Evolutionary Relationships of Three New Species of Enterobacteriaceae Living as Symbionts of Aphids and Other Insects (original) (raw)
Abstract
Ecological studies on three bacterial lineages symbiotic in aphids have shown that they impose a variety of effects on their hosts, including resistance to parasitoids and tolerance to heat stress. Phylogenetic analyses of partial sequences of gyrB and recA are consistent with previous analyses limited to 16S rRNA gene sequences and yield improved confidence of the evolutionary relationships of these symbionts. All three symbionts are in the Enterobacteriaceae. One of the symbionts, here given the provisional designation “Candidatus Serratia symbiotica,” is a Serratia species that has acquired a symbiotic lifestyle. The other two symbionts, here designated “Candidatus Hamiltonella defensa” and “Candidatus Regiella insecticola,” are sister groups to one another and together show a relationship to species of Photorhabdus.
Recent genomic and phylogenetic studies suggest that associations with arthropods have played a role in the evolution of the Enterobacteriaceae (see, e.g., Duchaud et al. [11]). Aphids, psyllids, scale insects, whiteflies, weevils, and other insects all harbor various maternally transmitted Enterobacteriaceae, distributed sporadically or ubiquitously within a particular host species (5, 20, 28, 29, 36, 37, 39). To date, very few of these symbionts have been successfully cultured and characterized outside of hosts, with the exceptions of Arsenophonus nasoniae, the facultative symbiont of a parasitoid wasp (15), and Sodalis glossinidius, the secondary symbiont of tsetse, a blood-feeding fly that vectors trypanosomes (6). (“Secondary” symbionts are so named because they coexist with a “primary” symbiont that appears to represent a more ancient infection of the same host lineage.) These symbiotic lineages represent a number of independent infections of arthropod hosts, with each cluster occurring in several diverse insect groups. For example, phylogenies derived from 16S rRNA gene sequences indicate that the closest known relatives of Sodalis are the primary symbionts of grain weevils in the genus Sitophilus, followed by facultative symbionts of sap-feeding insects (28). Likewise, the cluster corresponding to the genus Arsenophonus has representatives in parasitoid hymenopterans (A. nasoniae) and also in a wide variety of hemipterans such as assassin bugs, aphids, psyllids, and whiteflies (15, 17, 28, 31, 34, 37, 39, 46).
Aphids provide the best-studied instance in which Enterobacteriaceae form heritable symbioses that are facultative for the host insects. Almost all aphids contain a primary symbiont, Buchnera aphidicola, and many also contain secondary symbionts from one of three groups of Enterobacteriaceae, with members of each group showing near-identity of 16S rRNA gene sequences (29). All three types of symbiont are known from the pea aphid, Acyrthosiphon pisum, as well as from a wide variety of different species and subfamilies of aphids (16, 28, 29, 40), with a few records from other insect hosts. Although the mechanisms of transfer in nature are not known, the symbionts can be cured or transferred among hosts in the lab, allowing experimental studies of their effects on hosts (4, 9, 19, 26, 41). These studies have demonstrated major effects of these bacteria on aphid biology, including resistance to parasitoid wasps (26), tolerance to heat stress (3, 23), restoration of reproduction in aphids experimentally deprived of B. aphidicola (19), and changes in host plant range (41).
Currently, most of these symbionts are characterized only through 16S rRNA gene sequences. Use of additional genes can improve phylogenetic resolution within the Enterobacteriaceae (10, 32, 45). Furthermore, the nomenclature of these bacteria has been confusing, in part because early studies did not recognize them as distinct (2) and the initial sequence-based identification included only one of the three groups (43). Following discovery of the additional types (29), different informal labels were applied within different studies, referring either to isolates from a particular host species or to the whole clade (see, e.g., references 29 and 40).
Here we clarify the phylogenetic relationships of these three groups of bacteria using information from 16S rRNA genes and from partial sequences of recA and gyrB, the latter being a universally distributed gene for which many sequences from Enterobacteriaceae are available in databases (see, e.g., references 10, 14, and 45). We propose candidate names for these three symbionts and summarize the correspondence of published working names and accession numbers to the proposed names.
MATERIALS AND METHODS
Insect material.
New sequences for gyrB and recA were obtained from representative strains of the three symbiont types maintained continuously in stock A. pisum cultures in the Moran lab in Tucson at 20°C with a photoperiod of 16 h of light and 8 h of darkness (16L:8D) for periods ranging from 24 to 60 months. These lines are routinely screened for contamination or loss/gain of symbionts by using genomic markers for the aphids and diagnostic screens for the symbiont types (29). Each aphid line in culture contains zero or one secondary symbiont infection; these have never lost or gained a symbiont during at least 40 to 100 generations in culture. Some lines containing no secondary symbionts have been infected with symbionts through microinjection (as described in reference 26); once established for a generation, none of these artificial infections have been lost, as indicated by diagnostic sequencing of 16S rRNA genes. We used these stocks to provide template DNA for reactions amplifying 16S-23S rRNA genes, partial gyrB sequences, and partial recA sequences.
16S-23S rRNA genes.
For one representative clone containing each symbiont type, universal PCR primers, described by Thao and Baumann (36), were used to amplify a product of approximately 4,500 nucleotides that included most of the 16S rRNA and the 23S rRNA gene plus the intervening spacer region. Because B. aphidicola (the primary symbiont) does not have linked 16S and 23S rRNA genes, it yields no product with these primers. Isolates sequenced were “R type” from A. pisum clone 19F, “T type” from A. pisum clone 6-1, and “U type” from A. pisum clone 2a; each of these A. pisum clones was descended from a single founding female collected in Madison, Wis., in 1999.
gyrB.
Primers used initially were gyr-320 and rgyr-1260 from the work of Dauga (10), designed to amplify gyrB in Enterobacteriaceae. When checked against the B. aphidicola gene (30), these primers showed mismatches at the 3′ ends, excluding amplification of B. aphidicola gyrB. Reaction conditions were the same as those in the work of Dauga (10), with the following temperature cycle: 94°C (3 min); 35 cycles of 94°C (1 min), 55°C (1 min), and 72°C (2 min); then 72°C for 10 min. The U-type symbiont did not amplify with gyr-320 and rgyr-1260, and we used primers UP1 and 2Tr-SR1, described by Yamamoto and Harayama (45), with the same reaction conditions except that the annealing temperature was 52°C. The sequenced region was 920 bp for R- and T-type symbionts and 1,904 bp for U-type symbionts. Isolates sequenced were R-type 2BB, T-type 6-1, and U-type 2a, all from A. pisum hosts established from collections in Madison, Wis., in 1999.
recA.
