Notch1IC Partially Replaces EBNA2 Function in B Cells Immortalized by Epstein-Barr Virus (original) (raw)
Abstract
Immortalization of B cells by Epstein-Barr virus (EBV) depends on the virally encoded EBNA2 protein. Although not related by sequence, the cellular Notch protein and EBNA2 share several biochemical and functional properties, such as interaction with CBF1 and the ability to activate transcription of a number of cellular and viral genes. Whether these similarities are coincidental or exemplify EBNA2 mimicry of evolutionarily conserved cellular signaling pathways is unclear. We therefore investigated whether activated forms of Notch could substitute for EBNA2 in maintaining the immortalized phenotype of EBV-infected B cells. To address this question, we devised a transcomplementation system using EREB2.5 cells. EREB2.5 cells are immortalized by EBV expressing a conditional estrogen receptor EBNA2 fusion protein (EREBNA2), and cellular proliferation is dependent on the availability of estrogen. Withdrawal of estrogen results in inactivation of EREBNA2, leading to growth arrest and eventually to cell death. Transduction of EREB2.5 cells with a lentiviral vector expressing wild-type EBNA2 rescued EREB2.5 cells from the growth-inhibitory effects of estrogen deprivation, in contrast to transduction with the lentivirus vector alone. EREB2.5 cells were also rescued by enforced expression of human Notch1IC after estrogen starvation, but this effect was restricted to cells expressing high levels of the transcription factor. Compared to wild-type EBNA2-expressing EREB2.5 cells, the Notch-expressing cells expanded more slowly after estrogen starvation, and once established, they continued to display a lower proliferation rate. Analysis of viral and cellular gene expression from transduced EREB2.5 cells after estrogen withdrawal indicated that both wild-type EBNA2- and Notch1IC-positive cells expressed c-Myc at levels similar to those found in parental EREB2.5 cells. However, the latter cells expressed LMP-1 far less efficiently than cells transduced with the wild-type EBNA2 gene. Cells rescued by either wild-type EBNA2 or Notch1IC expressed surface CD21 and CD23 proteins, but not CD10, indicating that induction of relevant type III latency markers was maintained. The data imply that both Notch and EBNA2 activate an important subset of cellular genes associated with type III latency and B-cell growth, while EBNA2 more efficiently induces important viral genes, such as LMP-1. Thus, exploitation of conserved Notch-related signaling pathways may represent a key mechanism by which EBNA2 contributes to EBV-induced cell immortalization.
Epstein-Barr virus (EBV) infection is associated with several human malignancies, including Burkitt's lymphoma, Hodgkin's disease, nasopharyngeal carcinoma, and lymphomas in the immunosuppressed host (53). EBV latent infection of human B lymphocytes in vitro induces expression of B-cell activation markers, proliferation, and eventual outgrowth of continuously growing lymphoblastoid cell lines (LCLs) (36). LCLs phenotypically resemble physiologically activated primary B cells (36). The ability of EBV to stimulate B-cell growth independently of physiologic stimuli, such as stimulation by antigen and CD4+ T-cell help, is mediated by a subset of viral proteins. Uncovering the mechanisms by which these viral proteins function is essential to understanding EBV pathobiology, including the association with human malignancy, and may yield insight into the molecular mechanisms that govern the normal physiologic responses of B lymphocytes.
Efficient immortalization of B lymphocytes by EBV requires establishment of the type III latent gene expression program characterized by the expression of only a small subset of viral proteins (36). These include several EBV nuclear antigens (EBNAs)—EBNA1, EBNA2, EBNA3A and -3C, EBNA-LP—and an integral latent membrane protein, LMP-1. EBNA2 and EBNA-LP are the first viral proteins expressed upon infection of lymphocytes by EBV (1, 2). EBNA2 is essential for EBV-induced immortalization of B lymphocytes (8, 21). EBNA2 induces transcription of the LMP-1 and LMP-2A genes (13, 32, 66, 81) and stimulates expression of several cellular proteins, including c-Myc, which is likely to be crucial for immortalization (6, 9, 32, 70). EBNA2 also stimulates transcription from the major latency C promoter, which directs expression of all the EBNA genes (64, 75), establishing this nuclear protein as a key regulator of both viral and cellular gene expression during cell immortalization.
Although an efficient transcriptional activator, EBNA2 does not appear to bind DNA directly. One mechanism for promoter targeting is through an interaction with the DNA-binding transcription factor CBF1 (C promoter binding factor 1) (20, 23, 43, 69, 80). Recently, an additional cellular cofactor, SKIP, which interacts with both CBF1 and EBNA2, has been identified and appears to facilitate CBF1-EBNA2 interactions (79). Complex formation between EBNA2 and CBF1 is essential for EBV immortalizing activity (77). CBF1-mediated targeting of EBNA2 to viral promoters contributes to transcription activation, as EBNA2 masks the CBF1 transcriptional repressor domain and possesses a strong carboxy-terminal activation domain (7, 29, 44, 71). The CBF1 core DNA recognition sequence GTGGGAA is present in most EBNA2 responsive promoters characterized to date (67). Although multimerized CBF1 binding sites alone can confer EBNA2 responsiveness in transient transfection assays, other _cis_-acting elements, including those binding CBF2/AUF1 and Spi-1/PU.1, clearly influence whether EBNA2 can effectively stimulate a target promoter (17, 31, 40, 43). This suggests that EBNA2 may be involved in a complex network of individual promoter-specific protein-protein and protein-DNA interactions that mediate its immortalizing functions. Intriguingly, the N-terminal part of EBNA2, although not required for CBF1 or SKIP binding, is essential for immortalization, suggesting that there are other presently unknown mechanisms by which EBNA2 contributes to immortalization by EBV (76).
CBF1 is also an important component of Notch signaling pathways, which are highly conserved from invertebrates to vertebrates. CBF1 and its homologs in Drosophila melanogaster, Suppressor of Hairless [Su(H)], and in Caenorhabditis elegans, Lag-1, have been collectively designated CSL proteins [stands for CBF1, Su(H), Lag-1].
Several studies have demonstrated that activated cellular Notch proteins interact with CBF1 and can transactivate genes containing CBF1 binding sites in their promoters (25–28). Notch proteins are large membrane-bound proteins that possess an extracellular domain consisting of epidermal growth factor (EGF)-like repeats, a transmembrane domain, and a large cytoplasmic domain that contains several motifs, including CDC10 ankyrin repeats (3, 5, 47, 49). Mammalian cells express several related Notch proteins, Notch 1, 2, 3, and 4 (12, 39, 54, 73, 74), while Drosophila expresses only a single Notch protein. The remarkable evolutionary conservation of Notch proteins reflects their essential role in multiple stages of animal development. Notch signaling pathways are activated by interaction of the Notch extracellular domain with a ligand present on neighboring cells. Multiple Notch ligand genes have been cloned and characterized in both invertebrates and vertebrates (3, 5, 47, 49), and their products typically contain DSL motifs (Delta, Serrate, Lag-2) required for binding to the Notch extracellular domain. Once ligand binding takes place, the intracellular domain of Notch (Notch1 IC) is proteolytically cleaved, released into the cytoplasm, and transported to the nucleus, where it can activate some target gene promoters by a mechanism thought to involve CBF1-mediated recruitment, much in the manner of EBNA2 (26, 37, 57, 61, 63, 78). Like EBNA2, a significant amount of evidence also suggests that not all of Notch activities are mediated by CBF1 (4, 45, 48, 58, 72). Ectopic expression of Notch IC in cells appears to mimic the effects of ligand binding to Notch and therefore represents a constitutively active form of the protein. Enforced expression of Notch IC often results in effects consistent with its role in cell fate determination and differentiation inhibition during development (16, 38, 41, 62). Aberrant expression of Notch or Notch IC is also associated with several human neoplasms and is oncogenic in some animal model systems (12, 18, 50, 54).
