Coupling between Replication and Packaging of Flavivirus RNA: Evidence Derived from the Use of DNA-Based Full-Length cDNA Clones of Kunjin Virus (original) (raw)
Abstract
In order to study whether flavivirus RNA packaging is dependent on RNA replication, we generated two DNA-based Kunjin virus constructs, pKUN1 and pKUN1dGDD, allowing continuous production of replicating (wild-type) and nonreplicating (with a deletion of the NS5 gene RNA-polymerase motif GDD) full-length Kunjin virus RNAs, respectively, via nuclear transcription by cellular RNA polymerase II. As expected, transfection of pKUN1 plasmid DNA into BHK cells resulted in the recovery of secreted infectious Kunjin virions. Transfection of pKUN1dGDD DNA into BHK cells, however, did not result in the recovery of any secreted virus particles containing encapsidated dGDD RNA, despite an apparent accumulation of this RNA in cells demonstrated by Northern blot analysis and its efficient translation demonstrated by detection of correctly processed labeled structural proteins (at least prM and E) both in cells and in the culture fluid using coimmunoprecipitation analysis with anti-E antibodies. In contrast, when dGDD RNA was produced even in much smaller amounts in pKUN1dGDD DNA-transfected repBHK cells (where it was replicated via complementation), it was packaged into secreted virus particles. Thus, packaging of defective Kunjin virus RNA could occur only when it was replicated. Our results with genome-length Kunjin virus RNA and the results with poliovirus replicon RNA (C. I. Nugent et al., J. Virol. 73:427–435, 1999), both demonstrating the necessity for the RNA to be replicated before it can be packaged, strongly suggest the existence of a common mechanism for minimizing amplification and transmission of defective RNAs among the quasispecies in positive-strand RNA viruses. This mechanism may thus help alleviate the high-copy error rate of RNA-dependent RNA polymerases.
Flavivirus virions contain single-stranded positive-sense RNA of ∼11 kb encapsidated by the structural proteins C, prM, and E (18, 19). The mechanism ensuring selective packaging of only the flavivirus RNA into virions in virus-infected cells, as well as the required signals in the RNA and in the structural proteins involved in this process, have not been determined. We demonstrated previously in _trans_-encapsidation experiments using Kunjin virus (KUN) replicon RNA with deleted structural genes that only KUN replicon RNA was packaged into the secreted virus-like particles, while coreplicating Semliki Forest virus replicon RNA (used as a vector for expression of KUN structural genes) was not packaged (6). In addition, our _trans_-complementation experiments with KUN genomic RNAs containing deletions in the NS1 and NS5 genes, and _trans_-complementation experiments of others with yellow fever virus RNAs containing deletions in the NS1 gene, both using as helpers Sindbis virus replicons expressing corresponding wild-type flavivirus nonstructural genes, showed that only the flavivirus RNAs and not the coreplicating Sindbis virus replicon RNAs were packaged into secreted virions by the flavivirus structural proteins (9, 11, 12). These _trans_-encapsidation and _trans_-complementation experiments demonstrated clearly that packaging of flavivirus RNA occurs by a highly specific mechanism.
Our encapsidation studies with KUN replicon RNAs also showed that the most efficient packaging of replicon RNA into virus-like particles occurred at the time of maximum RNA replication (6, 16), suggesting that these two processes (replication and packaging) are closely related. Similarly, in flavivirus-infected cells the assembly and release of infectious virions coincided with the large increase in viral RNA synthesis at the end of the latent period (18). Interestingly, coupling between replication and packaging of poliovirus RNA as a mechanism ensuring its specific encapsidation was initially proposed by Baltimore (1) and recently demonstrated by Nugent et al. (15). The latter study showed that selective inhibition of replication of poliovirus replicon RNA by guanidine dramatically decreased the encapsidation efficiency of the accumulated replicon RNA by the poliovirus capsid proteins provided in trans by the coinfected guanidine-resistant mutant poliovirus. It was suggested by the authors that only actively replicating RNA (i.e., an RNA strand emerging from the replication complex) could be encapsidated by the structural proteins.
Since no selective inhibitors of flavivirus RNA replication have been reported, we decided to take a different approach to study the relationship between replication and packaging of flavivirus RNA. Our recently developed DNA-based KUN replicon constructs incorporate a mammalian expression promoter upstream and a simian virus 40 poly(A) signal downstream of the KUN cDNA sequence and allow production of authentic KUN RNAs in cells from transfected plasmid DNAs by cellular RNA polymerase II (17). We showed that both replication-competent and replication-deficient KUN replicon RNAs were produced in cells after transfection with the plasmid DNAs pKUNrep2 and pKUNrep2dGDD, respectively, with the latter having a deletion of the RNA polymerase active site GDD in the KUN NS5 gene. This replication-deficient RNA, however, was efficiently translated in cells, resulting in synthesis of the nonstructural proteins detectable by radioimmunoprecipitation analysis (17) and by immunofluorescence (IF) analysis (A. N. Varnavski and A. A. Khromykh, unpublished data). In the present study, we exploited the ability to continuously produce translation-competent but replication-deficient genome-length KUN RNA by nuclear transcription from transfected plasmid DNA, as well as our previously established _trans_-complementation system, to demonstrate the requirement of RNA replication for its packaging into virus particles.