Approximately 560 nucleotides of the coding region of recA were amplified using degenerate primers recAF and recAr; primers and reaction conditions were those described by Dale et al. (7). Amplification of B. aphidicola DNA was excluded because it lacks recA. Isolates sequenced were R-type Tucson (originating from a single aphid female collected in Tucson, Ariz., in 1999), T-type 4A, and U-type 2a (each of the latter from single females collected in Madison, Wis., in 1999).
Cloning and sequencing.
Amplified sequences were checked for length on agarose gels, purified from the gels, and then cloned using a TopoTA kit with the pCR 2.1 vector (Invitrogen). After verification of insert sizes, plasmids were sequenced at the University of Arizona Genomic Analysis and Technology Center. Sequencing primers for gyrB are listed in the work of Dauga (10) and Yamamoto and Harayama (45). Additionally, new sequencing primers were used for the U-type fragment (UgyrR4 [5′-CATGGCCACGTTCAATAATTTCG-3′], UgyrF4 [3′-TATAAAAGATGACATCGGTG-5′], UgyrF2 [5′-TTGCATGGTGTCGGTATATC-3′], UgyrF1 [5′-TAATTCCTATAAAGTCTCCG-3′], and UgyrR1 [5′-GGAAATACCCGAGTTAAGAA-3′]). Sequences were then checked using blastx, to verify that the closest matches were gyrB sequences from Enterobacteriaceae. Partial gyrB sequences (single reads) were obtained from two or three isolates of each symbiont type; the full sequence used in phylogenetic analyses was derived from a single symbiont isolate, from A. pisum lab stocks, and was based on at least one read in each direction.
For sequencing the 16S-23S rRNA genes, we used universal PCR primers, and we cloned and sequenced the product for a single isolate of each type from A. pisum, using the protocol and sequencing primers described by Thao and Baumann (36). For sequencing of recA, we used the PCR primers.
Verifying the sources of sequences.
For the 16S-23S rRNA gene sequence, we could verify the source of the amplified sequence by the perfect or near-perfect (<1% difference) identity of the 16S rRNA gene portion with the sequences previously obtained for each symbiont type. For gyrB, two approaches were used to ensure that the sequences obtained were from the symbiont corresponding to the 16S rRNA gene sequence and not from a contaminating bacterium (such as cells on the outer surface or gut of the host insect). First, after obtaining sequences of gyrB that appeared to correspond to each of the three secondary symbiont types, we designed diagnostic PCR screens for the new sequence types by aligning the new sequences against other gyrB sequences of Enterobacteriaceae, selecting diagnostic sites for the new sequence, and designing primers with 3′ positions at these diagnostic sites. To verify that a particular gyrB sequence was associated consistently and exclusively with the presence of a particular symbiont type, we used these primers to screen aphid lines that did and did not contain the designated symbiont type (as established through 16S rRNA gene screens), Diagnostic primer sequences for the R, T, and U types, respectively, were rgyrF2 (5′-AGCCGCAGGCACCGCTGAAG) and rgyrR1 (5′-TAGTCTGGGAAGAGAATTTTG-3′), tgyrF1 (5′-GTAGGGGTTTCAGTGGTG-3′) and tgyrR1 (5′-CCAGTTTTTCATTCATCAAG-3′), and UgyrF1 and UgyrR1 (also used as sequencing primers; for sequences, see “Cloning and sequencing” above). For the R-type diagnostic primers, we screened 27 aphid lines previously characterized for symbiont infections; of these, 11 lines were known to be infected with a total of seven R-type isolates. For the T-type diagnostic primers, we screened 16 lines, including 6 infected with a total of five isolates. For the U-type diagnostic primers, we screened 13 lines, of which 3 were infected with two isolates. Reaction conditions for diagnostic PCR were the same as for the previously described reactions except that the annealing temperature was 60°C. In addition to the diagnostic PCR, we obtained single-pass sequences from the ends of the diagnostic PCR products. These yielded identical, or near-identical, sequences from individuals sampled at different times from the same aphid stock and from at least two stocks known to be infected with the same symbiont type. In six instances, involving uninfected aphid hosts, the highly degenerate primers used for recA gave visible products that, following cloning and sequencing, corresponded to several disparate organisms outside the γ-3 Proteobacteria. These were considered to represent contaminants from the surfaces or guts of the aphids. Occasional amplification of contaminants is expected when one is using highly degenerate primers on universally distributed genes in the absence of a target template, as with the uninfected aphid lines. We consistently obtained a single sequence corresponding to the Enterobacteriaceae for each aphid that did possess a secondary symbiont and not for uninfected lines.
Taxon selection for phylogenetic analyses. (i) 16S rRNA gene.