Taken together, the above observations suggest a broad overlap between Notch and EBNA2 functions in vivo; however, it is unlikely that NotchIC can fully reproduce all EBNA2-mediated effects in the context of EBV-immortalized cell lines. The experiments described here test the hypothesis that EBNA2 possesses immortalization activities distinct from the evolutionarily conserved functions of the human NotchIC protein. This aim was pursued with a transcomplementation system that bypasses most of the events involved in the long, complex process of LCL generation from EBV-infected B lymphocytes.
MATERIALS AND METHODS
Cell culture.
EREB2.5 is a conditionally immortalized lymphoblastoid cell line and has been described previously (35). DG75 is an EBV-negative Burkitt's lymphoma cell line. Lymphoid cells were maintained in RPMI 1640 medium and 293T cells were maintained in Dulbecco's modified Eagle medium (high glucose) and grown in 5% CO2 at 37°C. The growth media were supplemented with 10% fetal bovine serum (Hyclone) and 15 mM HEPES, pH 7.4. For EREB2.5 cells the culture medium contained 1 μM 1,2-estradiol (Sigma) unless otherwise specified.
Transient transfections and reporter gene assays.
Transient transfections of DG75 cells were performed with the pJT123A firefly luciferase reporter plasmid as described before (51). pJT105 was generated by PCR amplification of a human Notch 1 cDNA (hN1) using primers 5′-CGATGAATTCCATCATGCGCAAGCGCCGGCGGCAG-3′ and 5′-CGATAGATCTTTACTTCTCATCGTCGTCCTTGTAGTCCTTGAACGCCTCCGGGAT-3′. The resulting amplified product also added sequences encoding a Flag epitope to the COOH terminus of the protein. The PCR product was directly cloned into a T-easy vector (Promega) and sequenced to confirm that no errors were introduced. pSG5-Notch1IC was created by subcloning the Notch1IC cDNA, generated by _Eco_RI/_Bgl_II digestion of pJT105, into the _Eco_RI/_Bgl_II sites in the eukaryotic expression vector pSG5 (Stratagene). pPDL176A is an EBNA2 expression plasmid (51).
Lentivirus vector construction and production.
The lentivirus vector pLIG (lentivirus-IRES-eGFP; plasmid pAG131) was derived from pHIV-AP E-F-V-T by replacing the 1.6-kb _Not_I/_Xho_I fragment, encoding the alkaline phosphatase gene, with the 1.4-kb IRES-eGFP cassette from pBS-IRES-eGFP (see Fig. 2) (65). Thus, expression of any cDNA inserted just 5′ of the IRES will be linked with green fluorescence protein (GFP) expression. pLIG.EBNA2 was generated by excising the _Eco_RI-_Bgl_II DNA fragment from pPDL176A, which was then blunt ended with Klenow (Gibco BRL) and ligated into _Xba_I-cleaved pLIG (see Fig. 2). pLIG.E2mut. was generated from pPDL152, which contained mutations that change amino acids (aa) 323 and 324 of EBNA2 from WW to SR in a similar manner (43). pLIG.hNotch1IC was created by purification of the _Eco_RI/_Bgl_II fragment from pJT105 that contained sequences encoding aa 1758 to 2555 from hNotch 1, blunt ending and cloning into pAG131 as described for pLIG.EBNA2. Virus was produced by transient transfection of 293T cells as described before (65). Briefly, 50% confluent 293T cells were transfected with a mixture of plasmid DNAs including proviral plasmid, an HIV-Tat-encoding plasmid (pBC12-Tat), and a vesicular stomatitis virus G protein-encoding plasmid (pME-VSVG) by the calcium phosphate coprecipitation method. The cells were incubated with the DNA precipitate for at least 8 h. Forty-eight hours later, supernatants were collected and cleared of cellular debris by centrifugation at 3,000 × g for 10 min and stored at −70°C until use.
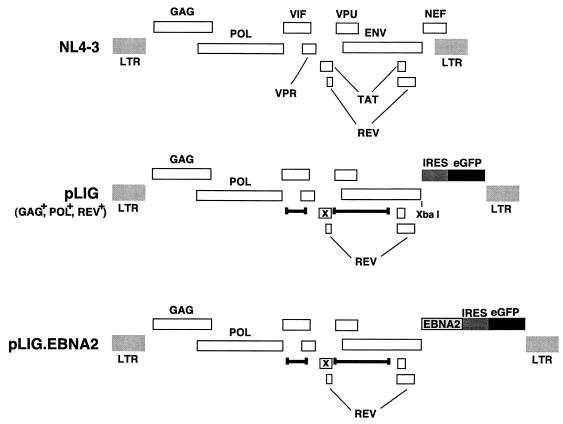
FIG. 2.
HIV vector constructions. A schematic of the wild-type parental virus genome NL4-3 is shown at the top with the indicated open reading frames. pLIG, which was derived from pHIV-AP E-F-V-T- is shown in the middle. Deletions are indicated by delimited bars, and each frameshift is indicated by an x. The IRES and eGFP elements of the cassette that was inserted are indicated by the shaded and black boxes, respectively. The provirus expresses Gag, Pol, and Rev but does not express Vif, Vpr, Vpu, Tat, Env, or Nef. pLIG.EBNA2 is shown at the bottom. The EBNA2 gene was cloned into the _Xba_I site in pLIG.
Lentivirus transduction of EREB2.5 cells and enrichment for GFP-positive cells.
For transduction, 5 × 105 log-phase EREB2.5 cells were resuspended in a 200-μl volume of culture medium by centrifugation and 2 ml of lentivirus stock supernatants was added. The concentration of estrogen was immediately adjusted to 1 μM. After incubation for 3 h at 37°C with occasional mixing, the cells were diluted with 3 ml of EREB2.5 conditioned culture medium. Five days posttransduction, the cells were analyzed for GFP expression by fluorescence-activated cell sorting (FACS). Enriched GFP-positive populations were established by positively sorting for the brightest 17.5% GFP-positive cells. Generally, 3.5 × 105 to 5 × 105 output cells were collected by FACS followed by expansion under standard culture conditions in the presence of antibiotic-antimycotic (Gibco BRL). Once sufficient numbers of cells were obtained, they were then maintained as described in “Cell culture” above.
Flow cytometry.
GFP-expressing and fluorescently labeled cells were sorted or analyzed using Becton Dickinson or Cytomation flow cytometers. For CD10, CD21, and CD23 surface expression analyses, phycoerythrin-conjugated anti-CD10 and anti-CD23 (DAKO) or phycoerythrin-conjugated anti-CD21 and isotype Simulitest Control γ1 (BD Pharmingen) monoclonal antibodies were used. Fluorescent staining was done according to the manufacturer's recommendations.
Immunoblotting.
Transiently transfected, transduced, or nontransduced cells were lysed in Laemmli sample buffer and boiled. The protein load per lane was equalized by quantification using Bradford's method (Bio-Rad's DC protein assay) according to the manufacturer's recommendations. Proteins were resolved by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 7.5% gel and transferred to nitrocellulose membrane. The membranes were blocked with 5% nonfat dried milk in phosphate-buffered saline. EBNA2 and EREBNA2 proteins were detected by incubation of the blots with monoclonal antibody PE2 (DAKO) followed by horseradish peroxidase-conjugated anti-mouse antibody (Amersham Pharmacia Biotech). Detection of EBNA-LP and LMP-1 with monoclonal antibodies JF186 and S12 has been described previously (52). c-Myc and EBNA3 were detected by polyclonal sheep anti-EBNA3 (Exalpha) and rabbit anti-c-myc (Santa Cruz) antibodies followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (Molecular Probes). After secondary antibody incubation, the proteins were visualized by an enhanced chemoluminescence kit (Pierce).