MATERIALS AND METHODS
Cells and plasmids.
Helper repBHK cells persistently expressing KUN replicon RNA were generated as described previously (7). Both BHK and repBHK cells were grown in Dulbecco's modification of minimal essential medium (Life Technologies) supplemented with 10% fetal bovine serum at 37°C in a CO2 incubator. Medium for growing repBHK cells also contained 1 mg of G418 Sulfate (Calbiochem) per ml.
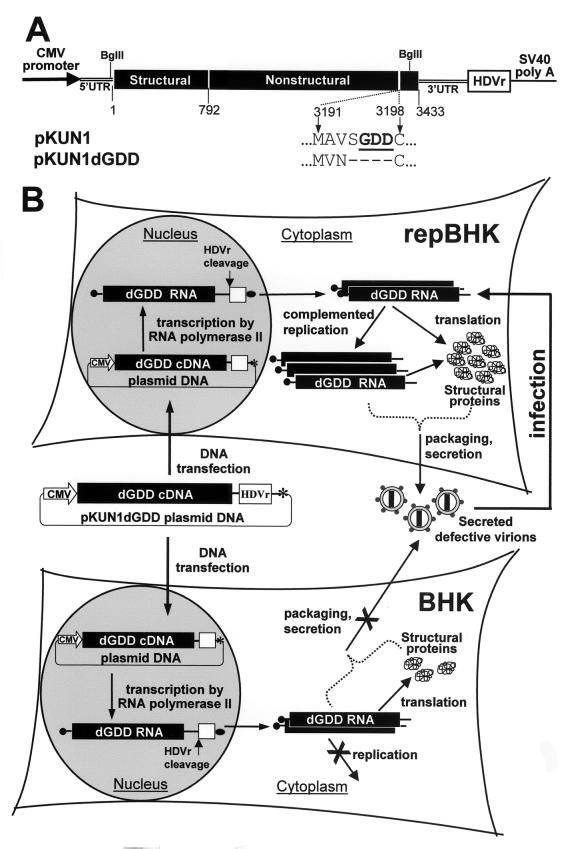
The pKUN1 and pKUN1dGDD plasmids (see Fig. 1A) containing cDNA copies of the replicating and nonreplicating (GDD deleted) full-length KUN genomes were constructed by replacing the fragment between the two Bgl_II restriction sites (situated at the 3′ end of the 5′ untranslated region (UTR) and at the 3′ end of the NS5 gene) in the pKUNrep1 plasmid (17) with the corresponding fragment derived from the FLSDX or FLd_GDD plasmid, respectively (7).
FIG. 1.
Schematic representation of the KUN plasmid DNA constructs (A) and of the complementation experiments (B) conducted with the normal BHK cells and with the helper BHK cells (repBHK) stably expressing the KUN replicon RNA. Filled boxes show the translated region of the KUN genome with numbers representing amino acid positions (A) and the KUN replicon sequence (B). Also shown are the 5′ and 3′ KUN UTRs, the cytomegalovirus early promoter-enhancer region (CMV promoter), and the antigenomic sequence of the hepatitis delta virus ribozyme (HDVr). The HDVr sequence ensures generation of the correct KUN 3′ terminus. SV40 poly(A) in panel A and the asterisk in panel B show the simian virus 40 polyadenylation signal; the filled oval (B) indicates a poly(A) sequence. dGDD refers to the deletion of the RNA polymerase motif GDD in the NS5 gene, shown in bold and underlined letters in pKUN1 and as dashes in pKUN1dGDD (A), and the filled circle in front of the KUN RNA (B) represents the cap structure. For the explanation of the experimental design shown (B), see the text.
DNA transfections.
For the transfection experiments shown in Fig. 2, ∼50%-confluent monolayers of cells on coverslips in the 24-well culture plate (Nunc) were transfected with 0.8 μg of pKUN1 or pKUN1dGDD DNAs mixed with 2 μl of FuGENE 6 reagent (Roche Biochemicals) essentially as described by the manufacturer. For other transfection experiments, cells were seeded in 35-mm dishes and transfected with various concentrations of DNA and Lipofectamine Plus reagents (Life Technologies) as described by the manufacturer. Two micrograms of DNA was transfected by mixing with 8 μl of Plus and 8 μl of Lipofectamine, 0.6 μg of DNA was transfected by mixing with 5 μl of Plus and 2 μl of Lipofectamine, 0.2 μg of DNA was transfected by mixing with 4 μl of Plus and 1 μl of Lipofectamine, and 0.1 μg of DNA was transfected by mixing with 1 μl of Plus and 0.5 μl of Lipofectamine.
