice site (Fig. 7A). Such a model also explains how the nonfunctional NRS mutants, such as UU/CC and RG11, are unable to suppress splicing (Fig. 7C). These mutant NRS sequences do not bind U1 snRNP (5), so they cannot form 50S complexes containing all five spliceosomal snRNPs to sequester the 3′ splice site. Thus, splicing occurs from the distal 5′ splice site. Finally, the UU/AA NRS mutant, which has a functional 5′ splice site, favors splicing from the proximal (NRS) 5′ splice site (Fig. 7B). This model for NRS function differs from other known mechanisms of splicing suppression. Rio and coworkers have identified an intronic pseudo-5′ splice site in Drosophila P element transcripts, which inhibits the binding of U1 snRNP to the functional 5′ splice site (41). This mechanism of splicing suppression does not necessitate the formation of an RNP complex that bridges the pseudo-5′ splice site and the downstream 3′ splice site. In contrast, NRS-mediated splicing suppression does not seem to prevent the binding of U1 snRNP to the upstream, functional 5′ splice site. If the NRS functioned by inhibiting U1 binding to the authentic 5′ splice site, nonfunctional NRS mutants would cause an overall increase in both viral alternatively spliced mRNAs. However, only splicing to the distal src 3′ splice site was increased in infections with RSV bearing mutant NRS sequences; the env mRNA level was not affected (37).
The pseudo 5′ splice site of the NRS is part of a hairpin structure, with the two adjacent nonconsensus U's at positions +3 and +4 present in a dynamic UUGU tetra-loop (5). The third nonconsensus U at position −2 is bulged out of the stem, causing it to be destabilized. Deletion of this U simultaneously strengthens the helix and reduces U1 snRNP binding (5). Further high-resolution structural investigations may provide more insight into the molecular interactions between a 5′ splice site RNA sequence, U1 snRNP, and Prp8, which appear to be critical to NRS-mediated splicing suppression and to splicing itself.
Acknowledgments
We thank Yingying Li for technical assistance, Tim Nilsen for helpful discussions, Massimo Caputi for aid with affinity selection, Melissa Moore for antibody against human Prp8, and Yun-Xing Wang for T7 polymerase.
This work was supported by National Institutes of Health research grant RO1 CA238796-16 to K.L.B. K.E.G. was supported in part by National Institutes of Health predoctoral training grant T32GM07231.
REFERENCES
- 1.Amendt, B. A., S. B. Simpson, and C. M. Stoltzfus. 1995. Inhibition of RNA splicing at the Rous sarcoma virus src 3′ splice site is mediated by an interaction between a negative cis element and a chicken embryo fibroblast nuclear factor. J. Virol. 69**:**5068-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8**:**4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ast, G., and A. M. Weiner. 1997. A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA 3**:**371-381. [PMC free article] [PubMed] [Google Scholar]
- 4.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome. .In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Cabello-Villegas, J., K. E. Giles, A. M. Soto, P. Yu, A. Mougin, K. L. Beemon, and Y. X. Wang. 2004. Solution structure of the pseudo-5′ splice site of a retroviral splicing suppressor. RNA 10**:**1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18**:**4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1848. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Cook, C. R., and M. T. McNally. 1999. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J. Virol. 73**:**2394-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5**:**1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11**:**1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419**:**86-90. [DOI] [PubMed] [Google Scholar]
- 12.Fogel, B. L., and M. T. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 275**:**32371-32378. [DOI] [PubMed] [Google Scholar]
- 13.Frendewey, D., and W. Keller. 1985. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42**:**355-367. [DOI] [PubMed] [Google Scholar]
- 14.Fu, X. D., R. A. Katz, A. M. Skalka, and T. Maniatis. 1991. The role of branchpoint and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 5**:**211-220. [DOI] [PubMed] [Google Scholar]
- 15.Gontarek, R. R. 1994. A Rous sarcoma virus intronic element negatively regulates splicing by binding snRNPs. Ph.D. thesis. The Johns Hopkins University, Baltimore, MD.