Proliferation assays.
To measure the proliferation of transduced and nontransduced EREB2.5 cells in the absence of estrogen, we washed 5 × 106 log-phase cells twice with 10 ml of estrogen-free medium and resuspended them in estrogen-free culture medium at 5 × 105 cells/ml. Cells were immediately plated in 200 μl of culture medium/well in estrogen-free culture medium in 96-well plates. Otherwise, 24 h later the cells were counted, washed once again, and seeded in 96-well plates in 200 μl of culture medium/well at the indicated concentrations. Proliferation was monitored by direct counting with a hemocytometer or by an MTT conversion assay (Cell Proliferation Kit I; Molecular Roche) as described by the manufacturer.
RESULTS
Human immunodeficiency virus-based defective lentiviruses efficiently transduce EBV-immortalized B lymphocytes.
To determine whether Notch could functionally replace EBNA2 in maintaining the viability and growth of EBV-immortalized cells, we developed a transcomplementation assay in the conditionally immortalized EREB2.5 cell line (35). Such cells require estrogen for proliferation and survival, since the EBNA2 protein they express is fused to the hormone-binding domain of the estrogen receptor, rendering its activity dependent on estrogen (34). Removal of estrogen results in the immediate growth arrest and apoptotic death of many cells, while the remainder die within 5 to 7 days. We hypothesized that enforced expression of certain genes (e.g., Notch1IC) in EREB2.5 cells by use of lentivirus vectors would result in continued proliferation of these cells in media without estrogen, provided that essential EBNA2 functions were supplied in trans (Fig. 1). A significant advantage of such a system over traditional recombinant virus approaches is that if Notch were unable to substitute for EBNA2, the cells could still be analyzed for viral and cellular gene expression or cellular phenotype, allowing identification of Notch-mediated effects in human B cells.
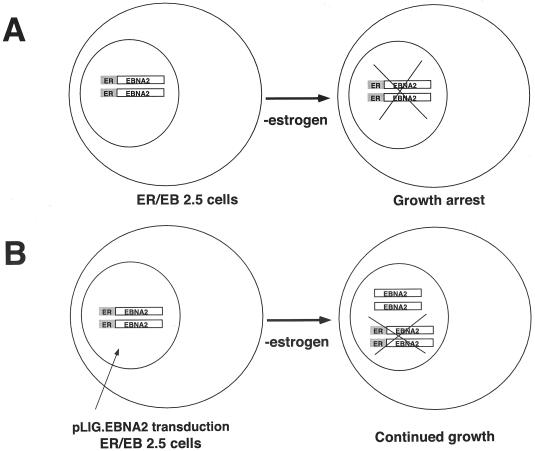
FIG. 1.
Transcomplementation assay strategy. (A) EREB2.5 cells grown in the absence of estrogen result in EREBNA2 molecules becoming inactivated. As a result, the cells are unable to survive. (B) EREB2.5 cells transduced with pLIG.EBNA2 virus express wild-type EBNA2. Withdrawal of estrogen results in inactivation of the EREBNA2 proteins; however, the cells survive since wild-type EBNA2 is provided in trans.
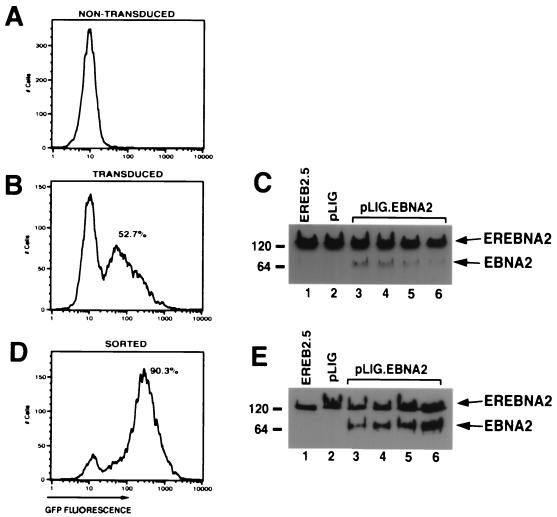
We therefore modified a previously described lentivirus vector, pHIV-AP E-F-V-T, by replacing the alkaline phosphatase gene with an IRES-GFP cassette to create pLIG (Fig. 2) (65). A unique _Xba_I site upstream of the IRES element allows cloning of any cDNA into the proviral vector, whose expression is also linked to GFP expression (Fig. 2). Using a pLIG.EBNA2 proviral plasmid, we generated vesicular stomatitis virus G-pseudotyped virus stocks (described in Materials and Methods) that were used to transduce EREB2.5 cells. Transduction efficiencies routinely ranged from 50 to 75% (Fig. 3B). Similar results were obtained with the pLIG vector alone (data not shown). To characterize the ability of the pLIG.EBNA2 lentivirus vector to direct synthesis of EBNA2 in the transduced cells, we analyzed EBNA2 expression in the cells by Western blotting. In four independently derived cell lines, wild-type EBNA2 was detectable, but at lower levels than endogenous EREBNA2 protein (Fig. 3C). EREB2.5 cells transduced with wild-type EBNA2 were then enriched for the brightest GFP-positive cells (top 17.5%) by FACS. After sorting, the cell pools were greater than 90% GFP positive and showed a greater-than-fivefold increase in mean GFP fluorescent intensity over unsorted cells (Fig. 3D). The sorted cells also synthesized similar levels of EBNA2 and endogenous EREBNA2 (Fig. 3E). GFP expression in the cells transduced with pLIG or pLIG.EBNA2 was stable for more than 2 months (data not shown).
FIG. 3.
Transduction and expression of EBNA2 into EREB2.5 cells. (A) FACS analysis of EREB2.5 cells for GFP fluorescence. (B) FACS analysis of GFP expression in EREB2.5 cells transduced with pLIG.EBNA2 virus. (C) Western blot analysis of cellular extracts from EREB2.5 cells (lane 1), pLIG-transduced EREB2.5 cells (lane 2), and four independently derived cell pools from EREB2.5 cells transduced with pLIG.EBNA2 virus (lanes 3 to 6). The blots were probed with the PE2 monoclonal antibody against EBNA2. Migration of both endogenous EREBNA2 and wild-type EBNA2 expressed from the pLIG vector are indicated on the right of the blot. (D) FACS analysis of pLIG.EBNA2-transduced EREB2.5 cell lines that have been sorted for the top 15% of GFP-positive cells as shown in panel B. (E) Western blot analysis of cellular extracts from EREB2.5 cells (lane 1), pLIG-transduced and sorted (as described in Materials and Methods) EREB2.5 cells, and four similarly derived pools of EREB2.5 cells transduced (as in panel C) with pLIG.EBNA2 virus and then sorted as described in Materials and Methods (lanes 3 to 6). The blots were probed with the PE2 monoclonal antibody against EBNA2. Migration of both endogenous EREBNA2 and wild-type EBNA2 expressed from the pLIG vector are indicated on the right of the blot.
Wild-type EBNA2 expression rescues EREB2.5 cell growth in the absence of estrogen.