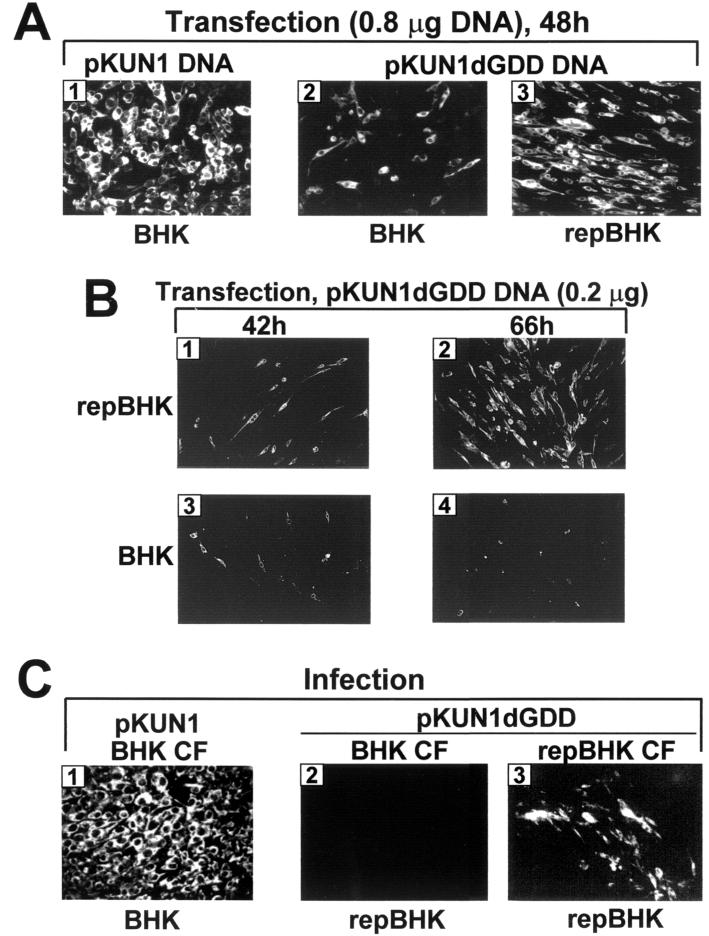
FIG. 2.
Evidence of KUN RNA production and translation in BHK and repBHK cells either transfected with pKUN1 or pKUN1dGDD DNAs (A and B) or infected with the recovered defective viruses (C). (A) pKUN1 DNA (0.8 μg) was transfected into BHK cells (panel 1), and 0.8 μg of pKUN1dGDD DNA was transfected into BHK and repBHK cells (panels 2 and 3, respectively), and cells were then analyzed for expression of the KUN E gene by immunofluorescence analysis with anti-E antibodies at 48 h after these transfections as described in Materials and Methods. (B) repBHK and normal BHK cells were transfected with 0.2 μg of pKUN1dGDD DNA and assayed for E expression at 42 h (panels 1 and 3, respectively) and 66 h (panels 2 and 4, respectively) after transfection. (C) BHK cells were infected with CF harvested at 36 h after transfection of normal BHK cells with 0.8 μg of pKUN1 DNA and analyzed for expression of KUN E at 31 h after infection (panel 1). Panels 2 and 3 show the expression of E at 48 h after infection of repBHK cells with CFs harvested at 48 h after transfection of either BHK or repBHK cells, respectively, with 0.8 μg of pKUN1dGDD DNA.
IF, Northern blot, and radioimmunoprecipitation analyses.
Cells on coverslips were fixed with acetone at −20°C for 30 s usually at 48 h either after transfection with DNAs or after infection with culture fluid (CF) collected from DNA-transfected cells. Fixed cells were then assayed for expression of KUN NS3 by indirect IF with anti-NS3 antibodies as described previously (5). Northern blot analysis of total cell RNA (10 μg) isolated at 48 h after DNA transfection was performed by hybridization with a 32P-labeled _Aat_II-_Cla_I fragment representing 568 nucleotides of the KUN prM-E region (KUN nucleotides 522 to 1089) (2, 4) as described previously (7). Radioimmunoprecipitation with anti-E antibodies of CFs collected from DNA-transfected cells was performed as described previously (6). Immunoprecipitates were then analyzed by polyacrylamide gel electrophoresis to identify precipitated radiolabeled proteins.
