Generation and Characterization of Rac3 Knockout Mice (original) (raw)
Abstract
Rac proteins are members of the Rho family of GTPases involved in the regulation of actin dynamics. The three highly homologous Rac proteins in mammals are the ubiquitous Rac1, the hematopoiesis-specific Rac2, and the least-characterized Rac3. We show here that Rac3 mRNA is widely and specifically expressed in the developing nervous system, with highest concentration at embryonic day 13 in the dorsal root ganglia and ventral spinal cord. At postnatal day 7 Rac3 appears particularly abundant in populations of projection neurons in several regions of the brain, including the fifth layer of the cortex and the CA1-CA3 region of the hippocampus. We generated mice deleted for the Rac3 gene with the aim of analyzing the function of this GTPase in vivo. Rac3 knockout animals survive embryogenesis and show no obvious developmental defects. Interestingly, specific behavioral differences were detected in the Rac3-deficient animals, since motor coordination and motor learning on the rotarod was superior to that of their wild-type littermates. No obvious histological or immunohistological differences were observed at major sites of Rac3 expression. Our results indicate that, in vivo, Rac3 activity is not strictly required for normal development in utero but may be relevant to later events in the development of a functional nervous system.
Members of the Rho family of small GTPases are key regulators of a broad range of cellular functions in eukaryotes, including cytoskeletal organization, membrane trafficking, transcription, cell growth, and development (17, 47). Each family member is believed to be involved in the regulation of multiple cellular processes, and there appears to be functional overlap between individual Rho family GTPases (21, 47). The Rho family includes about 20 Rho proteins that can roughly be divided into five groups, the Rho-like, Rac-like, Cdc42-like, Rnd, and RhoBTB subfamilies (7). The Rac subfamily consists of three genes: Rac1, Rac2, and Rac3/Rac1B (20, 29), herein referred to as Rac3. The corresponding polypeptides share about 90% protein identity. Rac proteins participate in a broad range of cellular functions, such as actin cytoskeletal reorganization (38), cell adhesion (21), cell growth (36), and superoxide formation (2, 24). The most obvious difference among the three proteins is their pattern of expression. Rac1 is ubiquitously expressed in most organs and is involved in the regulation of actin cytoskeletal reorganization (33). In contrast, Rac2 is specifically expressed in hematopoietic cells (15, 20, 40), while Rac3 has predominant expression in the developing nervous system and adult brain (20, 29).
Targeted disruption of the gene has shown that Rac1-null mice die early during development (42). The Rac1-deficient embryos show a high degree of cell death in the space between the embryonic ectoderm and endoderm at the primitive streak stage, and die before embryonic day 9.5 (E9.5). Investigation of the primary epiblast culture isolated from Rac1-deficient embryos indicated that Rac1 is involved in lamellipodium formation, cell adhesion, and cell migration in vivo, suggesting that Rac1-mediated cell adhesion is essential for the formation of three germ layers during gastrulation.
In mammals, Rac2 is restricted in expression to hematopoietic cells, where it is coexpressed with Rac1. Rac2-null mice develop normally and are fertile (39). Gross and histopathological examinations of nonhematopoietic organs are normal, although neutrophils isolated from Rac2-null mice demonstrate deficits in multiple functions, including cytoskeletal remodeling and superoxide production, with significant consequences for leukocyte trafficking and host defense, implicating Rac2 in the regulation of the functions of different hematopoietic cells (1, 5, 12, 13, 18, 19, 48, 51).
Rac3 has the strongest homology to Rac1 (29). The greatest divergence between Rac3 and Rac1 occurs at the carboxy-terminal hypervariable region (residues 180 to 192). This region is posttranslationally modified and is important for the specific intracellular localization and interaction of the GTPase with target proteins (4, 23). In vitro, overexpression of avian Rac3 specifically potentiates neuritogenesis and branching from cultured avian retinal neurons (3). In previous studies, we found that the transcript for avian Rac3 (cRac1B) is specifically expressed in the developing chicken peripheral and central nervous system (29). The levels of the transcript are developmentally regulated in the avian and mouse brain, with a peak corresponding to the time of intense neurite branching and synaptogenesis (3, 6). The highest Rac3 mRNA expression among human tissues studied is in brain (20), where Rac1 mRNA but not Rac2 mRNA is also present (15). Moreover, the in situ hybridization analysis presented here shows that Rac3 is widely and specifically expressed in the developing nervous system of wild-type mice.
The restricted expression pattern of Rac3 in the developing animal differs substantially from that of Rac1 and Rac2, suggesting specific biological roles for each gene. In this study, we have begun to examine the in vivo functions of Rac3 via the generation of mice with a targeted deletion of the coding region of the gene. The resultant homozygous mice are viable and fertile. The work reported here details the first characterization of Rac3 knockout mice and shows the existence of behavioral differences between wild-type and Rac3 mutant mice.
MATERIALS AND METHODS
Construction of the Rac3 targeting vector and generation of mutant mice.