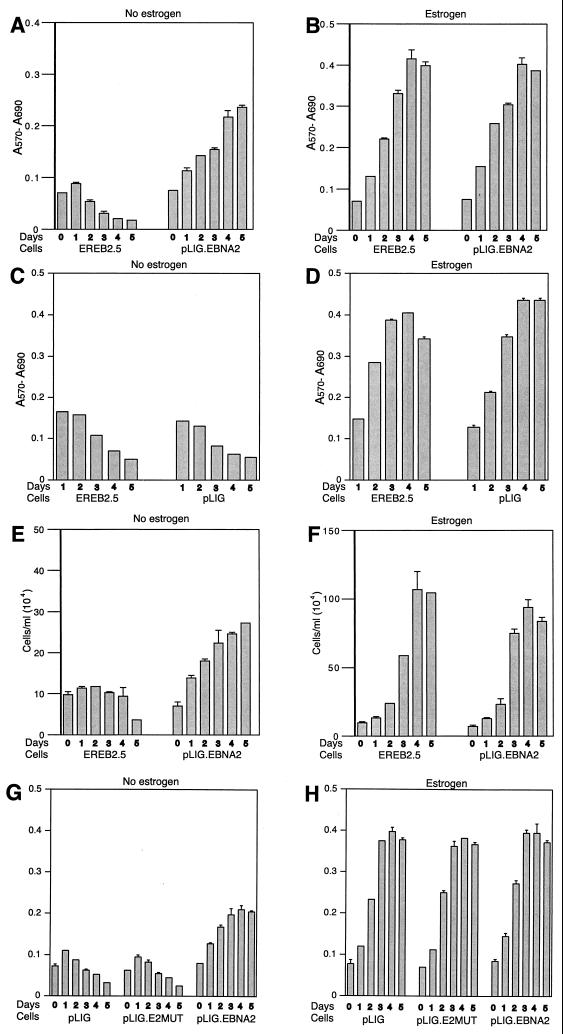
To demonstrate that wild-type EBNA2 expressed from pLIG.EBNA2 can functionally substitute for EREBNA2, we cultured the sorted, transduced cells (Fig. 3E) in the absence of estrogen. pLIG-transduced cells sorted in an identical manner served as controls. EREB2.5 cells transduced with pLIG.EBNA2 consistently showed the ability to proliferate after estrogen depletion, in contrast to nontransduced cells and those transduced with pLIG only (Fig. 4A, C, and E). Long-term cultures of pLIG.EBNA2-transduced EREB2.5 cells could also be readily established after estrogen removal from the growth medium (data not shown). Growth in the presence of estrogen did not differ appreciably among the three categories of experimental cells (Fig. 4B, D, and F), indicating that neither pLIG nor pLIG.EBNA2 inhibited the proliferation of EREB2.5 cells. Since nontransduced and pLIG-transduced cells had similar growth profiles in both the presence and absence of estrogen, we used the latter as a negative control in all further experiments. The experiments in Fig. 4 were also reproducible with several independently derived transduced cell lines (data not shown).
FIG. 4.
Proliferation of transduced EREB2.5 cells expressing wild-type EBNA2 after estrogen starvation. (A) Ten thousand cells/well of the indicated cells were plated into 96-well plates in growth medium without estrogen. Cell proliferation was monitored by MTT assays as described in Materials and Methods each day over the course of 5 days. (B) Same as for panel A, except that cells were grown in media containing estrogen. (C and D) Same as for panels A and B, except that the comparison was between EREB2.5 and pLIG-transduced EREB2.5 cells as indicated below the graphs. (E) The indicated cell lines were plated out as described for panel A, except that cells were removed, stained with trypan blue, and counted each day over a 5-day period. (F) Same as for panel E, except that cells were grown in media containing estrogen. (G and H) Same as for panels A and B, except that growth rate comparisons are between pLIG-, pLIGE2mut.-, and pLIG.EBNA2-transduced cell lines.
Experiments were then performed to compare the cells expressing EBNA2 or EBNA2 with a mutation in conserved region 6 (pLIG.EBNA2.PI326SR) which abolishes interaction with the cellular CBF1 protein (42, 43). Levels of mutant EBNA2 protein and endogenous EREBNA2 were similar in transduced cells sorted into discrete cell lines (data not shown). The EBNA2 mutant lacked any discernable effect on EREB2.5 cell growth in the presence of estrogen (Fig. 4H) and did not rescue cells from estrogen starvation (Fig. 4G). There was no outgrowth of pLIG.E2mut.-transduced cells even when they were plated at high concentrations after estrogen withdrawal (data not shown). Since wild-type EBNA2 can support the proliferation of EREB2.5 cells in the absence of estrogen but a mutant EBNA2 protein known to be transformation defective cannot, we concluded that our system would be useful in testing the ability of Notch1IC to replace EBNA2 function in immortalized B lymphocytes.
Properties of EREB2.5 cells expressing hNotch1IC.
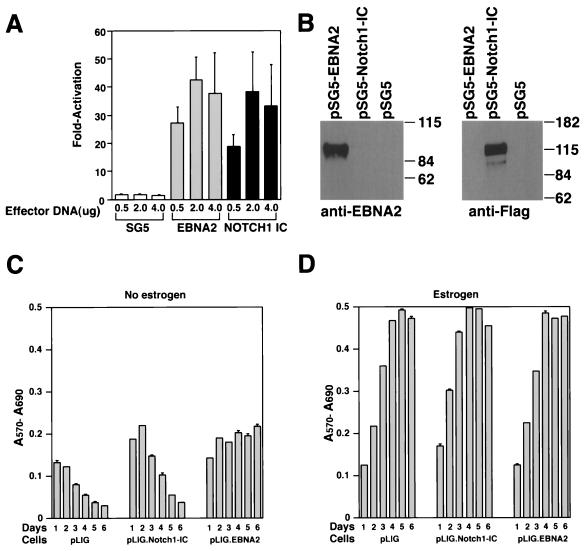
To determine whether the human Notch1IC domain (aa 1757 to 2555) displays its expected transcriptional activation function, we performed transient cotransfection experiments. Both EBNA2 and hNotch1IC activated a CBF1-responsive promoter with an efficiency comparable to that given in previously published data, resulting in high protein expression levels upon transient transfection of DG75 cells (Fig. 5A and B) (22, 42, 43, 51, 59). Subsequently, EREB2.5 cells were transduced with viruses derived from pLIG.Notch1IC and positively sorted for GFP expression, as described above. Comparative studies of differently transduced cell lines expressing similar amounts of GFP showed that Notch1 IC could not rescue EREB2.5 cells after estrogen depletion (Fig. 5C). Enforced Notch1 IC expression did not appear to have any adverse effect on the proliferation of these cells under normal growth conditions (Fig. 5D).
FIG. 5.
Proliferation of EREB2.5 cells constitutively expressing Notch1IC after estrogen starvation. (A) Notch1IC protein efficiently transactivates luciferase gene under the control of a promoter containing multimerized CBF1-binding sites. Either Notch1IC- or EBNA2-expressing effector plasmids were transiently cotransfected with the luciferase reporter plasmid pJT123 into DG75 cells. Luciferase activity was quantified for different amounts of the effector plasmids. T-bars indicate standard errors. (B) Western blot analyses of Notch1IC or EBNA2 proteins in cells transiently transfected with the respective effector plasmids (as in panel A). (C) Ten thousand cells/well of the indicated cells were plated into 96-well plates in growth medium without estrogen. Cell proliferation was monitored by MTT assays as described in Materials and Methods each day over the course of 6 days. (D) Same as for panel C, except that cells were grown in media containing estrogen.