RT-PCR analysis.
RNA from anti-E immunoprecipitates was isolated as described previously (6). The RNA samples were treated with RQ1 DNase (Promega) to eliminate any residual plasmid DNA from the initial transfection and subjected to a reverse transcription-PCR (RT-PCR) with the primers corresponding to the KUN prM-E region (KUN nucleotides 412 to 1511) (2, 4) using a SuperScript One-Step RT-PCR kit (Life Technologies) as described by the manufacturer. Reactions without RT were performed under the same conditions, except that the RT-Taq enzyme mixture was replaced by Taq polymerase only.
RESULTS
Experimental system.
In order to provide structural proteins in cis for KUN RNA packaging, we prepared two plasmid DNA constructs, pKUN1 and pKUN1dGDD, which allowed production in transfected cells of the full-length replication-competent and replication-deficient RNAs, respectively (Fig. 1A). The assumption was that transfection of pKUN1 DNA in BHK cells would result in production of replicating wild-type viral RNA by transcription from cDNA, followed by translation and replication and eventually assembly and secretion of wild-type virions. A similar scenario should occur after transfection of pKUN1dGDD DNA into the helper BHK cells persistently expressing KUN replicon RNA (repBHK), in which replication of the transcribed dGDD RNA would be complemented by the replication complex produced from the helper KUN replicon RNA (Fig. 1B) (7). As a result of this complemented replication, virus particles containing encapsidated dGDD RNA should be produced and secreted, but they will be noninfectious in normal BHK cells. In contrast, transfection of pKUN1dGDD DNA into normal BHK cells should result in transcription of translatable but replication-deficient RNA. If this RNA could be packaged by the structural proteins translated in cis, secreted defective virus particles should be produced (Fig. 1B). These secreted defective virus particles should be detectable by infection of repBHK cells with the recovered CF, followed by complementation and amplification of dGDD RNA (Fig. 1B). We showed previously that this complementation system operating via amplification of the defective RNA in repBHK cells allowed detection of very small amounts of defective virus particles by using IF analysis of the infected repBHK cells with anti-E antibodies (7–10). If, however, replication of viral RNA is essential for packaging, no defective virus particles will be produced and secreted in the CF of pKUN1dGDD-transfected BHK cells (Fig. 1B), and thus no E-positive cells will be detected after infection of repBHK cells with these CFs.
Replication-competent but not replication-deficient KUN RNA can be packaged into virus particles in BHK cells.
In order to test the ability of replication-deficient RNA to be packaged, we transfected the same amounts (0.8 μg) of pKUN1dGDD and pKUN1 (positive control) DNAs into BHK cells. As expected, most of the BHK cells were positive for expression of the KUN E gene by 48 h after transfection with pKUN1 DNA, while a smaller proportion of BHK cells (∼10 to 20%) transfected with pKUN1dGDD DNA were E positive (Fig. 2A, panels 1 and 2, respectively). Transfection of the same amount of pKUN1dGDD RNA into the helper repBHK cells resulted in detection of expression of E in most cells by 48 h (Fig. 2A, panel 3), thus indicating efficient complementation of dGDD RNA replication initially in transfected cells as well as the later spread of complemented virus. It was not possible to estimate the relative efficiencies of pKUN1dGDD DNA transfection in BHK and repBHK cells in this experiment due to the apparent spread of complemented virus by 48 h posttransfection. However, in a separate experiment with a smaller amount of transfected pKUN1dGDD DNA (0.2 μg), we observed a similar number of E-positive cells in both BHK and repBHK cells earlier in transfection (42 h), but repBHK cells showed a greater intensity of IF staining (Fig. 2B, panels 1 and 3, respectively), suggesting complementation of dGDD RNA replication in these cells. Later in transfection (66 h), the number of E-positive repBHK cells dramatically increased (Fig. 2B, panel 2), demonstrating the spread of complemented virus, while the number of E-positive BHK cells did not increase (Fig. 2B, panel 4), clearly indicating the absence of virus spread. In our previous experiments with the DNA-based KUN replicon construct pKUNrep2dGDD (involving no virus spread), the efficiencies of its transfection into BHK and repBHK cells were also similar (17).
