Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: Genetic requirements (original) (raw)
Abstract
Natural competence for genetic transformation is the best-characterized feature of the major human pathogen Streptococcus pneumoniae. Recent studies have shown the virulence of competence-deficient mutants to be attenuated, but the nature of the connection between competence and virulence remained unknown. Here we document the release, triggered by competent cells, of virulence factors (e.g., the cytolytic toxin pneumolysin) from noncompetent cells. This phenomenon, which we name allolysis, involves a previously undescribed bacteriocin system consisting of a two-peptide bacteriocin, CibAB, and its immunity factor, CibC; the major autolysin, LytA, and lysozyme, LytC; and a proposed new amidase, CbpD. We show that CibAB are absolutely required for allolysis, whereas LytA and LytC can be supplied either by the competent cells or by the targeted cells. We propose that allolysis constitutes a competence-programmed mechanism of predation of noncompetent cells, which benefits to the competent cells and contributes to virulence by coordinating the release of virulence factors.
Keywords: allolysis bacteriocin, competence, predation, virulence
The Gram-positive bacterium Streptococcus pneumoniae is a frequent colonizer of the nasopharynx in humans and a causative agent of otitis media, pneumonia, bacteraemia, and meningitis throughout the world (reviewed in ref. 1). S. pneumoniae is best known for its ability to enter naturally into a transient state, termed competence, in which it takes up and is transformed by external DNA (reviewed in ref. 2). Genetic transformation is believed to constitute a major source of genetic variability for S. pneumoniae, which enables this pathogen to cope with host defenses (3).
Competence probably represents the first example of a cell-density-dependent phenomenon controlled by a secreted signaling molecule in bacteria (4). The competence-stimulating peptide (CSP), a 17-aa peptide (5), accumulates in the medium and stimulates a two-component regulatory system, ComDE (6), to induce expression of a competence regulon (com). The pre-CSP, encoded by comC, belongs to a family of secreted peptides and small proteins synthesized with a double-glycine (GG) leader at their N termini (2). The 24-aa GG-leader is cleaved off during export across the cytoplasmic membrane (7) by a dedicated proteolytic ATP-binding cassette (ABC)-transporter, ComAB (8). ComD, a membrane-localized kinase, is the receptor of CSP and autophosphorylates upon binding the pheromone; it then transphosphorylates its cognate response regulator, ComE (2), which in turn induces the com regulon. The com genes are temporally regulated (9, 2, 10). ComE activates transcription of the early genes, including comABCDE, which leads to the rapid accumulation of CSP, and comX (11). The _comX_-encoded σX factor (12) activates transcription of the late genes. Although the entire com regulon comprises 105-124 CSP-responsive genes (13, 10), only 22 are necessary for transformation, and at least 70 are individually dispensable. This system thus appears to serve purposes other than genetic transformation.
Recent studies have shown the virulence of comB and comD mutants to be attenuated in bacteraemia and/or lung infection (refs. 14 and 15, reviewed in ref. 16). In addition, two late com genes, lytA and cbpD, have been implicated in virulence. The lytA gene (17) encodes the major autolytic amidase, which is known to be important to pneumococcal pathogenesis (1). This gene is expressed in noncompetent (NC) cells (18), but is also part of the CSP-inducible recA operon (19). The cbpD gene was first shown to be CSP-induced (20, 21) and was further identified as a virulence factor by signature-tagged mutagenesis (22). Although it seems to play a role in nasopharyngeal colonization (23), its function remained unknown. These observations suggest that part of the com regulon could be dedicated to virulence, but they do not indicate how competence and virulence are connected.
During a study of the regulation of comCDE expression, we characterized a G → T change within the transcription terminator of the tRNAArg located immediately upstream of comCDE (24). This point mutation, named RT hereafter, was observed to confer an unusual β-hemolytic phenotype on bloodagar plates (see Fig. 1_A_). S. pneumoniae normally forms colonies surrounded by α-hemolytic halos, which correspond to the “greening” of red blood cells. In contrast, β-hemolysis corresponds to true hemolysis (i.e., the lysis of red blood cells) and is usually associated with the release of intracellular pneumolysin (Ply), an important virulence factor of S. pneumoniae (25). Ply belongs to a group of proteins of pathogenic Gram-positive bacteria known as cholesterol-dependent cytolysins (1), but unlike other toxins, lacks an N-terminal signal sequence for export (26) and usually requires cell lysis for release. Therefore, we took advantage of the unusual β-hemolytic phenotype conferred by the RT mutation to conduct an in depth genetic dissection of the underlying phenomenon, with the aim of shedding light on a possible connection between competence and virulence. Here we present evidence that β-hemolysis results from Ply release and involves the lysis of NC cells triggered by competent cells, a phenomenon we refer to as allolysis. We show that allolysis requires the products of six genes that all, but one, belong to the com regulon, and provide evidence that it is a general property of the species. We discuss the biological implications of these observations.
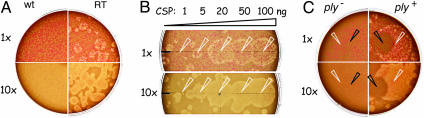
Fig. 1.