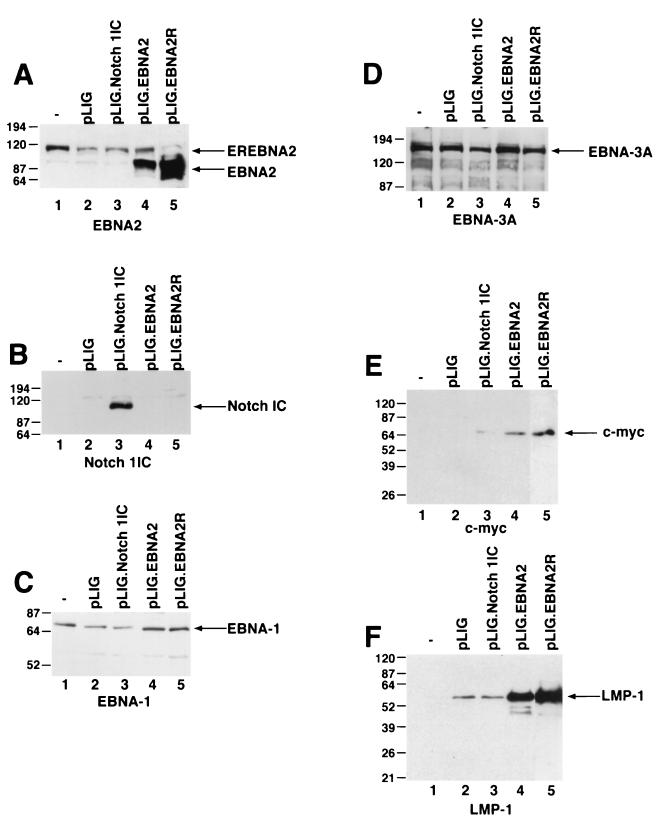
To account for the failure of Notch to sustain proliferation of EREB2.5 cells, we surveyed the expression of viral and cellular genes known to be induced by EBNA2. Western blot analyses of extracts from transduced EREB2.5 cells that had been starved for estrogen for 4 days showed expression of EBNA1, EBNA3A, and EBNA-LP in EREB2.5 cells transduced with pLIG, pLIG.EBNA2, and pLIG.Notch even after 4 days of estrogen withdrawal (Fig. 6C and D; data not shown). By contrast, only cells expressing EBNA2 expressed appreciable amounts of c-Myc (Fig. 6E). Although the oncoprotein was detected in Notch1IC-expressing cells, its levels were on average 14- to 30-fold lower than in cells expressing EBNA2 (Fig. 6E; Table 1). LMP-1 levels in Notch-transduced cells did not exceed those in cells transduced with the pLIG vector alone and were approximately 16- and 24-fold less than in pLIG.EBNA2 and pLIG.EBNA2.R cells, respectively (Fig. 6F; Table 1). Thus, one reason for the inability of Notch to rescue EREB2.5 cells in the absence of a functional EBNA2 protein is its failure to induce LMP-1, and possibly to induce c-Myc to physiologically relevant levels.
FIG. 6.
Detection of EBNAs, LMP-1, and c-myc in EBNA2- and Notch1IC-expressing EREB2.5 cells after estrogen withdrawal. (A) Western blot of cellular extracts derived from cells starved for estrogen for 4 days. The blots were probed with the PE2 monoclonal antibody directed against EBNA2. Both the EREBNA2 and wild-type EBNA2 genes can be detected and are indicated by arrows. The cell lines used are indicated above the blot. −, parental EREB2.5 cells were used as a control. pLIG.EBNA2R is a cell line expressing wild-type EBNA2 from pLIG and grown long-term in media lacking estrogen. (B) Same as for panel A, except that the blot was probed with an anti-Notch1 antisera. (C) Same as for panel A, except that the blot was probed with an anti-EBNA-1 antibody. (D) Same as for panel A, except that the blot was probed with an anti-EBNA-3A antibody. (E) Same as for panel A, except that the blot was probed with an anti-c-myc antibody. (F) Same as for panel A, except that the blot was probed with an anti-LMP-1 antibody. Migration of molecular weight markers is shown to the left of each blot, and the specific proteins detected are indicated by the arrows on the right.
TABLE 1.
Relative abundancea of protein expression in differently transduced cell lines
| Cell line | Relative abundance of: | |||
|---|---|---|---|---|
| EBNA1 | EBNA3A | c-Myc | LMP-1 | |
| pLIG | 1 | 1 | NDb | 1 |
| pLIG-NOTCH | 0.6 | 0.6 | 1 | 1 |
| pLIG-EBNA2 | 2 | 1.6 | 14 | 16 |
| pLIG-EBNA2.R | 2.6 | 0.9 | 30 | 24 |
| EREB2.5(−ES) | 1 | 1.25 |
Selection of EREB2.5 cells expressing high levels of hNotch1IC.
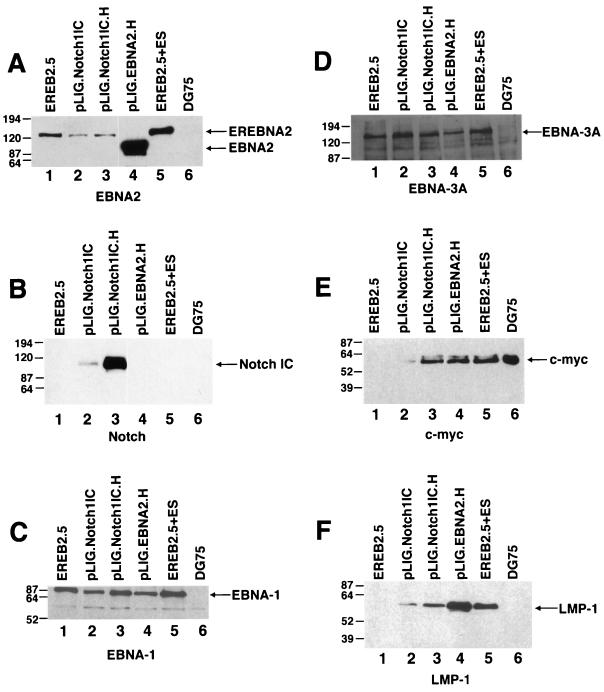
Using transient transfection assays, several groups have demonstrated that activated Notch1IC and EBNA2 stimulate similar repertoires of promoters (10, 25, 26, 55, 59, 60). Since Notch1IC was likely overexpressed or at least expressed at high levels in these studies, we undertook a second round of FACS of our pLIG-transduced populations (Fig. 4 and 5). The results indicated the feasibility of generating cell lines that express considerably higher levels of GFP than were seen after the first positive sorting procedure (data not shown). Thus, we selected for Notch1IC transduced EREB2.5 cells expressing >40-fold-higher levels of GFP. One of the resultant cell lines (pLIG.Notch1 ICH) expressed much higher levels of Notch than did our initial cell lines (see Fig. 8B). A second transduced cell line expressing significantly larger amounts of wild-type EBNA2 (pLIG.EBNA2.H) was also selected (see Fig. 8A). Although these cells retained the EREBNA2 gene, they did not express the endogenous EREBNA2 protein (data not shown).
FIG. 8.