- 16.Gontarek, R. R., M. T. McNally, and K. Beemon. 1993. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 7**:**1926-1936. [DOI] [PubMed] [Google Scholar]
- 17.Grabowski, P. J., S. R. Seiler, and P. A. Sharp. 1985. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42**:**345-353. [DOI] [PubMed] [Google Scholar]
- 18.Grabowski, P. J., and P. A. Sharp. 1986. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science 233**:**1294-1299. [DOI] [PubMed] [Google Scholar]
- 19.Hibbert, C. S., R. R. Gontarek, and K. L. Beemon. 1999. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA 5**:**333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., and Z. R. Liu. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277**:**12810-12815. [DOI] [PubMed] [Google Scholar]
- 21.Jurica, M. S., and M. J. Moore. 2002. Capturing splicing complexes to study structure and mechanism. Methods 28**:**336-345. [DOI] [PubMed] [Google Scholar]
- 22.Katz, R. A., and A. M. Skalka. 1990. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10**:**696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libri, D., F. Duconge, L. Levy, and M. Vinauger. 2002. A role for the Psi-U mismatch in the recognition of the 5′ splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J. Biol. Chem. 277**:**18173-18181. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22**:**5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lund, M., and J. Kjems. 2002. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8**:**166-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, H. R., G. A. Moreau, N. Levin, and M. J. Moore. 1999. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5**:**893-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova, O. V., E. M. Makarov, H. Urlaub, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23**:**2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malca, H., N. Shomron, and G. Ast. 2003. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 23**:**3442-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maroney, P. A., C. M. Romfo, and T. W. Nilsen. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6**:**317-328. [DOI] [PubMed] [Google Scholar]
- 30.Maroney, P. A., C. M. Romfo, and T. W. Nilsen. 2000. Nuclease protection of RNAs containing site-specific labels: a rapid method for mapping RNA-protein interactions. RNA 6**:**1905-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNally, L. M., and M. T. McNally. 1998. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 18**:**3103-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNally, L. M., and M. T. McNally. 1996. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70**:**1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNally, L. M., and M. T. McNally. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73**:**2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNally, M. T., and K. Beemon. 1992. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J. Virol. 66**:**6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally, M. T., R. R. Gontarek, and K. Beemon. 1991. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology 185**:**99-108. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, T. W. 1994. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell 78**:**1-4. [DOI] [PubMed] [Google Scholar]
- 37.O'Sullivan, C. T., T. S. Polony, R. E. Paca, and K. L. Beemon. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses src mRNA splicing and promotes polyadenylation. Virology 302**:**405-412. [DOI] [PubMed] [Google Scholar]
- 38.Paca, R. E., C. S. Hibbert, C. T. O'Sullivan, and K. L. Beemon. 2001. Retroviral splicing suppressor requires three nonconsensus uridines in a 5′ splice site-like sequence. J. Virol. 75**:**7763-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-261. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses, second ed. Cold Spring Harbor Laboratory Press, Woodbury, N.Y. [PubMed]
- 40.Reyes, J. L., P. Kois, B. B. Konforti, and M. M. Konarska. 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2**:**213-225. [PMC free article] [PubMed] [Google Scholar]
- 41.Siebel, C. W., L. D. Fresco, and D. C. Rio. 1992. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 6**:**1386-1401. [DOI] [PubMed] [Google Scholar]
- 42.Sorek, R., R. Shamir, and G. Ast. 2004. How prevalent is functional alternative splicing in the human genome? Trends Genet. 20**:**68-71. [DOI] [PubMed] [Google Scholar]
- 43.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92**:**315-326. [DOI] [PubMed] [Google Scholar]
- 44.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3**:**55-64. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9**:**31-44. [DOI] [PubMed] [Google Scholar]
- 46.Tarn, W. Y., and J. A. Steitz. 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84**:**801-811. [DOI] [PubMed] [Google Scholar]
- 47.Thanaraj, T. A., S. Stamm, F. Clark, J. J. Riethoven, V. Le Texier, and J. Muilu. 2004. ASD: the Alternative Splicing Database. Nucleic Acids Res. 32(Database issue)**:**D64-D69. [DOI] [PMC free article] [PubMed]
- 48.Turner, I. A., C. M. Norman, M. J. Churcher, and A. J. Newman. 2004. Roles of the U5 snRNP in spliceosome dynamics and catalysis. Biochem. Soc. Trans. 32**:**928-931. [DOI] [PubMed] [Google Scholar]
- 49.Wu, S., and M. R. Green. 1997. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 16**:**4421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyatt, J. R., E. J. Sontheimer, and J. A. Steitz. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 6**:**2542-2553. [DOI] [PubMed] [Google Scholar]