Competence-dependent induction of β-hemolysis involves Ply release. (A) R841 mutant (RT) but not R800 wild-type (wt) cells display spontaneous β-hemolysis on 5.5% horse blood CAT-agar (1×= 1.5 × 104 cells per plate). (B) CSP-induced β-hemolysis on RT mutant cells. One-microliter spots (indicated by white tips) containing 1-100 ng of CSP were deposited immediately after plating (1×= 2.5 × 104 cells per plate), and plates were incubated overnight at 37°C. (C) Inactivation of the ply gene or antibodies to Ply abolish the formation of HH. R1065 (RT ply+) but not R1130 (RT _ply_-) cells display spontaneous HH (1× = 2.5 × 104 cells per plate). CSP (1 μl, 100 ng, white tips) and PLY-4 monoclonal mouse antibody (5 μl, 4 μg, black tips) (53) were spotted immediately after plating. Note the zone of inhibition of β-hemolysis around the PLY-4 antibody spot.
Materials and Methods
Growth, Transformation, DNA Manipulations, and Analysis. S. pneumoniae strains, plasmids, and primers used in this study are listed in Table 3, which is published as supporting information on the PNAS web site. Growth and transformation, DNA manipulations (including mariner minitransposon mutagenesis), and analysis followed standard procedures (for details, see Supporting Text, which is published as supporting information on the PNAS web site). Cells were plated (90-mm-diameter plates) into CAT-agar (27) supplemented with 5.5% (vol/vol) horse blood and incubated overnight aerobically at 37°C.
Construction of Δ**cibABC,** Δ**cibAB, and** Δ**cibC Mutations.** Precise deletions of cibC and cibAB were generated by taking advantage of strain R1391 in which the cibABC operon was substituted with Janus (see Supporting Text). Janus is a kan_-rpsL+ cassette that confers resistance to kanamycin (KanR) and dominant sensitivity to streptomycin (SmS) in a streptomycin-resistant (SmR) background (28). Its replacement by an arbitrary segment of DNA during a second transformation restores SmR (and sensitivity to Kan). To delete only cibC, a PCR fragment in which cibC upstream and downstream regions were joined was generated and used to transform R1391 to SmR (see Supporting Text). A similar strategy was used to delete cibAB (see Supporting Text). The cibABC region of SmR transformants was sequenced to retain strain R1407 (Δ_cibC) and R1408 (Δ_cibAB_).
Results
Competence Development Is Responsible for the Induction of β-Hemolysis in the RT Mutant. Unlike its isogenic wild-type parent (R800, an R6 derivative), the RT mutant (R841) displayed ≈25 various-sized circular zones composed of β-hemolytic colonies per 104 cells plated (Fig. 1 A). Hereafter, we refer to them as “hemolytic halos” (HHs). Because the RT mutant displayed reduced spontaneous competence (hence the name RT, for reduced transformability; S.G., V. Hénard, B.M., and J.-P.C., unpublished data), we hypothesized that the formation of HH resulted from rare competence induction events. Inactivation of any of three genes essential for spontaneous competence development, comC, comA, and comE, abolished the spontaneous formation of HH in the RT mutant (data not shown), providing support to our hypothesis. Application of 1-μl spots containing >5 ng of CSP to the surface of plates was then shown to induce large HH on RT mutant cells (Fig. 1_B_), but to have no effect on RT comE mutant cells (data not shown). This finding suggested that CSP diffusion in agar contributed to the spreading of competence on plates and to the concomitant (or subsequent) appearance of HH. Altogether, these data established that competence induction and the formation of HH are causally related in the RT mutant.
Interestingly, 1-μl spots of CSP induced larger β-hemolytic zones on RT cells than on RT _comA_- cells (≈1.7-fold difference in diameter; data not shown). In the latter case, it is likely that the β-hemolytic zone matched the area of diffusion of synthetic CSP in agar because _comA_- cells are unable to export CSP. In contrast, comA+ cells have the ability to respond to externally added CSP by producing more CSP. Endogenous CSP production can therefore relay competence initiated by the addition of synthetic CSP, which would readily account for the formation of larger HH. This interpretation was confirmed by the observation that 1-μl CSP spots induced β-hemolytic zones similar in diameter to those induced on RT _comA_- cells (data not shown) on RT _comC_- cells, which do not produce CSP.
The observation that 1-μl spots containing 20-100 ng of CSP induced halos with similar diameters (Fig. 1_B_) suggested that propagation of competence through endogenous CSP production stops at about the same time throughout a plate. An experiment in which CSP was spotted at timed intervals revealed that the diameter of CSP-induced HH decreased regularly with time of CSP addition, from ≈25 mm (time 0) to ≈5 mm (6 h) (see Supporting Text and Fig. 5, which is published as supporting information on the PNAS web site). After 7-h incubation, deposition of CSP could no longer induce HH. In view of these data, we propose that the variation in size of spontaneous HH results from stochastic competence induction, followed by propagation of a wave of competence. These events can take place only during the first ≈7 h of incubation.