Infection of BHK cells with the 36-h CF from the pKUN1 DNA-transfected BHK cells resulted in detection of distinctive foci of E-expressing cells at 24 h postinfection (data not shown) which were enlarged with time, and the great majority of cells were E positive by 31 to 36 h postinfection (Fig. 2C, panel 1). Although we did not perform further characterization of the recovered virus, it was evident from the results of IF analysis (Fig. 2C) and from the later RT-PCR results (see Fig. 4B) that infectious KUN virus was indeed produced in cells transfected with pKUN1 plasmid DNA. Infection of repBHK cells with the 48-h CF from the pKUN1dGDD-transfected BHK cells, however, did not produce any E-positive cells (Fig. 2C, panel 2), while infection of repBHK cells with the 48-h CF from the pKUN1dGDD-transfected helper repBHK cells did produce a significant number of E-positive cells (Fig. 2C, panel 3). In our previous complementation experiments with FLdGDD RNA in transfected repBHK cells, we convincingly demonstrated that no recombination occurred between helper replicon RNA and complemented FLdGDD RNA, which would have led to the recovery of wild-type KUN virus detectable by infection of normal BHK cells (7). Although some individual BHK cells were E positive after infection with recovered CFs, it was concluded (7) that these cells were simultaneously coinfected with two types of virus particles, one containing encapsidated helper replicon RNA and another containing complemented dGDD RNA. This coinfection event would lead to complementation of dGDD RNA replication in these individual cells and subsequent detection of E expression. Importantly, further incubation of these cells did not lead to detection of any E-positive cell foci (7), demonstrating the absence of spreading self-replicating (recombinant) virus in complemented CFs. It was also demonstrated in these experiments that dGDD RNA encapsidated into complemented defective virions retained the introduced GDD deletion (7). Thus, after excluding the possibility of formation of recombinant self-replicating virus in pKUN1dGDD DNA-transfected repBHK cells, we concluded from the IF results described in this section that dGDD RNA was packaged into secreted virus particles when it was produced in repBHK cells where it was able to replicate via complementation. The equivalent dGDD RNA, however, was not packaged into secreted virus particles when produced in normal BHK cells where it was not replicating.
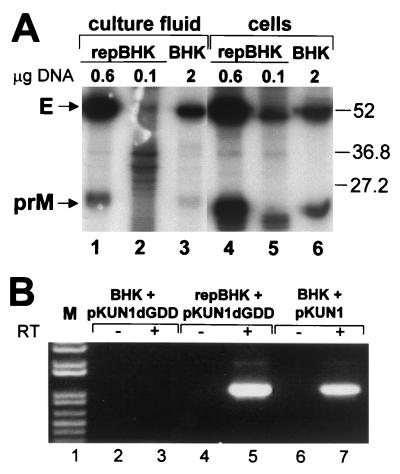
FIG. 4.
Analyses of KUN proteins in cells and in the culture fluid (A) and of KUN RNA in the secreted virions after transfection of BHK and repBHK cells with pKUN1 or pKUN1dGDD DNAs as indicated. (A) Autoradiograph of the polyacrylamide gel after electrophoresis of KUN proteins radiolabeled for 6 h immediately prior to being immunoprecipitated with KUN anti-E antibodies from the CFs (lanes 1 to 3) or lysates (lanes 4 to 6) of repBHK cells (lanes 1, 2, 4, and 5) or BHK cells (lanes 3 and 6) at 48 h after transfection with the indicated amounts of pKUN1dGDD DNA. Arrows indicate positions in the gel of the KUN E and prM proteins. Labeled bands between the prM and E in all lanes probably represent nonspecifically coprecipitated cell proteins. The left (lanes 1 to 3) and the right (lanes 4 to 6) halves of the gel were exposed to X-ray film for 3 weeks and 3 days, respectively. Numbers at the right side of the gel show positions of low-molecular-weight protein standards, given in thousands (Bio-Rad). (B) RT-PCR analysis of RNAs isolated from anti-E immunoprecipitates of 48-h CFs from BHK cells (lane 3) or repBHK cells (lane 5) transfected with 2 μg of pKUN1dGDD DNA and BHK cells transfected with 0.1 μg of pKUN1 DNA (lane 7). Lanes 2, 4, and 6 show the results of corresponding RT-PCRs with no RT added (negative controls). M, 1-kb Plus DNA ladder (Life Technologies).