Taxon sampling is densest for the 16S rRNA gene. We used only sequences of at least 1,300 bp. An initial phylogenetic analysis was performed to verify previous evidence that each of the three symbionts forms a tightly clustered clade and that each falls within the Enterobacteriaceae (28, 29). These analyses included 53 taxa: 25 secondary symbiont sequences (5, 12, and 8 sequences of the R, T, and U types, respectively) and 30 other sequences from Gammaproteobacteria. Sequences from the databases were chosen to include the most divergent representatives within each of the three symbiont groups, plus representative species in the Gammaproteobacteria, especially Enterobacteriaceae. Symbionts included in analyses were from several subfamilies of aphids from Asia, Europe, and North America and from insects other than aphids. Other gammaproteobacterial species used (with accession numbers in parentheses) were B. aphidicola APS (NC_002528), “Candidatus Blochmannia floridanus” (NC_005061), Erwinia salicis (AJ233419), Escherichia coli MG1655 (NC_000913), Haemophilus influenzae (NC_000907), Klebsiella pneumoniae (AJ233420), Morganella morganii (AB089246), Pantoea agglomerans (AJ233423), Pectobacterium carotovorum subsp. atrosepticum (NC_004547; previously placed under Erwinia), Photobacterium profundum (CR378663), Photorhabdus asymbiotica (AY280574), Photorhabdus luminescens (NC_005126), Photorhabdus temperata (AY296252), Proteus vulgaris (AJ233425), Providencia alcalifaciens (AJ300547), Pseudomonas aeruginosa (NC_002516), Salmonella enterica (NC_003198), Serratia entomophila (AJ233427), Serratia ficaria (AJ233428), Serratia fonticola (AY236502), Serratia liquefaciens (AY253924), Serratia marcescens (AY395011), Serratia rubidaea (AJ233436), Shewanella oneidensis (NC_004347), Shigella flexneri (NC_004741), Vibrio cholerae (NC_002505), Wigglesworthia glossinidia (NC_004344), Xenorhabdus nematophila (AY278674), Yersinia pseudotuberculosis (AF365945), and Yersinia pestis (NC_004088). Results of distance analyses in PAUP* (see below) indicated that each symbiont type constituted a very strongly supported clade (100% bootstrap support) with little variation among clade members, as previously documented (28). Furthermore, the Enterobacteriaceae contained all three symbiont groups and formed a strongly supported clade (100% bootstrap support), consistent with previous phylogenetic analyses. Further analyses, aimed at elucidating relationships of the three symbionts to other lineages of Enterobacteriaceae, were conducted using 39 taxa. In these analyses, we excluded species outside of Enterobacteriaceae except for V. cholerae, which was designated as the outgroup. The focus on Enterobacteriaceae was aimed at improving the resolution of relationships of the three symbiont lineages. We included (i) the free-living species of Enterobacteriaceae listed above, including the insect associates Photorhabdus and Xenorhabdus; (ii) representatives of the study taxa, including a single isolate each of the R type (AY296732) and U type (AY296734) (both from A. pisum), several sequences assigned to the more diverse T type, including a symbiont from A. pisum (AY296733), and sequences obtained from other insects, including a beetle (AJ272038), a whitefly (AY264676), and a psyllid (AY136145); and (iii) additional sequences obtained from insects and presumed to represent symbionts previously found to fall within the Enterobacteriaceae but not known to be allied to these three symbionts. The last category included representatives of eight clusters derived from mealybugs and psyllids (AF263556, AF263558, AF263561, AF077607, AF476099, AF476109, AF476107, and AF476110 [38]), a primary symbiont of Sitophilus weevils (AF548137), a secondary symbiont of tsetse flies, Sodalis glossinidius (AF548135), and A. nasoniae, a heritable symbiont of wasps (AY264674).
(ii) gyrB.
After the 16S rRNA gene, the gyrB molecule is the most densely sampled across species of Enterobacteriaceae. Analyses included a single sequence from each symbiont type, since single sequence reads from different isolates indicated no appreciable variation among isolates. Initially we carried out distance and parsimony analyses of a peptide alignment to examine the broader placement of the three symbiont sequences within the Gammaproteobacteria. All three were found to fall within the Enterobacteriaceae with strong bootstrap support (>80%), as concluded in earlier studies on 16S rRNA genes (28, 29). This initial analysis, which is not presented, included GyrB sequences from B. aphidicola (NC_002528), “Ca. Blochmannia floridanus” (NC_005061), Enterobacter cloacae (AB084013), Pectobacterium carotovorum subsp. atrosepticum (NC_004547), E. coli (NC_000913), H. influenzae (NC_000907), Photobacterium profundum (CR378663), Photorhabdus luminescens (NC_005126), Photorhabdus temperata (AY278513), Photorhabdus asymbiotica (AY278496), Photorhabdus sp. (AY278514), Providencia alcalifaciens (AJ300547), Pseudomonas aeruginosa (NC_002516), Salmonella enterica (NC_003198), Shigella flexneri (NC_004741), Serratia ficaria (AJ300540), Serratia entomophila (AJ300543), Serratia marcescens (AB014946), Serratia liquefaciens (AJ300537), Serratia fonticola (AJ300539), Shewanella oneidensis (NC_004347), V. cholerae (NC_002505), W. glossinidia (NC_004344), Yersinia frederiksenii (AJ639639), Yersinia enterocolitica (AB084023), and Y. pestis (NC_004088). In subsequent analyses, we eliminated the more distantly related species, retaining only members of the Enterobacteriaceae and three outgroups, and we used DNA sequences which contain additional phylogenetic signals for resolving these relatively close relationships.
(iii) recA.
We included a single partial sequence for each symbiont type and performed all analyses on DNA sequences, because amino acid sequences show little variation for this highly conserved gene. Other species (and accession numbers) were Erwinia chrysanthemi (AY208918), E. coli (NC_000913), H. influenzae (NC_000907), Pantoea agglomerans (L03291), Pantoea ananatis (AY219005), Pectobacterium carotovorum subsp. atrosepticum (NC_004547), Photorhabdus luminescens (NC_005126), Proteus vulgaris (X55555), Salmonella enterica (NC_003198), Serratia marcescens (M22935), Shewanella oneidensis (NC_004347), Shigella flexneri (NC_004741), Sitophilus oryzae symbiont, or “SOPE” (AY148456), Sodalis glossinidius (AY148454), V. cholerae (NC_002505), W. glossinidia (NC_004344), Y. enterocolitica (AY332973), Y. frederiksenii (AY332975), and Y. pestis (AE013691).
Phylogenetic analyses.
For all genes, alignments were built using ClustalX (18) followed by manual adjustment in MacClade 4 (21). For rRNA genes, regions of ambiguous alignment were removed prior to phylogenetic analysis; alignments were unambiguous for both protein-coding genes due to an almost complete lack of insertions or deletions. Phylogenetic analyses were performed using parsimony, distance (neighbor-joining), and maximum-likelihood (ML) methods within PAUP* (35). For distance analyses, the Kimura two-parameter model of substitution was used. For parsimony and distance analyses, we performed 1,000 bootstrap replicates to determine support for individual nodes. For ML analyses, we used the General Time Reversible model of substitution with an empirically derived rate matrix, gamma distribution with an empirically derived alpha parameter for among-site rate variation, and empirically determined base frequencies. No molecular clock was enforced, and zero-length branches were collapsed during searches.
In situ hybridization.
Aphid embryos were dissected from adult aphids, fixed overnight in Carnoy's solution (chloroform:ethanol:acetic acid, 6:3:1), and subjected to fluorescent in situ hybridization (FISH) as described elsewhere (42). The following probes were used for detection of 16S rRNA genes of the symbionts: ApisP2a-Cy5 (5′-Cy5-CCTCTTTGGGTAGATCC-3′) for B. aphidicola, PASSisR-Cy3 (5′-Cy3-CCCGACTTTATCGCTGGC-3′) for the R type, T16-Cy3 (5′-Cy3-GCCGACATGAACTCAGTAAA-3′) for the T type, and U16-Cy3 (5′-Cy3-GTAGCAAGCTACTCCCCGAT-3′) for the U type. To confirm the specificity of the detection, no-probe and RNase-digested control experiments were conducted. The nuclei of the host cells were counterstained with 4′,6′-diamino-2-phenylindole (DAPI).