The Formation of HH Requires an Intact ply Gene. Our working hypothesis that Ply was responsible for competence-induced β-hemolysis was verified by showing that both spontaneous and CSP-induced formation of HH required an intact ply gene (Fig. 1_C_). These observations were confirmed by using an RT strain harboring a different _ply_- mutation (data not shown). To further demonstrate that β-hemolysis depended on the release of Ply into the medium, we used antibodies to Ply. Complete inhibition of HH by monoclonal antibodies demonstrated the direct involvement of released Ply in β-hemolysis (Fig. 1_C_). We next investigated the genetic requirements for the release of Ply.
Three Cell-Wall Hydrolases Are Involved in CSP-Induced Ply Release. Because cell lysis is the usual mechanism for Ply release, we investigated whether inactivation of the major autolysin, LytA, and the autolytic lysozyme, LytC (29), would affect the formation of HH. Inactivation of lytA or lytC had no detectable effect (Table 1, compare types 2 and 3 to 1), but the simultaneous inactivation of both lytA and lytC resulted in a clear reduction of HH (Table 1, compare type 7 to 1; see Fig. 2_A_ Left). [Note that a set of isogenic strains combining different insertions in lytA (and in cbpD or cibAB; see below) was used throughout this study (see Table 3).]
Table 1. Genetic requirements for competence-induced Ply release.
| Genotype* | |||||||
|---|---|---|---|---|---|---|---|
| Experiment type no. | lytA† | lytC‡ | cbpD§ | cibA¶ | cibB¶ | cibC∥ | Hemolysis intensity |
| 1 | + | + | + | +++** | |||
| 2 | − | + | + | +++ | |||
| 3 | + | − | + | +++ | |||
| 4 | + | + | − | +++ | |||
| 5 | + | − | − | +++ | |||
| 6 | − | + | − | +++ | |||
| 7 | − | − | + | +†† | |||
| 8 | − | − | − | 0 | |||
| 9 | + | + | + | +++** | |||
| 10 | − | + | + | + | |||
| 11 | + | − | + | + | |||
| 12 | − | − | + | + | |||
| 13 | − | − | − | + | |||
| 14 | + | + | − | ++++ |
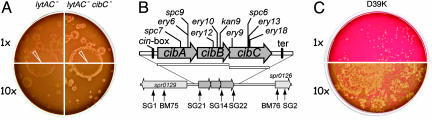
Fig. 2.
Genetic requirements for the release of Ply. (A) The absence of both LytA and LytC in strain R1332 (_lytAC_-) reduced HH formation (Left), whereas inactivation of cibC in its derivative R1414 (_lytAC_- _cibC_-) exacerbated it (Right). The 1× concentration corresponds to 2.5 × 105 cells per plate. CSP (1 μl, 100 ng) was deposited immediately after plating (indicated by white tips). (B) Organization of the bacteriocin system operon, cibABC. Open bars indicate the limits of cibAB and cibC precise deletions. mariner minitransposon insertions are also indicated (only cotranscribed orientations of inserted minitransposons with respect to the targeted ORF were retained to minimize polar effects). Insertions occurred, respectively, after positions 108, 142, 142, 262, 280, 296, 347, 424, 427, and 430 with respect to the _A_TG of cibA. Arrows indicate primers used for mariner mutagenesis and for transcriptional studies. (C) Strain D39K, the parent of the laboratory strain R800, also forms HH. The 1× concentration corresponds to 8 × 102 cells per plate.
Residual HH were nevertheless detected in strains lacking LytA and LytC, suggesting the possible involvement of a third hydrolytic activity. This led us to consider the implication of CbpD, a protein containing the CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) amidase domain (30). Inactivation of cbpD alone or in combination with a lytA or a lytC mutation had no detectable effect on HH formation (Table 1, see types 4-6). However, HH were completely abolished in cbpD lytA lytC triple mutants (Table 1, see type 8). To rule out the possible occurrence and contribution of unselected mutations, individual reversions to wild type of each mutation in a triple mutant were generated by transformation (see Supporting Text). Every single reversion restored the phenotype expected from the corresponding double mutant (data not shown).
These data strongly suggest that CbpD is one of three cell-wall hydrolases involved in CSP-induced release of Ply and makes it unlikely that a fourth cell-wall hydrolase is required. The specific expression of cbpD at competence (20, 21) fits such a role. Interestingly, although LytA or LytC alone (i.e., in the absence of the other two) could promote formation of normal HH (Table 1, see types 5 and 6, respectively), CbpD seemed less efficient (Table 1, see type 7), suggesting a reduced hydrolytic activity of the latter protein compared to that of LytA or LytC.
CibAB, a Competence-Induced Two-Peptide Bacteriocin, Triggers Ply Release. At this stage, two of the genes (cbpD and lytA) implicated in the process of Ply release were known to be part of the competence regulon. However, their simultaneous inactivation in the RT background did not reduce competence-induced HH (Table 1, see type 6), suggesting that another competence-induced agent(s) was responsible for triggering cell lysis. We focused our attention on two small ORFs, previously identified among the late com genes as _orf62_-orf51 (20) with the potential to encode bacteriocins with GG leaders (2, 21). We renamed them CibAB (for competence induced bacteriocin). The mature peptides would be 36 and 28 residues long, respectively. A precise deletion of cibAB as well as different minitransposon insertions in cibA or cibB (Fig. 2_B_) reduced HH to an extent similar to that observed with a lytA lytC double mutant (Table 1, compare types 10-12 to 9). These data provided evidence for the involvement of CibAB in competence-induced HH. Further evidence for their involvement was obtained in mixed culture experiments (see below). The finding that inactivation of cibA, cibB, or cibAB conferred the same phenotype suggests that CibA and CibB act together and thus belong to the family of two-peptide bacteriocins (ref. 31, reviewed in ref. 32).