Detection of EBNAs, LMP-1, and c-Myc in EREB2.5 cells expressing high levels of Notch1IC after estrogen withdrawal. (A) Western blot of cellular extracts derived from cells starved for estrogen for 4 days. The blot was probed with the PE2 monoclonal antibody directed against EBNA2. Both EREBNA2 and wild-type EBNA2 genes can be detected and are indicated by arrows. The cell lines used are indicated above the blot. Lane 5, EREB2.5 cells grown in media containing estrogen; lane 6, DG75 cells. (B) Same as for panel A, except that the blot was probed with an anti-Notch1 antisera. (C) Same as for panel A, except that the blot was probed with an anti-EBNA-1 antibody. (D) Same as for panel A, except that the blot was probed with an anti-EBNA-3A antibody. (E) Same as for panel A, except that the blot was probed with an anti-c-myc antibody. (F) Same as for panel A, except that the blot was probed with an anti-LMP-1 antibody.
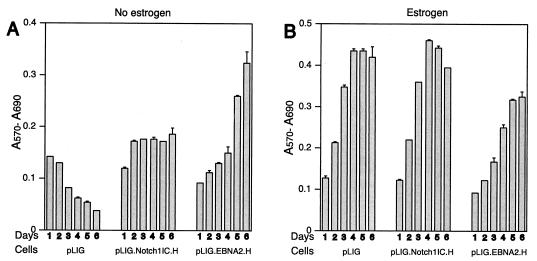
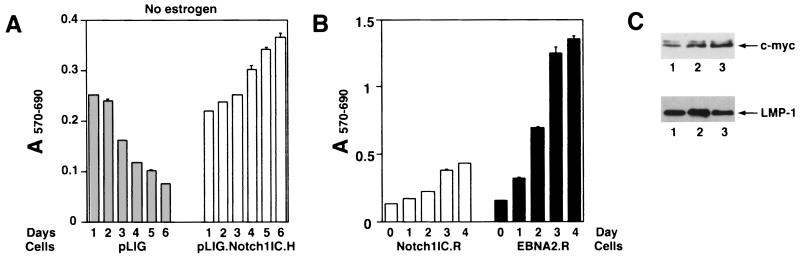
In contrast to cells transduced with pLIG only, both the pLIG.Notch1IC.H and pLIG.EBNA2.H cell lines retained viability over 6 days following estrogen withdrawal (Fig. 7A). Expression of large amounts of Notch1IC or wild-type EBNA2 from pLIG did not appear to have major effects on EREB2.5 cell proliferative capacity under normal growth conditions (Fig. 7B).
FIG. 7.
Proliferation of EREB2.5 cells constitutively expressing high levels of Notch1IC after estrogen starvation. (A) Ten thousand cells/well of the indicated cells were plated into 96-well plates in growth medium without estrogen. Cell proliferation was monitored by MTT assays as described in Materials and Methods each day over the course of 6 days. (B) Same as for panel A, except that the cells were grown in media containing estrogen. T-bars indicate standard errors.
Comparison of levels of gene expression in EBNA2.H- and Notch1IC.H-transduced cells showed comparable c-myc levels (Fig. 8E; Table 2), with lower levels of LMP-1 detected in the latter cell line (Fig. 8F). EBNA-1, EBNA-LP, and EBNA3A were uniformly detected in the transduced and nontransduced EREB2.5 cells (Fig. 8C and D and data not shown).
TABLE 2.
Relative abundancea of protein expression in differently transduced cell lines
| Cell line | Relative abundance of: | |||
|---|---|---|---|---|
| EBNA1 | EBNA3A | c-Myc | LMP-1 | |
| pLIG-NOTCH | 1 | 1 | 1 | 1 |
| pLIG-NOTCH.H | 3 | 0.6 | 17 | 4.5 |
| pLIG-EBNA2.R | 1.5 | 0.3 | 20 | 21 |
| EREB2.5(+ES) | 4 | 1.3 | 25 | 12 |
Expression of high levels of Notch1IC ultimately rescues EREB2.5 cells after EBNA2 inactivation.
Since the pLIG.Notch1IC.H cell line retained viability after estrogen starvation, we asked whether the cells could survive for extended times. pLIG.Notch1IC.H cells grown without estrogen for 6 days were resuspended in fresh estrogen-free medium, and their growth was monitored by MTT assay. Surprisingly, after day 7 of estrogen starvation (day 2 in Fig 9A), cell proliferation was still apparent (Fig. 9A) and some cell lines selected by this procedure continued to grow for several months (Fig. 9B and data not shown). Comparison of gene expression in rescued pLIG.Notch1IC.H cells versus pLIG.EBNA2 and parental EREB2.5 cells revealed no differences in EBNA-1, EBNA-LP, EBNA3, or c-myc expression (data not shown). LMP-1 expression levels were likewise similar among the cell lines (Fig. 9C), in contrast to findings at 4 days after estrogen withdrawal, when cells were not actively proliferating (Fig. 7A and 8F). Why the rescued Notch1IC.H cells (Notch1 IC.R) express high levels of LMP-1 is unclear, but it may be related to secondary changes occurring after transduction or to selection of a subpopulation of cells that are more permissive for LMP-1 expression. An open question is whether endogenous EREBNA2 in these cells contributed to their rescue by Notch in the absence of estrogen. We consider this possibility unlikely, since EREBNA2 levels in estrogen-starved cells were only a fraction of levels observed when estrogen was present. Also, the absolute amount of EREBNA2 protein decreases with time in Notch-rescued cells (data not shown).
FIG. 9.
Proliferation of pLIG.Notch1IC.H cell lines after long-term estrogen withdrawal. (A) pLIG.Notch1IC.H-expressing cells were allowed to continue growing after 6 days without estrogen (as for Fig. 7A). After that, the cells were resuspended in fresh estrogen-free medium at 3 × 104 per well and cell proliferation was monitored daily for an additional 6 days by MTT assay. (B) pLIG.hNotch1IC and pLIG.EBNA2 cells were allowed to grow without estrogen for 4 weeks. The resulting outgrowing rescued cells lines, Notch1IC.R and EBNA2.R, were plated at 2 × 104 cells per well in the estrogen-free medium, and cell proliferation was monitored daily for 4 days as described for panel A. (C) Western blots of cellular extracts derived from cell lines grown continously without estrogen. Lane 1, pLIG.Notch1IC-rescued cells (Notch1IC.R in panel B); lane 2, pLIG.EBNA2-rescued cells (EBNA2.R in panel B); lane 3, EREB2.5 cells grown with estrogen. The upper panel shows a blot probed with a c-Myc antibody, and the lower panel shows a blot probed with an LMP-1 antibody. c-Myc and LMP-1 proteins are indicated by the arrows.
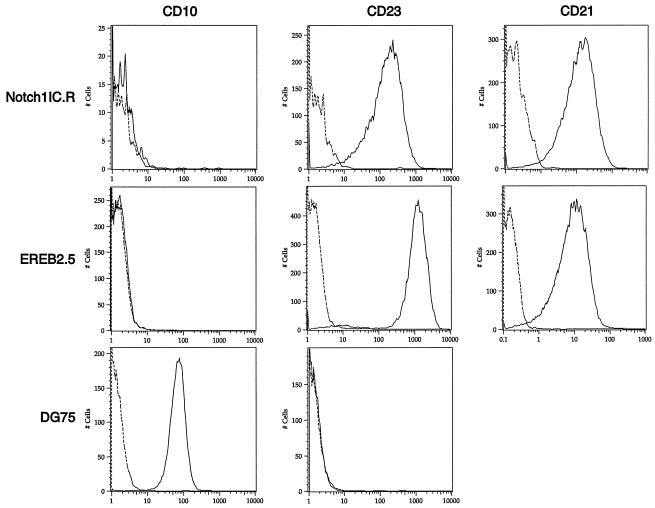
Finally, to determine whether a cellular state similar to the classic type III latency induced by EBV is maintained in pLIG.Notch1IC.R cells, we monitored the cells for CD23, CD21, and CD10 expression. Both Notch- and EBNA2-rescued cells expressed CD23 (Fig. 10), although in the Notch-expressing cells, levels of this differentiation marker were six- fold lower than in EBNA2-expressing cells. Whether LMP-1 contributes to or is entirely responsible for CD23 activation in Notch-positive cells cannot be determined from our analysis. Neither cell line expressed the CD10 marker, which is expressed on the Burkitt's lymphoma cell line DG75 but is characteristically absent on LCLs (Fig. 10). Both the Notch1IC.R cell line and parental EREB2.5 cells grown under normal conditions expressed similar levels of CD21 (Fig. 10), a known direct target of EBNA2 (32) and a recently reported target of Notch (59). Thus, high levels of Notch1 IC expression were associated with the activation of immortalization-related B-cell developmental markers.