Electron microscopy (EM).
Transmission electron micrographs were obtained for each symbiont type in different A. pisum strains. The methods used have been described by Sandström et al. (29). The isolates represented (with 16S rRNA gene accession numbers given in parentheses) are from A. pisum clones 2a (U type) (AY296732), 2BA (T type) (AF293616), and 2BB (R type) (AY136139).
Nucleotide sequence accession numbers.
New GenBank accession numbers for the three symbiont types are AY707922 to AY707924 (recA), AY729881 to AY729883 (gyrB), and AY296732 to AY296734 (16S-23S rRNA genes).
RESULTS
Determining sequences and associating them with symbiont isolates.
For each of the three symbiont types, sequences of the cloned 16S-23S rRNA gene fragments corresponded to previously determined 16S rRNA gene sequences. Combined with other evidence, including the known source of the isolate from established host lab stocks, this indicated that they came from the same organism. For gyrB, we verified that the amplified and cloned sequences were derived from the symbionts (and not from contaminants) in two ways: (i) by obtaining identical or near-identical sequences from multiple individuals and stocks infected by a given type and (ii) by PCR screening with diagnostic primers designed for gyrB for each type. Each diagnostic primer pair was used on a set of aphid lines, as summarized in Materials and Methods. For a given diagnostic primer pair, all lines infected with a given type gave a product with the corresponding primers, and uninfected aphid lines or lines infected with other symbiont types (based on rRNA gene typing) gave no product under the same PCR conditions. In each case of a positive screen, sequences were determined directly from the PCR product; these were perfect or near-perfect (>99.5%) matches to the expected sequence type, as determined from the cloned sequences used for the primer design. In a few cases, the highly degenerate recA primers gave products from contaminant sequences falling outside of the Enterobacteriaceae; however, aphids with known infections yielded sequences showing the highest similarity to sequences from Enterobacteriaceae, and these were >99% identical among isolates within each type.
Phylogenetic relationships. (i) 16S rRNA gene.
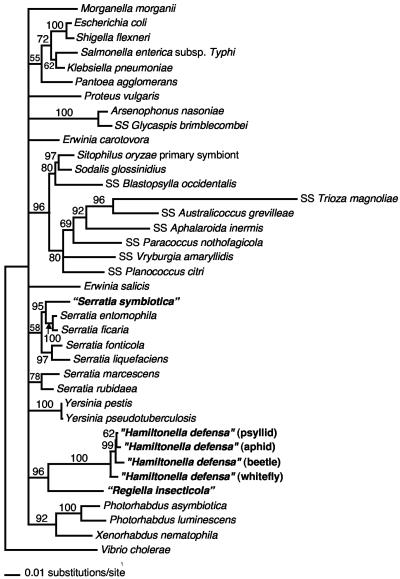
Initial phylogenetic analysis was conducted on 50 Enterobacteriaceae including 5, 12, and 8 isolates of the three symbiont types (R type/PASS [pea aphid secondary symbiont], T type/PABS [pea aphid _Bemisia_-like symbiont], and U type/PAUS [pea aphid U-type symbiont], respectively), 8 members of the Arsenophonus cluster, and numerous outgroups to Enterobacteriaceae (see Materials and Methods). This tree gave 100% bootstrap support for each of the three symbiont clusters, consistent with the observation that divergence within each cluster is <2%, and in agreement with previous results (28, 29). The tree also gave strong (90% bootstrap) support for a clade corresponding to Enterobacteriaceae with each of these symbionts included in that clade, again in agreement with previous results (28, 29, 43). Further analyses included a single representative of each of the 3 symbionts plus 11 other insect symbionts previously placed in Enterobacteriaceae, additional representatives of Enterobacteriaceae, and V. cholerae as an outgroup. Results from the parsimony, distance, and ML methods were in general agreement, and only distance trees are presented (Fig. 1). The R type/PASS is consistently nested within a clade consisting of Serratia species, with strong support (95% bootstrap) for a close relationship to Serratia ficaria and Serratia entomophila, two species associated with insects (1). The relationship to these two species was also supported by parsimony and ML analyses. In all analyses of 16S rRNA genes, Serratia species (including the R-type symbiont) did not form a single clade, a result also obtained in 16S rRNA gene analyses conducted by Dauga (10). However, the results did not conflict with monophyly of Serratia species but simply failed to resolve this node, along with several others.
FIG. 1.
16S rRNA gene-based phylogeny for relationships of the three symbiont types to other Enterobacteriaceae (distance tree with bootstrap support indicated). Nodes with <50% bootstrap values are collapsed.
The other two symbiont types (T type/PABS and U type/PAUS) are always sister groups, regardless of the method of analysis, with 96% bootstrap support in the distance tree (Fig. 1) and strong support in the parsimony and ML analyses. Despite strong support for a relationship, they do show considerable divergence (∼8% [see also references 28 and 29]). The closest relatives of this combined clade were not well resolved with any of the methods. In general, as in previous studies (10, 45, 38), the 16S rRNA gene gave incomplete resolution for basal nodes within Enterobacteriaceae.
None of the other insect symbionts within the Enterobacteriaceae, including lineages that are secondary symbionts of insects related to aphids, are sister groups to the R-type, T-type, and U-type symbionts (Fig. 1). This result is in agreement with the phylogenetic analysis of the 16S rRNA gene by Thao et al. (38).
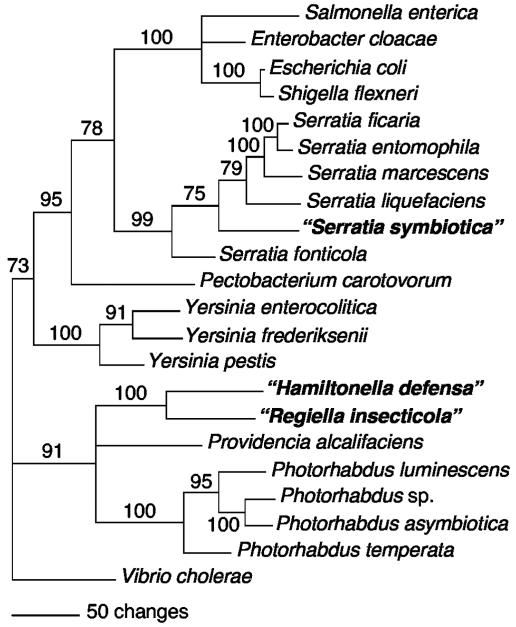
(ii) gyrB.