Immunity to CibAB. In search of a possible immunity factor for CibAB, we examined the cibAB region and detected the presence of a third ORF (Fig. 2_B_), potentially encoding a 65 residue-long peptide, CibC, with two predicted transmembrane segments. Evidence was obtained for the existence of a cibABC competence-specific transcript. In good agreement with the predicted distance between the σX competence promoter (_cin_-box; ref. 33) and the putative transcription terminator (ter) (Fig. 2B), a ≈600-nt-long transcript was detected in CSP-induced cells, but not in NC cells, using probes specific for cibC (SG14-SG22 PCR fragment) and cibABC (SG21-SG22 PCR fragment). As predicted, the size of the competence-specific transcript was increased by ≈1,100 nt in a strain harboring the cibA::_spc7_C minitransposon insertion (data not shown). These data prompted us to generate a precise deletion of as well as insertions in cibC (Fig. 2_B_). Inactivation of cibC was found to reinforce hemolysis, including in a _lytA_-lytC double mutant (Fig. 2 A), but only in the presence of CibAB (Table 1, compare types 14 to 9 and 13 to 10-12). These observations are consistent with CibC being the immunity factor to CibAB in competent cells. The presence of CibC readily accounts for the finding that wild-type cells display no HH despite full competence on blood agar plates.
Competent Cells Promote Lysis of Noncompetent Cells. If CibC confers immunity to CibAB, it ensues that NC cells are the target for CibAB. The observation that HH are sometimes more intense at the border of CSP diffusion zones (e.g., Fig. 2 A) further suggests that Ply release could be prompted when competent cells encounter NC cells. To check whether competent cells trigger the lysis of NC cells, a mixture of RT _ply_- cells and of comE mutant cells (i.e., cells that produce Ply but cannot become competent), was plated in horse blood CAT-agar; 1-μl spots of CSP were deposited on the surface. The detection of a ring of β-hemolysis around each spot of CSP (Fig. 3_A_ and Table 2, see type 1) provided direct evidence for the release of Ply from (and therefore the lysis of) _comE_- cells triggered by the RT _ply_- competent cells. We refer to this phenomenon as allolysis.
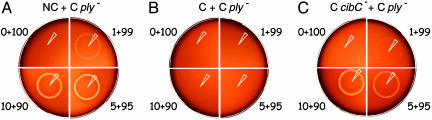
Fig. 3.
Competent cells cause noncompetent cells to release Ply. Mixtures (2 × 107 total cells) of R1130 (indicated as C _ply_-), a _ply_- derivative of the RT strain R1065, and 1, 5, or 10% (indicated as 1 + 99, 5 + 95, and 10 + 90, respectively) of either the competence-deficient and ply+ strain R1240 (NC) (A), the RT and ply+ strain R1065 (C) (B), or the RT and ply+, but cibC mutant strain R1135 (C _cibC_-) cells (C) were plated on 5.5% horse blood CAT-agar. CSP (1 μl, 100 ng) was deposited immediately after plating (white tips).
Table 2. Genetic requirements for allolysis.
| Experiment type no. | Genotype* | Allolysis intensity | ||||
|---|---|---|---|---|---|---|
| lytA† | lytC† | cbpD† | cibA‡ | cibB‡ | ||
| 1 | + | + | + | + | + | +++§ |
| 2 | + | + | + | −¶ | + | 0 |
| 3 | + | + | + | + | −¶ | 0 |
| 4 | + | + | + | −¶ | −¶ | 0 |
| 5 | − | + | −¶ | + | + | + |
| 6 | − | − | + | + | + | 0 |
The ring-shaped β-hemolytic halos could be accounted for if the number of _comE_- cells (0.2-2 × 105 ml-1) was too low when CSP was spotted to result in detectable β-hemolysis at the initial spot. Because competence can propagate on plate for ≈7 h (Fig. 5), division of _comE_- cells can occur and allow detection of β-hemolysis at the periphery of the zone of competence propagation, which corresponds to the highest cell-density.
Allolysis Can Use Cell-Wall Hydrolases from the Targeted Cells. In examining the genetic requirements for allolysis, we first observed that _cibA_-, _cibB_-, or _cibAB_- derivatives of the RT _ply_- strain were unable to induce the release of Ply from _comE_- cells (Table 2, see types 2-4), consistent with an essential role of CibAB in allolysis. Simultaneous inactivation of LytA (in both strains; see below) and of CbpD resulted in a clear reduction of allolysis, indicating that LytA and CbpD were also involved in the process (Table 2, see type 5). In contrast, a _lytA_- _lytC_- (RT _ply_-) strain could promote full allolysis (data not shown). This observation was puzzling because inactivation of LytA and LytC clearly reduced HH formation in single culture platings (Table 1, see type 7). To account for this finding, we hypothesized that the two cell-wall hydrolases from the targeted cells could participate to allolysis. In full agreement with this hypothesis, allolysis was completely abolished when both the RT _ply_- and the targeted ply+ _comE_- strains lacked LytA and LytC (Table 2, see type 6; note that inactivation of lytA and lytC only in the ply+ comE mutant had no effect). These data demonstrated that LytA and LytC are required for allolysis, but can be provided by either the competent or targeted cells. We conclude from these observations that competent cells can activate autolytic enzymes from the targeted cells by a mechanism requiring the presence of CibAB.