FIG. 10.
CD23 and CD21 expression in Notch1IC expressing EREB2.5 cells grown without estrogen. CD10, CD23, and CD21 expression was detected on EREB2.5 and DG75 cells grown under normal conditions and on EREB2.5 cells expressing large amounts of Notch1IC grown without estrogen for 4 weeks (Notch1IC.R) by flow cytometry. Dotted lines, cells stained with an isotype control antibody; solid lines, cells stained with the antibody indicated above the graphs. Cell lines are indicated to the left of the graphs.
DISCUSSION
This study shows that enforced expression of human Notch1IC in EBV-immortalized cells is sufficient to maintain the immortalized phenotype when EBNA2 is inactivated. Although Notch1IC had to be overexpressed to achieve this effect, our results suggest that EBNA2 maintains EBV-induced B-cell immortalization by mimicking Notch signaling activities.
These data were acquired with a transcomplementation assay that allowed us to express Notch1IC in LCLs under conditions where EBNA2 activity could be inactivated (Fig. 1). The evidence that Notch1IC can faithfully substitute for EBNA2 in EBV-infected cells was obtained by monitoring Notch1IC-rescued EREB2.5 cells for the three major traits of EBV-immortalized cells: (i) ability to proliferate in vitro without feeder cells, (ii) expression of viral latency III proteins (e.g., EBNAs, LMP-1), and (iii) expression of certain immortalization-associated cellular genes and B-cell developmental markers (e.g., c-myc, CD21, CD23). We also demonstrated that EBNA2 does not have to be in the regulatory context of the EBV genome to support the state of immortalization. Neither the lentivirus vector itself nor an EBNA2 mutant protein unable to interact with CBF1 was capable of rescuing EREB2.5 cells, demonstrating that the functions specifically provided by a wild-type EBNA2 protein are required for this effect. The transcomplementation approach also allowed testing of the ability of Notch1IC expression in EREB2.5 cells to compensate for inactivation of the EREBNA2 fusion protein. In the absence of estrogen, EREB2.5 cell lines expressing low levels of Notch1IC were unable to proliferate (Fig. 5) and did not express LMP-1 or appreciable levels of c-Myc, although viral EBNAs were detected in similar concentrations (Fig. 6). At higher Notch expression levels (>40-fold), the test cells grew readily in estrogen-depleted media, although the rate was considerably lower than that of cells expressing wild-type EBNA2 or parental EREB2.5 cells grown under normal conditions (Fig. 9). Moreover, they not only continued to express all of the EBNA proteins but also began to express LMP-1, CD21, and c-Myc at levels comparable to those in cells expressing wild-type EBNA2. The CD10 marker, characteristic of germinal center cells and Burkitt's lymphoma cells, was absent. Thus, EBV-infected cells expressing Notch1IC in the absence of a functional EBNA2 protein display all of the hallmarks of the type III latent gene expression program. However, since EBNA2 was more efficient than Notch1IC in providing necessary immortalizing changes, such as increased cell proliferation capacity and LMP-1 expression, we conclude that Notch only partially substitutes for the loss of EBNA2 activity in LCLs.
The relative inability of Notch1IC to support LMP1 expression in the expanded cell populations immediately after estrogen withdrawal agrees with a previous study's findings for an EBV-positive Burkitt's lymphoma cell line expressing a conditional ERNotch1IC protein (59). In that study, addition of estrogen to the culture medium resulted in high levels of cellular CD21 as well as viral LMP-2A transcription. LMP-1 transcripts could not be detected until cycloheximide was added to the medium, suggesting that ERNotch could activate the LMP-1 promoter, albeit to low levels. As expected, an EREBNA2 protein stimulated LMP-1 expression without cycloheximide treatment (59). The same study also indicated that Notch was unable to effectively induce CD23 expression and is in agreement with our observations (Fig. 10) (59). mNotch1IC also downregulated the μ-enhancer, a feature it has in common with EBNA2 (30, 59). While informative, this study of Burkitt's lymphoma cells assessed Notch function in cells expressing high levels of c-Myc. Since the c-myc gene is controlled by regulatory elements within the immunoglobulin locus, as a result of the chromosomal rearrangement typical of Burkitt's lymphomas, the introduction of Notch would be expected to downregulate c-myc, leading to effects on cell viability and growth phenotype. Finally, although it is 87% homologous to the human Notch1IC domain, mouse Notch1 IC may not fully recapitulate the full complement of interactions with human cellular cofactors that mediate Notch function when expressed in human cells.
While this paper was in review, a study describing the effects of murine Notch1IC expression in EREB2.5 cells was published (24). In EREB2.5 cells, mNotch1IC did not have a detectable effect on either proliferation or expression of LMP-1 and c-myc in the absence of functional EBNA2. However, in a lymphoblastoid cell line expressing LMP-1 independently of EBNA2, mNotch1IC could transiently maintain proliferation after EBNA2 inactivation. While mNotch1IC was able to maintain expression of CD21 in this system upon withdrawal of estrogen, it failed to upregulate c-myc and CD23. The general observations of inefficient or lack of LMP-1 and CD23 induction by Notch are similar to our results. However, the apparent ability of Notch to activate c-myc and to eventually rescue EREB2.5 cells after estrogen withdrawal is clear in our experiments. Potential species-specific interactions of human Notch IC with cellular cofactors versus that of murine Notch or the high levels of protein expression achievable in our system could account for the latter differences. Despite being functional in several assays, the murine Notch IC used in these studies also lacks the carboxy-terminal 238 aa (11, 56).
An intriguing aspect of our work is the implication of Notch in regulation of c-myc expression. In our system, Notch is expressed at ostensibly nonphysiological levels. It is thought that physiologically relevant functions of Notch are mediated by much lower levels of in vivo-processed Notch1IC. In this case it is worth noting that HES-1, a documented transcriptional target of Notch, is transiently induced by many growth factors which also induce c-myc (14). Therefore, it is not unlikely that growth factor- and Notch-induced events may overlap or cooperate with each other. It is tempting to speculate that Notch may function to either modify or substitute for growth factor receptor signaling. Supporting the former possibility is the fact that the oncogenic potential of Notch in in vitro transformation assays depends on the activity of ERK2 kinase, a member of MAP kinase family activated by many mitogenic signals (15). In addition, Jagged1, a Notch ligand, stimulates hematopoietic cell proliferation in vitro (33). Recently, it has also been shown that cytokine-dependent hematopoietic stem cells can be immortalized by constitutive Notch 1 signaling (68). Evidence is also emerging that Notch may directly control cell proliferation during development in Drosophila. Therefore, part of Notch signaling may be interaction with parts of cellular signal transduction machinery affecting growth-associated genes. Overexpression of Notch may compensate for the lack of surface growth factor receptor activity regarding c-myc upregulation. While Notch may have evolved to cooperate with cellular signaling processes, we would suggest that EBNA2 has perhaps evolved to activate Notch pathways without a requirement for cooperating signals. Whether this is due to EBNA2 possessing more robust transcriptional activating activity than Notch does or to EBNA2 utilizing additional cellular cofactors remains to be elucidated.