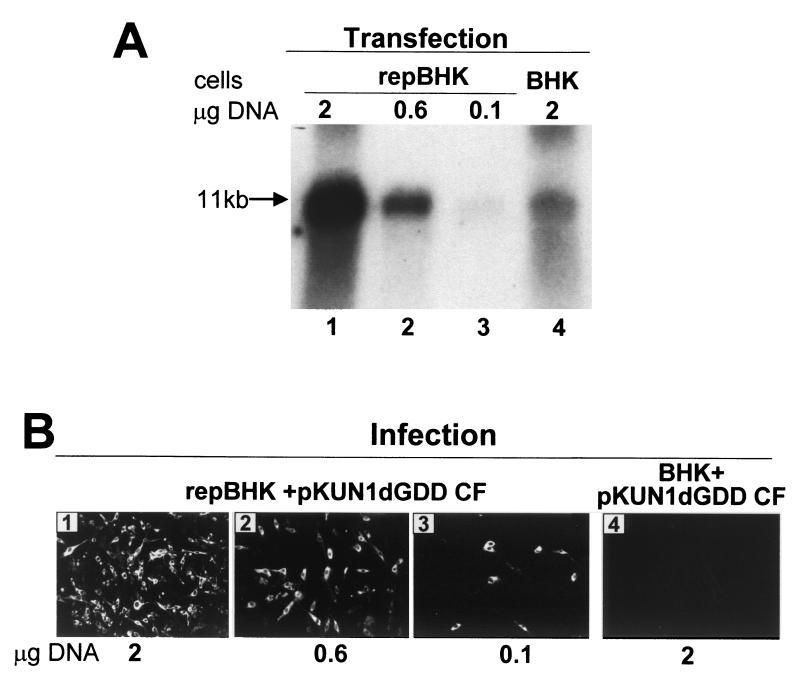
Comparative analyses of accumulation of KUN RNA, structural proteins, and virus particles in pKUN1dGDD-transfected repBHK and BHK cells.
We showed previously for KUN replicons that approximately sixfold more RNA was produced in BHK cells from transfected pKUNrep2 DNA than from the same amount of transfected pKUNrep2dGDD DNA (17). To compensate for this difference in RNA synthesis between cells producing nonreplicating and replicating full-length RNAs, smaller amounts of pKUN1dGDD DNA were transfected into repBHK cells than into BHK cells. Northern blot analysis of total cell RNA using KUN-specific labeled cDNA probe showed that in order to accumulate similar amounts of KUN RNA by 2 days after transfection, BHK cells required transfection with approximately four- to fivefold more pKUN1dGDD DNA than did repBHK cells (Fig. 3A). We next examined CFs from these transfected cells for the presence of infectious secreted defective virus particles by infecting repBHK cells and performing IF analysis with anti-E antibodies. IF-positive cells were detected in repBHK cells infected with each CF harvested from pKUN1dGDD-transfected repBHK cells (Fig. 3B, panels 1 to 3), including the repBHK cells transfected with the smallest amount of DNA (0.1 μg) (Fig. 3B, panel 4) and producing barely detectable amounts of KUN RNA (Fig. 3A, lane 3). For the same reasons enunciated in the previous section, the possibility of formation of wild-type infectious virus via recombination between helper replicon RNA and complemented dGDD RNA was excluded. No E-positive repBHK cells were detected after infection with CF collected from BHK cells transfected with the largest amount (2 μg) of pKUN1dGDD DNA (Fig. 3B, panel 4) and producing relatively high yields of KUN RNA (Fig. 3A, lane 4). Thus, despite the production and accumulation of readily detectable amounts of (nonreplicating) KUN RNA in BHK cells via DNA-to-RNA transcription, this RNA was not packaged into secreted virus particles, while even much smaller amounts of the same RNA produced in the helper repBHK cells, where its replication was complemented, were packaged into secreted virus particles.
FIG. 3.
Accumulation of the defective (dGDD) KUN RNA in repBHK and BHK cells (A) and of the defective virus in their culture fluid (B) after transfection with different amounts of pKUN1dGDD DNA. (A) Northern blot analysis with a radiolabeled cDNA probe representing the KUN structural region of the total cellular RNA isolated from repBHK or BHK cells at 48 h after transfection with the indicated amounts of pKUN1dGDD DNA. (B) Results of IF analysis with anti-NS3 antibodies of repBHK cells at 48 h after infection with CFs harvested at 48 h after transfection of either repBHK cells (panels 1 to 3) or BHK cells (panel 4) with the indicated amounts of pKUN1dGDD DNA.
In a separate experiment, we analyzed production, processing, and secretion of KUN structural proteins in lysates and CFs of pKUN1dGDD-transfected and radiolabeled BHK and repBHK cells by radioimmunoprecipitation with anti-E antibodies. The results demonstrated that at least structural proteins E and prM were produced and correctly processed in both repBHK and BHK cells and were secreted into the CFs (Fig. 4A). prM in cell lysates appeared to electrophorese slightly faster than in CFs, probably due to incomplete glycosylation. Core protein should also be detectable in the immunoprecipitates with anti-E antibodies, but in this experiment it apparently ran off the bottom of the gel. Although the amounts of secreted and cell-associated E and prM proteins produced from nonreplicating KUN RNA in BHK cells transfected with the largest amount (2 μg) of pKUN1dGDD DNA were relatively small (Fig. 4A, lanes 3 and 6), they were greater than those produced from replicating KUN RNA in repBHK cells transfected with the smallest amount (0.1 μg) of pKUN1dGDD DNA (Fig. 4A, lanes 2 and 5). Note that in other experiments, coprecipitated cell proteins migrating just below the gel positions of E and prM were prominent in cell lysates of mock-infected BHK cells, as is apparent in lane 5 of Fig. 4A. These results demonstrate that replication-deficient dGDD RNA transcribed in normal BHK cells transfected with pKUN1dGDD DNA directed production, correct processing, and secretion of structural proteins in amounts which should have been sufficient for packaging of dGDD RNA into secreted virus particles. However, immunofluorescence assays, as in previous experiments (Fig. 2C and 3B), clearly indicated that no secreted virus particles containing packaged dGDD RNA were produced in pKUN1dGDD DNA-transfected BHK cells in this experiment (data not shown).