Allolysis Versus Autolysis. The finding that allolysis takes place does not per se rule out the possibility that autolysis of competent cells occurs simultaneously. To evaluate the possible contribution of autolysis to Ply release, _comE_- (ply+) cells were replaced by the same number of RT (com+ ply+) cells in mixed culture platings. CSP-induced HH became almost undetectable (Fig. 3_B_). This experiment clearly showed that, in contrast to competence-deficient cells, competent cells are protected from lysis.
HH was readily detected only when RT _cibC_- (com+ ply+) cells were substituted for RT (com+ ply+) cells (Fig. 3_C_), further demonstrating that CibC confers immunity to CibAB in competent cells. Nevertheless, this experiment did not establish the occurrence of autolysis because Ply release could result either from autolysis of competent _cibC_- cells or from allolysis triggered by the RT _ply_- cells. Therefore, we regard residual HH detected in pure culture platings in the absence of CibAB (Table 1, see types 10-13) as the only evidence for autolysis, because allolysis was not detected when CibAB were missing (Table 2, see types 2-4).
Allolysis, a General Property of S. pneumoniae. Wild-type (R800) cells (15-μl spot containing ≈3.75 × 106 cells) could substitute for CSP in inducing HH on RT mutant plates (data not shown). This finding indicated that R800 cells become fully competent and produce CSP on horse blood CAT agar. However, R800 cells did not form spontaneous HH (Fig. 1 A), and spots of CSP did not induce β-hemolysis on them (data not shown). Therefore, we hypothesized that the formation of HH reflects population heterogeneity regarding competence induction and predicted that other strains displaying reduced spontaneous competence (compared to our wild-type laboratory strain, R800) should also form HH. This prediction was verified with several strains carrying mutations or constructs that reduced spontaneous competence induction (unpublished observations). Interestingly, the clinical isolate from which R800 was derived, D39, was found to behave similarly to the RT mutant in displaying spontaneous and CSP-induced HH (Fig. 2_C_). Therefore, we propose that allolysis is a general property of the species.
We attribute the failure to detect Ply release in the R800 laboratory strain to selection for better synchronization of competence during its domestication. As a consequence, R800 cultures do not consist of the mixture of competent and NC cells favoring the detection of allolysis. The observation that strain D39 is less competent than R800 (unpublished observations) is fully consistent with this interpretation.
Discussion
Recent experiments aimed at elucidating the question of the origin of pneumococcal transforming DNA in nature suggested that part of the com regulon could be involved in cell lysis. Although a coincidence of peaks of chromosomal DNA release and competence was reported more than four decades ago (34, 35), evidence that DNA release and competence were causally related was obtained only recently (36, 37). The concomitant release of DNA and of a cytoplasmic β-galactosidase suggested the occurrence of cell lysis (36). Consistent with this interpretation, we showed that the simultaneous inactivation of the major autolysin, LytA, and the autolytic lysozyme, LytC, almost completely abolished DNA release (37). However, these data did not discern between autolysis of competent cells and lysis of NC cells promoted by competent cells. The latter was demonstrated to occur during cocultivation of a competence-deficient strain with its competence-proficient parent (38). The genetic requirements for this phenomenon, termed heterolysis, were not established.
By monitoring on blood agar plates the release of Ply accompanying the propagation of competence, we obtained independent evidence that competent cells of S. pneumoniae display the ability to trigger the lysis of NC cells. We name this phenomenon allolysis to reserve heterolysis to the lysis of other species. The observation that the clinical isolate D39 release Ply on blood agar plates suggests that allolysis is a general property of S. pneumoniae. It involves a bacteriocin system (cibABC) and three cell-wall hydrolases (cbpD, lytA, and lytC). The genes, except lytC, belong to the late com class. Because LytA and LytC are required for the competence-dependent release of Ply on plates as well as of DNA in planktonic cultures (37), it is likely that these phenomena represent two facets of the same process.
Model for Allolysis. The proposed two-peptide bacteriocin CibAB is absolutely required for allolysis (Table 2, see type 6). However, the finding that Ply release was abolished when the three cell-wall hydrolases were missing (Table 1, see type 8) clearly indicates that CibAB cannot promote cell lysis on their own. This situation is not unprecedented. The bacteriocin produced by Lactococcus lactis IFPL105, which is bactericidal against several Lactococcus and Lactobacillus strains, was shown to require the activity of the major autolysin AcmA (39). Addition of the bacteriocin to logarithmic-phase cultures caused effective lysis of acmA+ but not of _acmA_- cells (39).