Overall, our data imply that EBNA2 contributes to B-cell immortalization by EBV through mechanisms common to the Notch signaling pathway (activation of cellular genes), while possessing the additional capacity to efficiently activate viral targets such as LMP-1. It is tempting to speculate that evolutionary pressures drove EBNA2 to exploit the same nuclear protein machinery used by Notch to gain access to the regulatory regions of cellular genes. The inability of Notch to efficiently regulate viral promoters such as LMP1 could reflect the much greater evolutionary plasticity of viral promoters compared to cellular ones. Hence, EBNA2 interactions not shared by Notch may have become essential during the rapid coevolution of viral promoters.
EBNA2 lacks homology to any known cellular proteins, posing a major obstacle to analysis of the signaling pathways activated by this protein. One possibility, suggested by our data linking EBNA2 immortalizing functions with Notch regulation of physiologically important cellular genes, would be to consider pathways that are Notch regulated and also drive processes required for cell transformation. For example, Notch has been shown to be involved in antiapoptotic and proliferative as well as differentiation processes (3, 19, 46). The findings that EBNA2 may usurp one or more Notch-activated pathways indicate that its robust transcriptional activity might prove useful in delineating the downstream biochemical cascades activated by Notch signaling.
ACKNOWLEDGMENTS
We thank Jeff Scott and Mike Cubbage for help with FACS and also A. J. Capabianco for human Notch cDNAs. We also thank Wolfgang Hammerschmidt for critical reading of the manuscript.
This work was supported by NIH grant CA69437 to P.D.L. and by Deutsche Forschungsgemeinschaft SFB455 to G.W.B. and B.K.
REFERENCES
- 1.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 2.Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 4.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 5.Bray S. Notch. Curr Biol. 2000;10:R433–R435. doi: 10.1016/s0960-9822(00)00549-2. [DOI] [PubMed] [Google Scholar]
- 6.Calender A, Billaud M, Aubry J P, Banchereau J, Vuillaume M, Lenoir G M. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc Natl Acad Sci USA. 1987;84:8060–8064. doi: 10.1073/pnas.84.22.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordier M, Calender A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G, Lenoir G M. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter M, Callahan J, Aster J, Robertson E. Intracellular forms of human NOTCH1 functionally activate essential Epstein-Barr virus major latent promoters in the Burkitt's lymphoma BJAB cell line but repress these promoters in Jurkat cells. J Virol. 2000;74:1486–1494. doi: 10.1128/jvi.74.3.1486-1494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumont E, Fuchs K P, Bommer G, Christoph B, Kremmer E, Kempkes B. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene. 2000;19:556–561. doi: 10.1038/sj.onc.1203352. [DOI] [PubMed] [Google Scholar]
- 12.Ellisen L W, Bird J, Wasr D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 13.Fahraeus R, Jansson A, Ricksten A, Sjoblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fambrough D, McClure K, Kazlauskas A, Lander E S. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- 16.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 17.Fuentes-Panana E M, Peng R, Brewer G, Tan J, Ling P D. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J Virol. 2000;74:8166–8175. doi: 10.1128/jvi.74.17.8166-8175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 19.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 20.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 22.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 24.Höfelmayr H, Strobl L J, Marschall G, Bornkamm G W, Zimber-Strobl U. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. J Virol. 2001;75:2033–2040. doi: 10.1128/JVI.75.5.2033-2040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Höfelmayr H, Strobl L J, Stein C, Laux G, Marschall G, Bornkamm G W, Zimber-Strobl U. Activated mouse notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J Virol. 1999;73:2770–2780. doi: 10.1128/jvi.73.4.2770-2780.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh J J-D, Nofziger D E, Weinmaster G, Hayward S D. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signaling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 29.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Jochner N, Eick D, Zimber-Strobl U, Pawlita M, Bornkamm G W, Kempkes B. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 1996;15:375–382. [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen E, Koh E, Mosislos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm G W, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karanu F N, Murdoch B, Gallacher L, Wu D M, Koremoto M, Sakano S, Bhatia M. The notch ligand jagged-1 represents a novel growth factor of human hematopoietic stem cells. J Exp Med. 2000;192:1365–1372. doi: 10.1084/jem.192.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kempkes B, Pawlita M, Zimber-Strobl U, Eissner G, Laux G, Bornkamm G W. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-Jκ in a conditional fashion. Virology. 1995;214:675–679. doi: 10.1006/viro.1995.0084. [DOI] [PubMed] [Google Scholar]
- 35.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart J W, Kremmer E, Delecluse H J, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 107–172. [Google Scholar]
- 37.Kopan R, Goate A. A common enzyme connects Notch signaling and Alzheimer's disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 38.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 39.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 40.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 42.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJκ. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuno K, Go M J, Sun X, Eastman D S, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- 46.Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Milner L A, Bigas A. Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood. 1999;93:2431–2448. [PubMed] [Google Scholar]
- 48.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osborne B, Miele L. Notch and the immune system. Immunity. 1999;11:656–663. doi: 10.1016/s1074-7613(00)80140-5. [DOI] [PubMed] [Google Scholar]
- 50.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng R, Gordadze A V, Fuentes Panana E M, Wang F, Zong J, Hayward G S, Tan J, Ling P D. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol. 2000;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng R S, Tan J, Ling P D. Mutational analysis of conserved regions in the Epstein-Barr virus leader protein: identification of distinct domains required for transcriptional cooperation with EBNA2 and nuclear localization. J Virol. 2000;74:9953–9963. doi: 10.1128/jvi.74.21.9953-9963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 54.Robbins J, Blondel B J, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroeder T, Just U. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 58.Shawber C, Nofziger D, Hsieh J J-D, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 59.Strobl L J, Hofelmayr H, Marschall G, Brielmeier M, Bornkamm G W, Zimber-Strobl U. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J Virol. 2000;74:1727–1735. doi: 10.1128/jvi.74.4.1727-1735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strobl L J, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-Jκ. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 61.Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 62.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 63.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–555. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 64.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sutton R E, Wu H T M, Rigg R, Bohnlein E, Brown P O. Human immunodeficiency virus type I vectors efficiently transduce human hematopoietic stem cells. J Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varnum-Finney B, Xu L, Brashem-Stein C, Nourigat C, Flowers D, Bakkour S, Pear W S, Bernstein I D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive notch1 signaling. Nat Med. 2000;6:1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- 69.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human J kappa recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang F, Gregory C D, Rowe M, Rickinson A B, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Younger-Shepherd S, Jan L Y, Jan Y N. Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless. Development. 1997;124:4435–4446. doi: 10.1242/dev.124.22.4435. [DOI] [PubMed] [Google Scholar]
- 73.Weinmaster G, Roberts V J, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 74.Weinmaster G, Roberts V J, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 75.Woisetschlaeger M, Jin X W, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yalamanchili R, Harada S, Kieff E. The N-terminal half of EBNA2, except for seven prolines, is not essential for primary B-lymphocyte growth transformation. J Virol. 1996;70:2468–2473. doi: 10.1128/jvi.70.4.2468-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 78.Ye Y, Lukinova N, Fortini M E. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 79.Zhou S, Fujimuro M, Hsieh J J, Chen L, Hayward S D. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]