To provide a possibly more sensitive assay for detection of packaged RNAs in secreted virus particles, we employed RT-PCR analysis (using primers specific for the prM-E region) of RNA isolated from virus particles immunoprecipitated with anti-E antibodies. In accord with the results obtained using complementation assays, RT-PCR analysis failed to detect any KUN RNA in particles precipitated from the CF harvested at 48 h from BHK cells transfected with 2 μg of pKUN1dGDD DNA (Fig. 4B, lane 3). In contrast, a prominent PCR band was detected in RT-PCRs with RNA recovered from particles precipitated from the CF harvested at 48 h from repBHK cells transfected with only 0.1 μg of pKUNdGDD DNA (Fig. 4B, lane 5) or from BHK cells transfected with 2 μg of the control (replicating) pKUN1 DNA (Fig. 4B, lane 7).
DISCUSSION
We have recently described construction and characterization of DNA-based KUN replicon plasmid constructs allowing continuous transcription by cellular RNA polymerase II and accumulation of the replication-competent (wild-type) and replication-deficient (GDD deleted) KUN replicon RNAs in transfected cells (17). In this study, we constructed the DNA-based KUN plasmids pKUN1 and pKUN1dGDD, each containing a genome-length cDNA copy of KUN RNA; these plasmids allow transcription and accumulation in transfected cells of replicating and nonreplicating full-length KUN RNAs, respectively. We first demonstrated that transfection of pKUN1 DNA into BHK cells resulted in production of replicating KUN RNA and subsequently of secreted infectious KUN virions. Having established the validity of the DNA-based approach for generation of fully functional viral RNA, we then applied this approach to achieve the main goal of these studies, which was to establish whether or not replication was a prerequisite for packaging of flavivirus RNA. We devised an experimental system allowing production in parallel of defective (dGDD) KUN genomic RNA either in normal BHK cells, where this nonreplicating RNA is produced and accumulates only via transcription from plasmid DNA, or in helper repBHK cells, where its replication is rescued by complementation. The data described in this report clearly show that despite the production and accumulation in normal BHK cells of sufficient amounts of dGDD RNA and of properly processed and secreted KUN structural proteins, this RNA could not be packaged into secreted virus particles. In contrast, the same RNA could be packaged into secreted virus particles when its replication was restored by the wild-type replicative proteins expressed from the helper replicon RNA (in repBHK cells), even when the defective viral RNA (Fig. 3A) and the structural proteins (Fig. 4A) were produced in much smaller amounts. These results represent the first demonstration of functional coupling between replication and packaging of flavivirus RNA.
Two scenarios to explain our data are possible. Results of previous complementation experiments (9) suggest that NS5 with a deletion of GDD was retained in a defective replicase complex bound to the 3′ UTR after translation in cis and was unable to copy its template but could exchange with wild-type NS5 provided by a helper RNA. Normal copying of the (defective) RNA could then ensue, followed by formation of the double-stranded RNA template and sequestering of the complex in induced membranes or vesicle packets (13, 21), leading to synthesis of progeny RNA(+) strands. In this first scenario, the defective progeny RNA molecules could be displaced from the replicase complex and subsequently encapsidated by the normal assembly process. In the absence of helper RNA (as in normal BHK cells), nucleus-transcribed defective KUN RNA and the translated viral proteins would continue to accumulate but without encapsidation, as observed. In essence, this defective transcribed RNA would remain “locked up” with a defective nonprocessive complex bound to the 3′ UTR, which prevents its release and hence any opportunity for subsequent packaging. An alternative scenario is that for encapsidation to occur, the viral RNA must be replicated in a membrane-associated site and be able to subsequently relocate to a (probably linked) membrane assembly site (9, 14). In cells lacking such sites, which are normally induced during replication of viral RNA, nucleus-transcribed defective KUN RNA cannot be encapsidated, as observed. These scenarios differ from the proposal that direct physical interaction between the RNA replication complex (RC) and the assembling virus particles is essential to provide the link between replication and packaging of poliovirus RNA (15). Relevant to this notion, recently synthesized poliovirus RNA appears to be exposed on the surface of virus-induced membranes, forming rosettes which can be reversibly dissociated and continue to synthesize plus-strand RNA (3), whereas the KUN RC is sequestered within vesicle packets, as noted above.