Therefore, we propose that CibAB acts as a trigger factor for allolysis (Fig. 4). It could do so by inserting into the membrane of sensitive cells and depleting cellular energy (40). Cell lysis caused by LytA, LytC, and CbpD would then occur as a secondary effect of CibAB action. Interestingly, examination of the cibABC region in five sequenced S. pneumoniae genomes [representing types 2, 4, 19F, 23F (www.sanger.ac.uk/Projects/S_pneumoniae, and 6B (www.tigr.org/tdb/mdb/mdbinprogress.html)] revealed up to two conservative changes in the predicted protein sequences (unpublished observations), strongly suggesting that this bacteriocin system is conserved in the species.
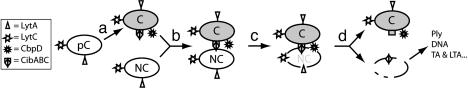
Fig. 4.
Proposed model for predation of noncompetent cells through allolysis in S. pneumoniae. (a) Precompetent (pC) cells differ from competent (C) cells by the presence of the CbpD amidase, the two-peptide bacteriocin, CibAB, and its immunity factor, CibC. (b) We propose that cell-to-cell contacts between C and NC cells allow CibAB to interact with the latter, whereas competent cells are protected by CibC. (c) CibAB then triggers the action of CbpD, as well as LytA and LytC (two hydrolases present at the surface of both NC and C cells) on the cell wall of NC cells. (d) Cell wall disruption results in release of cell components, including Ply, TA, LTA, and chromosomal DNA.
In an attempt to get an insight into how CibABs work, we harvested competent cultures and used supernatants to induce HH formation on ply+ _comE_- cells, but were unable to detect any effect (data not shown). Mature CibA and CibB synthetic peptides (used either singly or as mixtures with varied ratios) also failed to induce HH (data not shown). Thus, we believe that cell-to-cell contacts are required for CibAB to interact with the NC cells (Fig. 4). The transient agglutination properties of competent cells, a long-standing observation (41), could play an essential role in the process. Further work is needed to establish whether agglutination and allolysis are intimately connected.
CibAB is the only competence-induced component crucial for allolysis, because LytA can be provided by the targeted cells. The latter finding indicates that competence-specific induction of lytA is not critical, which is not really surprising because lytA is known to be expressed from at least two competence-independent promoters, in addition to the σX promoter (19). CbpD, the proposed competence-induced new amidase, is also not essential for Ply release on plates. Our previous observation that inactivation of both LytA and LytC abolished competence-induced DNA release in liquid cultures (37) already suggested that CbpD did not play a major role in the process, at least as an amidase.
Biological Significance of Allolysis. It was proposed that competence-triggered DNA release has been evolved to ensure coordination in time and space between release and uptake by competent cells, thus favoring genetic exchange (36, 38). However, the finding that DNA release continued for several hours after competence had disappeared (37) suggests that genetic exchange could not be the only purpose of allolysis. An obvious additional role is sheer competition among strains. Allolysis can also constitute an example of programmed cell death (42). Competent cells could benefit from the nutrients thereby released, thus mimicking the cannibalism of sporulating cells of Bacillus subtilis (43). In both cases, differentiating (sporulating or competent) cells synthesize a factor that kills genetically identical but nondifferentiated (i.e., nonsporulating or noncompetent) cells in the culture rather than other species.
Competence-induced cell lysis can also be potentially important for the release of Ply and of any other virulence factors requiring cell disruption for release, such as teichoic and lipoteichoic acid and chromosomal DNA. The former are well known inflammatory compounds (44), whereas bacterial DNA can cause septic shock (45), elicit inflammatory cytokine production, and stimulate lymphocyte proliferation (46, 47). Therefore, our data provide a plausible mechanism for the connection between competence and virulence in S. pneumoniae (16). They also refine our view of the regulation of autolysis during infection, until now considered unclear (16). An additional, more speculative but intriguing possibility is that pneumococcal proteins that are surface associated but have no obvious mechanism of secretion, such as glyceradehyde-3-phosphate dehydrogenase (48) and enolase (49), do not come from inside the cells harboring them but are “scavenged” from other “apoptosed” cells.
Allolysis could also reinforce the virulence of S. pneumoniae in the course of mixed infections in humans, by provoking the simultaneous release of complementary virulence factors carried by different pneumococcal strains. In fact, a ply mutant had already been reported to exhibit wild-type growth kinetics only in mice coinfected with its wild-type parent, which implied complementation of ply_- cells by extracellular Ply (50), although it was not established whether Ply release by wild-type cells depended on competence. The existence of different competence pherotypes in S. pneumoniae (reviewed in ref. 2) could favor such “_in vivo complementation,” because cells belonging to the most abundant pherotype would enter competence first (through specific CSP-mediated quorum-sensing) and provoke lysis of other pherotypes. Competence has long been considered only as favoring long-term genetic plasticity by transformation-mediated exchanges (3), taking advantage of the high degree of genomic variation within the species (51). We propose that competence can also provide nonheritable variability by allowing competent cells to benefit transiently from a pool of virulence factors through predation of NC cells during mixed infections.