Serratia species and the R type always formed a single clade when gyrB sequences were analyzed, regardless of the method used to construct the trees, with the distance analyses yielding 99% bootstrap support for a clade containing all the Serratia species plus the R type (Fig. 2). As was the case for 16S rRNA gene trees, the R type is nested within Serratia (not basal). Although all analyses supported the inclusion of this symbiont within the Serratia clade, its phylogenetic placement within Serratia varied with the method of analysis. The other two symbiont types (T type and U type) were sisters to one another in gyrB trees based on all methods of analysis, with 100% bootstrap support using the distance method (Fig. 2). Furthermore, the T-plus-U clade was related to Photorhabdus species and Providencia alcalifaciens by all methods, again with 91% bootstrap support in the distance analyses (Fig. 2).
FIG. 2.
Phylogenetic tree based on partial gyrB sequences for the three symbiont types and other Enterobacteriaceae (distance tree with bootstrap support indicated). Nodes with <50% bootstrap values are collapsed.
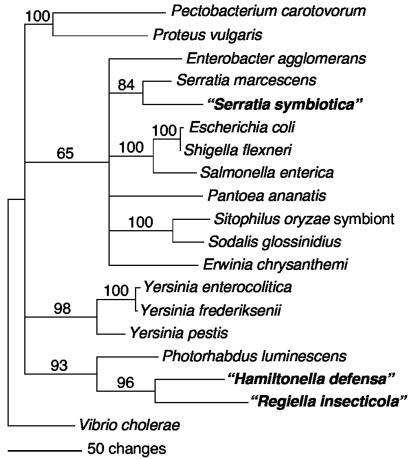
(iii) recA.
The recA trees, which included fewer taxa, also supported a relationship of the R type with the only Serratia species represented, Serratia marcescens (84% bootstrap for distance analyses), a sister relationship between the T and U types (96% bootstrap), and a relationship between the T-plus-U clade and Photorhabdus (93% bootstrap) (Fig. 3). All three of these relationships were supported by both parsimony and ML analyses, although the basal branching in Enterobacteriaceae differed in other respects, depending on the method.
FIG. 3.
Phylogenetic tree based on partial recA sequences for the three symbiont types and other Enterobacteriaceae (distance tree with bootstrap support indicated). Nodes with <50% bootstrap values are collapsed.
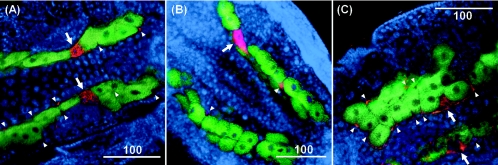
Localization in vivo.
All of the symbiont types are preferentially found in three locations within their aphid hosts: secondary bacteriocytes, sheath cells, and hemocoel (Fig. 4) (13, 19, 42). Secondary bacteriocytes are a small number of large cells that are intercalated between primary bacteriocytes that harbor B. aphidicola (Fig. 4). Sheath cells are small, flat cells located at the peripheries of the primary bacteriocytes (Fig. 4). The symbionts in the hemolymph are readily visualized with light microscopy by sampling clear hemolymph from a host, placing the sample on a slide, and staining with DAPI or another DNA stain (e.g., references 8 and 42). The presence of the symbionts in the hemolymph has also been confirmed by artificial infection experiments: injection of hemolymph from infected aphids into uninfected aphids readily results in the establishment of a heritable infection by the symbiont in the recipients (3, 4, 19, 23, 26, 41).
FIG. 4.
FISH of the three symbiont types in A. pisum embryos. (A) “Ca. Serratia symbiotica”; (B) “Ca. Hamiltonella defensa”; (C) “Ca. Regiella insecticola.” Bars, 100 μm. Arrows and arrowheads indicate secondary bacteriocytes and sheath cells, respectively.
Fine structure.
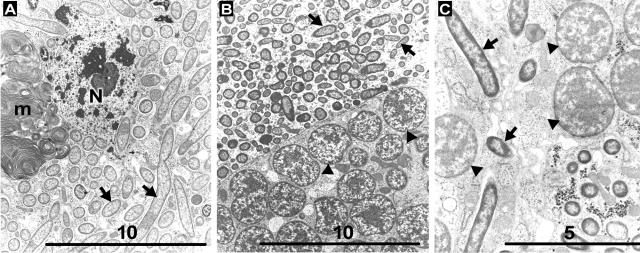
All of the symbiont types are rod-shaped or elongate cells with a diameter of approximately 0.6 to 0.7 μm and a length of 1.5 to 10 μm (Fig. 5). Some variation existed among hosts. In all cases, symbionts were present within bacteriocytes, within sheath cells of bacteriocytes, and within the hemocoel, as previously described (13, 22, 29, 42).
FIG. 5.
Electron micrographs of representatives of each symbiont type. (A) “Ca. Serratia symbiotica.” Bar, 10 μm. (B) “Ca. Hamiltonella defensa.” Bar, 10 μm. (C) “Ca. Regiella insecticola.” Bar, 5 μm. Arrows and arrowheads indicate secondary symbiouts and B. aphidicola, respectively. m, membranes from degenerated secondary symbionts; N, nucleus of host bacteriocyte.
DISCUSSION
Phylogenetic results for all three genes are consistent with the following conclusions. First, the R type belongs to the genus Serratia, based on all three genes analyzed. The evidence for this conclusion is the strong support for its placement within a larger clade consisting of Serratia species (for gyrB and the 16S rRNA gene) or as a sister group of the only available Serratia species (for recA). Second, the T and U types are sister groups to one another among known bacterial species, as supported by analyses of all three genes. Third, all three symbionts belong to the Enterobacteriaceae, based on all three genes, and are independent of other reported symbiont lineages, based only on the 16S rRNA gene. It is possible that the T and U types together are related to Photorhabdus and Xenorhabdus; some support for this was obtained from both protein-coding genes and from the 16S rRNA gene by ML analysis (not shown). Thus, members of three distinct clades of the Enterobacteriaceae routinely infect aphids. Members of the T type can also infect other insects, including psyllids and whiteflies (5, 28); so far, distribution of the other two clades is restricted to aphids, although this may partly reflect sampling efforts. Each symbiont cluster shows very low 16S rRNA gene sequence variation among isolates (<2% divergence; lower if database sequences of apparent poor quality are excluded). Thus, each of these three clusters appears to correspond roughly to units typically named as bacterial species (27, 33). For each, different isolates may be ecologically and genetically distinct, as suggested by molecular and ecological data (28; J. Russell, unpublished data). Phylogenetic placement based on both gyrB and recA is consistent with that based on the 16S rRNA gene. All three symbiont types may be related to other arthropod-associated taxa: in one case, to insect-associated Serratia species; in the other, to Photorhabdus and Xenorhabdus species, which infect insects during certain life cycle stages.