The necessity for viral RNA to be replicated before it can be packaged suggests the existence of a mechanism for minimizing amplification and transmission of defective viral RNA among the viral quasispecies, which arise because of the high-copy error of RNA-dependent RNA polymerase. Mutated nonstructural proteins translated in cis from defective RNA may prevent assembly of the RC or, if assembly does proceed, prevent processivity of the polymerase on the RNA template and thus exclude or eliminate this RNA from packaging. We have discussed previously how large deletions in KUN NS5 (8) or point mutations in NS1 (9) may still permit correct assembly and processivity of the KUN RC. We believe that those mechanisms of complementation proposed for NS1 and NS5 may also be applicable to complementations of NS1 and NS3, respectively, when both have large lethal deletions (10). The challenge that remains is to explain why RNAs with deletions in NS3 could not be packaged into secreted virus particles despite the relatively efficient complementation of their replication (10) and why replication of RNAs with deletions and mutations in the remaining components (the small hydrophobic proteins NS2A and NS4A) of the consensus KUN RC (13) could not be complemented (10).
The links between flavivirus RNA synthesis, translation, and packaging require further exploration before the complete process of replication can be defined. Recently we showed that late in infection, continuing translation of KUN RNA was not required for viral RNA synthesis and release of infectious virus (22). KUN RNA radiolabeled prior to application of a complete translation block could be incorporated in progeny virions during chase periods (A. A. Khromykh and E. G. Westaway, unpublished data). At present we are attempting to establish the relationship between the membrane sites of KUN virus replication and assembly, the sites of viral RNA synthesis, and the role(s) of cell marker proteins in virus-induced membranes (13, 14, 20–22). Such data are essential to further illuminate the still grey areas of flavivirus replication and assembly.
In addition to the demonstrated coupling between KUN RNA replication and packaging, the results presented here also show that generation of an infectious flavivirus in vivo directly from plasmid DNA is possible. This advance should facilitate further development of genetically engineered flavivirus vaccines based on attenuated full-length cDNA clones. Using the DNA-based approach will significantly improve the stability and simplify the preparation and testing of flavivirus vaccines by eliminating the cumbersome, time-consuming, and expensive preparation of labile RNA and the need to generate vaccine virus in vitro.
ACKNOWLEDGMENTS
We are grateful R. Hall for supplying KUN anti-E monoclonal antibodies.
This work was supported by grant N981442 from the National Health and Medical Research Council of Australia.
Footnotes
†
Publication no. 127 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Baltimore D. The replication of picornavaruses. In: Levy H B, editor. The biochemistry of viruses. New York, N.Y: Marcel Dekker, Inc; 1969. pp. 101–176. [Google Scholar]
- 2.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Egger D, Pasamontes L, Bolten R, Boyko V, Bienz K. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J Virol. 1996;70:8675–8683. doi: 10.1128/jvi.70.12.8675-8683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khromykh A A, Varnavski A N, Westaway E G. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin structural proteins in trans. J Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khromykh A A, Sedlak P L, Westaway E G. trans-complementation analysis of flavivirus Kunjin NS5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol. 1999;73:9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khromykh A A, Sedlak P L, Guyatt K J, Hall R A, Westaway E G. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999;73:10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khromykh A A, Sedlak P L, Westaway E G. cis- and trans-acting elements in Flavivirus RNA replication. J Virol. 2000;74:3253–3263. doi: 10.1128/jvi.74.7.3253-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindenbach B D, Rice C M. Trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindenbach B D, Rice C M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie J M, Jones M K, Westaway E G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in Flavivirus-infected cells. J Virol. 1999;73:9555–9567. doi: 10.1128/jvi.73.11.9555-9567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varnavski A N, Khromykh A A. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology. 1999;255:366–375. doi: 10.1006/viro.1998.9564. [DOI] [PubMed] [Google Scholar]
- 17.Varnavski A N, Young P R, Khromykh A A. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin replicon vectors. J Virol. 2000;74:4394–4403. doi: 10.1128/jvi.74.9.4394-4403.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westaway E G. Replication of flaviviruses. In: Schlesinger R W, editor. Togaviruses. New York, N.Y: Academic Press; 1980. pp. 531–581. [Google Scholar]
- 19.Westaway E G. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- 20.Westaway E G, Khromykh A A, Kenney M T, Mackenzie J M, Jones M K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 21.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westaway E G, Khromykh A A, Mackenzie J M. Nascent flavivirus RNA co-localized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]