The phenomenon of competence-programmed lysis of NC cells may not be restricted to S. pneumoniae. Production of a bacteriocin (STH1) active against NC cells concomitant with competence development was documented >30 years ago in Streptococcus sanguis (52). It was recently reported that STH1 production and competence development were causally related.§ It is therefore tempting to speculate that this represented another example of allolysis and that a predation strategy is conserved among transformable streptococci.
Supplementary Material
Supporting Information
Acknowledgments
We thank Agamemnon Carpousis and Dave Lane for critical reading of the manuscript; Sandy Gruss, Leiv Håvarstein, Tarek Msadek, and an anonymous reviewer for suggestions that improved the manuscript; Pedro García (Centro de Investigaciones Biológicas Ramiro de Maeztu 9, Madrid) for kindly providing us with plasmids pLCC14 and pGL80; David Villa for continuing help with photographs throughout this work; and Chantal Granadel for expert technical assistance. This research was financed in part by European Union Grant QLK2-CT-2000-00543. S.G. was the recipient of an EMBO Short Term Fellow-ship (2001; ASTF 9988).
Author contributions: B.M. and J.-P.C. designed research; S.G. performed research; T.J.M. contributed new reagents/analytic tools; S.G., B.M., and J.-P.C. analyzed data; and J.-P.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CSP, competence-stimulating peptide; ABC, ATP-binding cassette; NC, noncompetent; HH, hemolytic halo.
See Commentary on page 8401.
Footnotes
§
Heng, N. C. & Tagg, J. R. (2002) in Sixth ASM Conference on Streptococcal Genetics, Asheville, April 14-17, 2002 (Am. Soc. Microbiol. Press, Washington, DC), pp. 68-69 (abstr.).
References
- 1.Jedrzejas, M. J. (2001) Microbiol. Mol. Biol. Rev. 65 187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claverys, J. P. & Håvarstein, L. S. (2002) Frontiers Biosci. 7 1798-1814. [DOI] [PubMed] [Google Scholar]
- 3.Claverys, J. P., Prudhomme, M., Mortier-Barrière, I. & Martin, B. (2000) Mol. Microbiol. 35 251-259. [DOI] [PubMed] [Google Scholar]
- 4.Tomasz, A. (1965) Nature 208 155-159. [DOI] [PubMed] [Google Scholar]
- 5.Håvarstein, L. S., Coomaraswamy, G. & Morrison, D. A. (1995) Proc. Natl. Acad. Sci. USA 92 11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova, E. V., Håvarstein, L. S. & Morrison, D. A. (1996) Mol. Microbiol. 21 853-864. [DOI] [PubMed] [Google Scholar]
- 7.Håvarstein, L. S., Diep, D. B. & Nes, I. F. (1995) Mol. Microbiol. 16 229-240. [DOI] [PubMed] [Google Scholar]
- 8.Hui, F. M., Zhou, L. & Morrison, D. A. (1995) Gene 153 25-31. [DOI] [PubMed] [Google Scholar]
- 9.Alloing, G., Martin, B., Granadel, C. & Claverys, J. P. (1998) Mol. Microbiol. 29 75-84. [DOI] [PubMed] [Google Scholar]
- 10.Peterson, S., Sung, C. K., Cline, R., Desai, B. V., Snesrud, E., Luo, P., Walling, J., Li, H., Mintz, M., Tsegaye, G., et al. (2004) Mol. Microbiol. 51 1051-1070. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M. S. & Morrison, D. A. (1999) J. Bacteriol. 181 5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, P. & Morrison, D. A. (2003) J. Bacteriol. 185 349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagkessamanskaia, A., Moscoso, M., Hénard, V., Guiral, S., Overweg, K., Reuter, M., Martin, B., Wells, J. & Claverys, J. P. (2004) Mol. Microbiol. 51 1071-1086. [DOI] [PubMed] [Google Scholar]
- 14.Bartilson, M., Marra, A., Christine, J., Asundi, J. S., Schneider, W. P. & Hromockyj, A. E. (2001) Mol. Microbiol. 39 126-135. [DOI] [PubMed] [Google Scholar]
- 15.Lau, G. W., Haataja, S., Lonetto, M., Kensit, S. E., Marra, A., Bryant, A. P., McDevitt, D., Morrison, D. A. & Holden, D. W. (2001) Mol. Microbiol. 40 555-571. [DOI] [PubMed] [Google Scholar]
- 16.Hava, D. L., LeMieux, J. & Camilli, A. (2003) Mol. Microbiol. 50 1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García, P., García, J. L., García, E. & López, R. (1986) Gene 43 265-272. [DOI] [PubMed] [Google Scholar]
- 18.Díaz, E. & García, J. L. (1990) Gene 90 157-162. [DOI] [PubMed] [Google Scholar]
- 19.Mortier-Barrière, I., de Saizieu, A., Claverys, J. P. & Martin, B. (1998) Mol. Microbiol. 27 159-170. [DOI] [PubMed] [Google Scholar]
- 20.Rimini, R., Jansson, B., Feger, G., Roberts, T. C., de Francesco, M., Gozzi, A., Faggioni, F., Domenici, E., Wallace, D. M., Frandsen, N., et al. (2000) Mol. Microbiol. 36 1279-1292. [DOI] [PubMed] [Google Scholar]