Nomenclature.
Currently there are no clear distinguishing features for these three symbionts other than molecular sequences. Earlier microscopy studies on secondary symbionts of aphids cannot be reliably linked to one of these species, each of which occurs in a variety of host species, with overlap in the use of the same host species.
We propose the following candidate names for these organisms, none of which has been cultured successfully to date. These provisional names are in keeping with the recommendations for bacterial taxa that have not been cultured or fully characterized but for which information beyond a single 16S rRNA gene sequence is available (25).
“Candidatus Serratia symbiotica,” new species.
Phylogenetic results from all three genes indicate that the R type, some isolates of which have been called “PASS” and “S-sym,” belongs to the genus Serratia. This species corresponds to the first secondary symbiont of aphids for which DNA sequences were obtained (43). Several lines of evidence indicate that it is transferred among host species in nature, but transmission observed in the lab has been strictly maternal with high fidelity (4, 9, 28, 29).
(i) Diagnostic features.
The alignment of 16S rRNA gene sequences from five isolates of this symbiont plus representatives of other Proteobacteria indicates distinctive residues, including the following (numbering based on the E. coli sequence): GGUAAUGUGUUAA at positions 454 to 466, GUUUG at 598 to 602, UUGAGGGGUGGCUUCCUGA at 842 to 860, and ACCUCACAAA at 1280 to 1289. Previously deposited sequences for 16S rRNA genes of organisms corresponding to “Ca. Serratia symbiotica” have the following accession numbers: M27042, AB033777, AB033778, AB033779, AF293617, AF293624, AY136137, AY136139, AY136140, AY136144, AY136146, AY136151, AY136155, AY136157, and AY136159. Sequences reported here are AY296732 (16S-23S rRNA genes), AY707924 (recA), and AY729881 (gyrB). Also available are partial sequences of groE (AB063612) and dnaK (AB063613) from “Ca. Serratia symbiotica” infecting laboratory-grown clones, as verified through 16S rRNA gene sequences (19).
This symbiont lives within the cytoplasm of bacteriocytes (the cells that normally contain B. aphidicola), the cytoplasm of sheath cells associated with the bacteriocytes, and also extracellularly in the hemolymph within the body cavity of the aphid. FISH studies of an isolate within A. pisum show cellular tropism in large bacteriocytes and small sheath cells (19) (Fig. 4A). Electron micrographs show that this is a rod-shaped bacterium, with cells approximately 0.6 μm in width and 2 to 9 μm in length (Fig. 5A). The lengths vary among populations inhabiting different host cells within an individual host and also among different hosts. Fukatsu et al. (13) published a detailed EM study of an isolate of this symbiont within A. pisum. The sequence features show some deviation that might reflect the symbiotic habit of “Ca. Serratia symbiotica.” Many symbiont genomes show a shift in nucleotide base composition toward a higher A+T content, especially in intergenic spacers and silent positions of coding sequences. “Ca. Serratia symbiotica” contains 34.9% and 34.7% A+T in gyrB and recA, respectively, in contrast to 17.6 to 18.8% A+T in gyrB of Serratia ficaria, Serratia entomophila, and Serratia marcescens and 25.3% A+T in recA of Serratia marcescens.
(ii) Hosts.
This species occurs in many species of aphids from different subfamilies, including A. pisum, Cinara cupressi, Cinara maritimae, Cinara tujafilina, Macrosiphoniella helichrysi, Macrosiphum rosae, Pemphigus betae, Periphyllus bulgaricus, Pterochloroides persicae, Symthurodes betae, and Uroleucon caligatum. It is known from localities in Europe, North America, and Japan (4, 9, 16, 28, 29, 40, 43). Phylogenetic relationships of the host insects conflict with the phylogenetic relationships of isolates of these symbionts, as derived from the limited variation found in 16S rRNA genes (28). Thus, occasional horizontal transmission must occur, although the mechanisms in nature are not identified and such horizontal transmission has not been observed to occur spontaneously in laboratory colonies. This symbiont has been reported to provide host aphids with resistance to high temperatures (3, 23) and to confer some resistance to attack by endoparasitoid wasps (26). Infection with this symbiont can partially restore the reproduction of aphids from which B. aphidicola has been experimentally eliminated (19).
(iii) Nomenclature.
The specific epithet refers to the symbiotic habit of these bacteria. Although some other Serratia species are associated with insects, this species is the only member of the genus for which intracellular location and regular vertical transmission from mother to progeny have been documented.
“Candidatus Hamiltonella defensa,” new genus and new species.
Our phylogenetic results indicate that this symbiont, usually referred to as T type, or “PABS,” is divergent from other known members of Enterobacteriaceae, with its closest relative being the U-type symbiont.
(i) Diagnostic features.
The alignment of 16S rRNA gene sequences, from 12 isolates of this symbiont plus representatives of other Proteobacteria, indicates distinctive residues, including the following (numbering based on the E. coli sequence): CGAUAAAUGC(G/C)AA at positions 454 to 466, G(C/U)GAG at 598 to 602, UUGAACUGUGGCGUCCGGA at 842 to 860, and AACUCAGAAA at 1280 to 1289. Previously deposited sequences for 16S rRNA genes of organisms corresponding to “Ca. Hamiltonella defensa” have the following accession numbers: Z11926, AF293616, AF293621, AF293622, AF293626, AF400470, AF400471, AF400472, AF400473, AF400476, AF400477, AF400479, AJ272038, AJ297720, AY136136, AY136141, AY136145, AY136148, AY136156, AY136161, AY136162, AY136163, AY136164, AY136166, AY371187, AY429614, AY429615, AY429616, AY462101, AY264676, and AY264675. Sequences reported here are AY296733 (16S-23S rRNA genes), AY707922 (recA), and AY729882 (gyrB).
The cells (Fig. 5B) (29) are similar in size and shape to those of “Ca. Serratia symbiotica,” as described above, and they show a similar distribution in the hosts, with both intracellular and extracellular locations (Fig. 4B). As demonstrated by Russell et al. (28), “Ca. Hamiltonella defensa” shows accelerated sequence evolution compared to other members of the Enterobacteriaceae. This is a common feature of symbiont lineages (24). Another feature typical of many symbiotic species is the somewhat low G+C content of the coding sequences, with gyrB and recA fragments having G+C contents of 44.1 and 40.3%, respectively.