- 21.Peterson, S., Cline, R. T., Tettelin, H., Sharov, V. & Morrison, D. A. (2000) J. Bacteriol. 182 6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hava, D. L. & Camilli, A. (2002) Mol. Microbiol. 45 1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 23.Gosink, K. K., Rodgers, M. E., Guglielmo, C., Tuomanen, E. I. & Masure, H. R. (2000) Infect. Immun. 68 5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, B., Prudhomme, M., Alloing, G., Granadel, C. & Claverys, J. P. (2000) Mol. Microbiol. 38 867-878. [DOI] [PubMed] [Google Scholar]
- 25.Berry, A. M., Yother, J., Briles, D. E., Hansman, D. & Paton, J. C. (1989) Infect. Immun. 57 2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, J. A., Allen, R. L., Falmagne, P., Johnson, M. K. & Boulnois, G. J. (1987) Infect. Immun. 55 1184-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter, R. D. & Guild, W. R. (1976) J. Virol. 19 659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung, C. K., Li, H., Claverys, J. P. & Morrison, D. A. (2001) Appl. Environ. Microbiol. 67 5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García, P., González, M. P., García, E., García, J. L. & López, R. (1999) Mol. Microbiol. 33 128-138. [DOI] [PubMed] [Google Scholar]
- 30.Rigden, D. J., Jedrzejas, M. J. & Galperin, M. Y. (2003) Trends Biochem. Sci. 28 230-234. [DOI] [PubMed] [Google Scholar]
- 31.Nissen-Meyer, J., Holo, H., Håvarstein, L. S., Sletten, K. & Nes, I. F. (1992) J. Bacteriol. 174 5686-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eijsink, V. G., Axelsson, L., Diep, D. B., Havarstein, L. S., Holo, H. & Nes, I. F. (2002) Antonie Leeuwenhoek 81 639-654. [DOI] [PubMed] [Google Scholar]
- 33.Campbell, E. A., Choi, S. Y. & Masure, H. R. (1998) Mol. Microbiol. 27 929-939. [DOI] [PubMed] [Google Scholar]
- 34.Ottolenghi, E. & Hotchkiss, R. D. (1960) Science 132 1257-1258. [PubMed] [Google Scholar]
- 35.Ottolenghi, E. & Hotchkiss, R. D. (1962) J. Exp. Med. 116 491-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinmoen, H., Knutsen, E. & Håvarstein, L. S. (2002) Proc. Natl. Acad. Sci. USA 99 7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscoso, M. & Claverys, J. P. (2004) Mol. Microbiol. 54 783-794. [DOI] [PubMed] [Google Scholar]
- 38.Steinmoen, H., Teigen, A. & Håvarstein, L. S. (2003) J. Bacteriol. 185 7176-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Cuesta, M. C., Kok, J., Herranz, E., Peláez, C., Requena, T. & Buist, G. (2000) Appl. Environ. Microbiol. 66 3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Belkum, M. J., Kok, J., Venema, G., Holo, H., Nes, I. F., Konings, W. N. & Abee, T. (1991) J. Bacteriol. 173 7934-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomasz, A. & Zanati, E. (1971) J. Bacteriol. 105 1213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engelberg-Kulka, H. & Hazan, R. (2003) Science 301 467-468. [DOI] [PubMed] [Google Scholar]
- 43.González-Pastor, J. E., Hobbs, E. C. & Losick, R. (2003) Science 301 510-513. [DOI] [PubMed] [Google Scholar]
- 44.Nau, R. & Eiffert, H. (2002) Clin. Microbiol. Rev. 15 95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparwasser, T., Miethke, T., Lipford, G. B., Borschert, K., Häcker, H., Heeg, K. & Wagner, H. (1997) Nature 386 336-337. [DOI] [PubMed] [Google Scholar]
- 46.Krieg, A. M., Yi, A. K., Matson, S., Waldschmidt, T. J., Bishop, G. A., Teasdale, R., Koretzky, G. A. & Klinman, D. M. (1995) Nature 374 546-549. [DOI] [PubMed] [Google Scholar]
- 47.Chatellier, S. & Kotb, M. (2000) Eur. J. Immunol. 30 993-1001. [DOI] [PubMed] [Google Scholar]
- 48.Bergmann, S., Rohde, M. & Hammerschmidt, S. (2004) Infect. Immun. 72 2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergmann, S., Rohde, M., Chhatwal, G. S. & Hammerschmidt, S. (2001) Mol. Microbiol. 40 1273-1287. [DOI] [PubMed] [Google Scholar]
- 50.Benton, K. A., Everson, M. P. & Briles, D. E. (1995) Infect. Immun. 63 448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakenbeck, R., Balmelle, N., Weber, B., Gardes, C., Keck, W. & de Saizieu, A. (2001) Infect. Immun. 69 2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel, R. & Slade, H. D. (1973) J. Bacteriol. 115 655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de los Toyos, J. R., Mendez, F. J., Aparicio, J. F., Vazquez, F., Mar Garcia, S. M., Fleites, A., Hardisson, C., Morgan, P. J., Andrew, P. W. & Mitchell, T. J. (1996) Infect. Immun. 64 480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information