PCR screens from several studies (28, 29; J. A. Russell and N. A. Moran, unpublished data) indicate that all aphids harboring this symbiont also contain gene sequences with close homology to regions of the genome of bacteriophage APSE-1, which was described from an unidentified secondary symbiont of A. pisum in The Netherlands (44). Thus, “Ca. Hamiltonella defensa” is the likely host of the original isolate of APSE-1.
(ii) Hosts.
To date, most hosts are aphids, including the following species from several subfamilies: A. pisum, Aphis craccivora, Aphis fabae, Geopemphigus sp., Macrosiphum euphorbiae, Pemphigus spyrothecae, Periphyllus bulgaricus, Uroleucon ambrosiae, Uroleucon astronomus, Uroleucon atripes, Uroleucon nigrotuberculatum, Uroleucon pieloui, Uroleucon reynoldense, and Uroleucon rudbeckiae (28). “Ca. Hamiltonella defensa” is also known from whiteflies (Bemisia tabaci, Bemisia argentifolii [5, 8, 39]) and psyllids (Cacapsylla pyri [28]). In most of the host species listed, only one or a few individuals were tested, so no conclusions can be made about the universality of the infection within the species. In one widely sampled host (A. pisum), the presence varies among individuals; in another (U. ambrosiae), infection was universal in samples from all over North America and from several years of collecting (29). “Ca. Hamiltonella defensa” is known from A. pisum and other host species in North America and Europe (8, 16, 28, 29) but was absent from A. pisum populations in Japan (40).
Some isolates have been found to provide aphid hosts with defense against parasitoid wasps, based on experiments using artificially infected aphid lines, with genetically identical, uninfected aphids as controls (26), and also based on screens of naturally infected A. pisum with and without this symbiont (12). The occurrence of this symbiont in other insect groups raises the question of whether this defensive effect occurs in other insect hosts.
(iii) Nomenclature.
The generic name was chosen to honor the evolutionary biologist William D. Hamilton, who made major contributions to the understanding of host-pathogen coevolution, among other topics. The specific epithet refers to the role of these symbionts in defending hosts against natural enemies.
“Candidatus Regiella insecticola,” new genus and new species.
We propose a separate candidate genus containing a single species for the U type (29), some isolates of which have been called “PAUS” (40, 41). Although this lineage is clearly related to “Ca. Hamiltonella defensa” based on strong support in analyses of all three genes, a separate genus name is proposed. The decision to use a distinct genus name is ultimately arbitrary but is rationalized here by (i) the clear separation of the two clusters despite intensive sampling of both from many host species in several continents and (ii) the high degree of divergence of their 16S rRNA gene sequences (8%), which exceeds that commonly separating genera within the Enterobacteriaceae (e.g., 16S rRNA gene divergence is 2 to 8% for pairwise combinations of represented species in Escherichia, Salmonella, Pectobacterium, Photorhabdus, Yersinia, Serratia, and Xenorhabdus).
(i) Diagnostic features.
The alignment of 16S rRNA gene sequences, from eight isolates of this symbiont plus other bacteria, indicates diagnostic residues, including the following (numbering based on the E. coli sequence): GCG(G/A)UAAGAGUAA at positions 454 to 466, GUAAG at 598 to 602, GGGUAGCA at 773 to 780, UAGUGUUAUGGCGUC(U/C)GAA at 841 to 860, ACCUCAUAAA at 1280 to 1289, and UGGGCGG at 1420 to 1426. Previously deposited sequences for 16S rRNA genes of organisms corresponding to “Ca. Regiella insecticola” have the following accession numbers: AF293618, AF293619, AF293620, AF293623, AF293627, AY136141, AY136143, AY136149, AY136150, AY136152, AY136154, AY136165, AY136167, AY462102, and AB112788 to AB112793. Sequences reported here are AY296734 (16S-23S rRNA genes), AY707923 (recA), and AY729883 (gyrB).
As noted for “Ca. Hamiltonella defensa,” the coding sequences are somewhat low in G+C content: 40.4 and 47.1% for gyrB and recA fragments, respectively. Also, the 16S rRNA gene sequences have been shown to display accelerated evolution compared to related, nonsymbiotic bacteria (28).
Distributions within hosts, as well as cell morphology, are similar to those of “Ca. Serratia symbiotica” and “Ca. Hamiltonella defensa” (see Fig. 4C and 5C). Tsuchida et al. (42) published a detailed FISH and EM study of an isolate of this symbiont within A. pisum, showing cellular tropism in large bacteriocytes and small neighboring “sheath” cells (42).
(ii) Hosts.
To date, all hosts are aphids, including species from several subfamilies (A. pisum, Aphis citricola, Aphis nerii, Brachycaudus cardui, Chaitophorus populeti, Colopha kansugei, Macrosiphoniella ludovicianae, Macrosiphum avenae, M. euphorbiae, P. betae, Uroleucon aenum, Uroleucon helianthicola, U. astronomus, Uroleucon giganteus, U. rudbeckiae, Uroleucon solidaginis) and from Europe, North America, and Japan (16, 28, 29, 42). The main phenotypic effect associated with this symbiont so far is an enhanced ability to use specific host plants (41). It may also confer an increase in resistance to fungal infection (12).
(iii) Names.
The generic name is in honor of the entomologist Reginald F. Chapman, known as “Reg.” Through his research, writing, and teaching, he made outstanding contributions to the study of the functioning of insects, particularly adaptations by herbivorous species for exploiting particular host plants. The specific name refers to the hosts.
For each of these three groups, it is likely that strains in individual hosts will show genomic differences, as do strains of other bacterial species. Therefore, it is important that strain designations be given in studies of these symbionts, recognizing that strains may differ in gene content and other biological features (as for named species such as E. coli or Salmonella enterica). Since the symbionts have not been cultured outside of hosts, living strains currently can be maintained only by rearing within a host.
Acknowledgments
We thank Joshua White and David Bentley for electron microscopy assistance, Jean Tsai and Helen Dunbar for assistance with the lab work, Kerry Oliver for samples, Kim Hammond for help with aphid cultures, Robert Hershoff for advice on Latin nomenclature, and Becky Nankivell for preparing the manuscript and figures.
Financial support was from NSF grant 0313737 (to N.A.M.). J.A.R. was supported by a fellowship from the NSF-IGERT training grant in evolutionary genomics at the University of Arizona.
REFERENCES
- 1.Anahory, T., H. Darbas, O. Ongaro, H. Jean-Pierre, and P. Mion. 1998. Serratia ficaria: a misidentified or unidentified rare cause of human infections in fig tree culture zones. J. Clin. Microbiol. 36**:**3266-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Wiley Interscience, New York, N.Y.
- 3.Chen, D. Q., C. B. Montllor, and A. H. Purcell. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95**:**315-323. [Google Scholar]
- 4.Chen, D. Q., and A. H. Purcell. 1997. Occurrence and transmission of facultative endosymbionts in aphids. Curr. Microbiol. 34**:**220-225. [DOI] [PubMed] [Google Scholar]
- 5.Clark, M. A., L. Baumann, M. A. Munson, P. Baumann, B. C. Campbell, J. E. Duffus, L. S. Osborne, and N. A. Moran. 1992. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodoidea) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr. Microbiol. 25**:**119-123. [Google Scholar]
- 6.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49**:**267-275. [DOI] [PubMed] [Google Scholar]
- 7.Dale, C., B. Wang, N. A. Moran, and H. Ochman. 2003. Loss of DNA recombinational repair enzymes in the initial stages of genome degeneration in mutualistic bacterial endosymbionts Mol. Biol. Evol. 20**:**1188-1194. [DOI] [PubMed] [Google Scholar]
- 8.Darby, A. C., L. M. Birkle, S. L. Turner, and A. E. Douglas. 2001. An aphid-borne bacterium allied to the secondary symbionts of whitefly. FEMS Microbiol. Ecol. 36**:**43-50. [DOI] [PubMed] [Google Scholar]
- 9.Darby, A. C., and A. E. Douglas. 2003. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 69**:**4403-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauga, C. 2002. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int. J. Syst. Evol. Microbiol. 52**:**531-547. [DOI] [PubMed] [Google Scholar]
- 11.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21**:**1307-1313. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, J., A. C. Darby, T. J. Daniell, H. C. J. Godfray, and A. E. Douglas. 2004. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29**:**60-65. [Google Scholar]
- 13.Fukatsu, T., N. Nikoh, R. Kawai, and R. Koga. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 66**:**2748-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukushima, M., K. Kakinuma, and R. Kawaguchi. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 40**:**2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gherna, R. L., J. H. Werren, W. Weisburg, R. Cote, C. R. Woese, L. Mandelco, and D. J. Brenner. 1991. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int. J. Syst. Bacteriol. 41**:**563-565. [Google Scholar]
- 16.Haynes, S., A. C. Darby, T. J. Daniell, G. Webster, F. J. Van Veen, H. C. J. Godfray, J. I. Prosser, and A. E. Douglas. 2003. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 69**:**7216-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hypša, V., and C. Dale. 1997. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47**:**1140-1144. [DOI] [PubMed] [Google Scholar]
- 18.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 23**:**403-405. [DOI] [PubMed] [Google Scholar]
- 19.Koga, R., T. Tsuchida, and T. Fukatsu. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B 270**:**2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefevre, C., H. Charles, A. Vallier, B. Delobel, B. Farrell, and A. Heddi. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21**:**965-973. [DOI] [PubMed] [Google Scholar]
- 21.Maddison, D. R., and W. P. Maddison. 2000. MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland, Mass.
- 22.Mira, A., and N. Moran. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 44**:**137-143. [DOI] [PubMed] [Google Scholar]
- 23.Montllor, C. B., A. Maxmen, and A. H. Purcell. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27**:**189-195. [Google Scholar]
- 24.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93**:**2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, R. G. E., and K. H. Schleifer. 1994. Taxonomic notes: a proposal for recording the properties of putative taxa of prokaryotes. Int. J. Syst. Bacteriol. 44**:**174-176. [DOI] [PubMed] [Google Scholar]
- 26.Oliver, K. M., J. A. Russell, N. A. Moran, and M. S. Hunter. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 100**:**1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25**:**39-67. [DOI] [PubMed] [Google Scholar]
- 28.Russell, J. A., A. L. LaTorre, B. Sabater-Munoz, A. Moya, and N. A. Moran. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12**:**1061-1075. [DOI] [PubMed] [Google Scholar]
- 29.Sandström, J. P., J. A. Russell, J. P. White, and N. A. Moran. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 10**:**217-228. [DOI] [PubMed] [Google Scholar]
- 30.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407**:**81-86. [DOI] [PubMed] [Google Scholar]
- 31.Spaulding, A. W., and C. D. von Dohlen. 2001. Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol. Biol. 10**:**57-67. [DOI] [PubMed] [Google Scholar]
- 32.Sproer, C., U. Mendrock, J. Swiderski, E. Lang, and E. Stackebrandt. 1999. The phylogenetic position of Serratia, Buttiauxella, and some other genera of the family Enterobacteriaceae. Int. J. Syst. Evol. Bacteriol. 49**:**1433-1438. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Bacteriol. 52**:**1043-1047. [DOI] [PubMed] [Google Scholar]
- 34.Subandiyah, S., N. Nikoh, S. Tsuyumu, S. Somowiyarjo, and T. Fukatsu. 2000. Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zool. Sci. 17**:**983-989. [Google Scholar]
- 35.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 40b10. Sinauer Associates, Sunderland, Mass.
- 36.Thao, M. L., and P. Baumann. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 70**:**3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thao, M. L., M. A. Clark, L. Baumann, E. B. Brennan, N. A. Moran, and P. Baumann. 2000. Secondary endosymbionts of psyllids have been acquired multiple times. Curr. Microbiol. 41**:**300-304. [DOI] [PubMed] [Google Scholar]
- 38.Thao, M. L., P. J. Gullan, and P. Baumann. 2002. Secondary (γ-Proteobacteria) endosymbionts infect the primary (β-Proteobacteria) endosymbionts of mealybugs multiple times and coeolve with their hosts. Appl. Environ. Microbiol. 68**:**3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thao, M. L., and P. Baumann. 2004. Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae) Curr. Microbiol. 48**:**140-144. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchida, T., R. Koga, H. Shibao, T. Matsumoto, and T. Fukatsu. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11**:**2123-2135. [DOI] [PubMed] [Google Scholar]
- 41.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303**:**1989. [DOI] [PubMed] [Google Scholar]
- 42.Tsuchida, T., R. Koga, X.-Y. Meng, T. Matsumoto, and T. Fukatsu.. Characterization of a facultative endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum. Microb. Ecol., in press. [DOI] [PubMed]
- 43.Unterman, B. M., P. Baumann, and D. L. McLean. 1989. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J. Bacteriol. 171**:**2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. van den Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262**:**104-113. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, S., and S. Harayama. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61**:**1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zchori-Fein, E., and J. K. Brown. 2002. Diversity of prokaryotes associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 95**:**711-718. [Google Scholar]