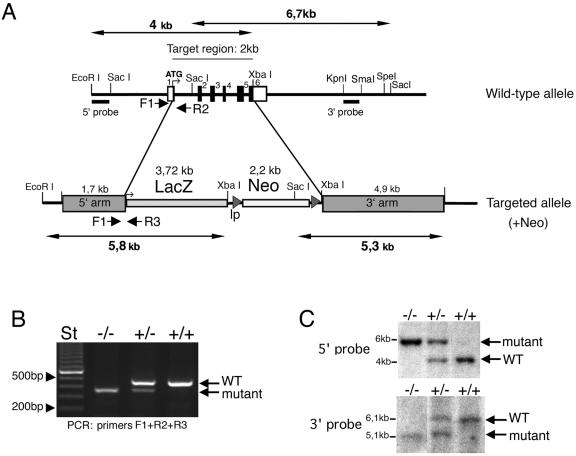
Genomic clones including the region with the six exons encoding the full coding region were obtained from a mouse 129SVJ lambda FIX II genomic library (Stratagene). Restriction analysis and sequencing were used to identify suitable regions for subcloning into the targeting vector. A 4.9-kb XbaI/SpeI fragment was blunted and inserted between the neomycin and thymidine kinase cassettes of the pPNT-loxP vector (with the neomycin cassette derived from the pFlox vector) as the 3′ arm. A unique XhoI site was then inserted by ligation of paired oligonucleotides between the NotI and SalI sites just upstream of the neomycin cassette. The final targeting vector (pRac3.5) included a 1.7-kb fragment ob-tained by PCR, corresponding to the region just upstream of the Rac3 ATG, as 5′ arm, followed by a 3.7-kb fragment coding for bacterial β-galactosidase gene (lacZ).
The β-galactosidase cDNA was knocked into the Rac3 locus to allow Rac3 expression to be monitored in vivo. The β-galactosidase insertion sites were analyzed by direct sequencing of the recombinant construct. The β-galactosidase cDNA was inclusive of the full coding region and of the polyadenylation sequence. The position of the starting ATG of the β-galactosidase coincided with that of the ATG of the deleted Rac3. Homologous recombination of the targeting construct with the wild-type gene results in deletion of exons 1 to 6, as depicted in Fig. 1A.
FIG. 1.
Targeted disruption of the mouse Rac3 gene. (A) Targeting strategy. The structures of the wild-type allele and the disrupted allele are shown. In the wild-type allele, exons are numbered 1 to 6 and are represented by black boxes. The primers used for routine genotyping are represented by arrows. (B) PCR genotyping with primers for wild-type and recombinant alleles. A 0.46-kb fragment was generated from the wild-type allele (primers F1 and R2), and a 0.37 kb fragment was generated from the mutant allele (primers F1 and R3). (C) Genomic DNA prepared from tail biopsies was used for Southern blot analysis after digestion with EcoRI and XbaI (upper blot), or with SacI (lower blot). Filters were probed with the 0.25-kb EcoRI/SacI fragment generated from sequence immediately upstream of the 5′ arm (5′ probe, upper blot), and with the 0.4-kb KpnI/SmaI fragment from the 3′ arm (3′ probe, lower blot). ATG, start methionine; lp, loxP sites; +/+, wild-type; +/−, heterozygous; −/−, homozygous knockout.
The targeting construct was linearized with XhoI and electroporated into R1 (129/SvJ × 129/Sv-CP) embryonic stem (ES) cells. Resistant cells were selected in the presence of G418 and ganciclovir. DNA was isolated from a total of 683 clones. A 0.25-kb EcoRI/SacI fragment generated from sequence immediately upstream of the 5′ arm was used to screen ES cell genomic DNA for homologous recombination by Southern blotting and hybridization. Two independent ES cell lines were injected into C57BL/6 blastocysts, which were subsequently transferred into pseudopregnant females to generate chimeric offspring. Chimeras were bred with either 129/SvPasIco (129/Sv) or C57BL/6 female mice to produce heterozygotes. All procedures were carried out in accordance with institutional policies following approval from the Animal Ethical Committee of the San Raffaele Scientific Institute.
Genotyping of mutant mice.
The genotypes of mutant mice were determined by PCR and confirmed by Southern blot analysis of genomic DNA from tail biopsies. Briefly, tail samples were incubated overnight in lysis buffer (50 mM Tris, pH 8.0, 100 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate, 1 mg of proteinase K ml−1), followed by isopropanol precipitation. Routine genotyping through PCR was performed with genomic DNA with specific oligonucleotide primers for the Rac3 wild-type allele (F1, 5′-CATTTCTGTGGCGTCGCCAAC-3′, and R2, 5′-CACGCGGCCGAGCTGTGGTG-3′). For the targeted allele, a primer was designed from within the lacZ gene (R3, 5′-TTGCTGGTGTCCAGACCAAT-3′). The three primers were used in a multiplex PCR with LA Taq (Takara) with the following amplification conditions: 94°C for 1 min and 30 cycles of 98°C for 20 s, 66°C for 1 min, and a 10-min incubation at 72°C at the end of the run. Amplification products were resolved on a 1.6% agarose gel. Genotyping of animals was confirmed by Southern blotting on genomic DNA probed with the 0.25-kb EcoRI/SacI fragment generated from sequence immediately upstream of the 5′ arm, and with a 0.4-kb KpnI/SmaI fragment from the 3′arm.
Reverse transcription-PCR and Northern blot analysis.
Total cellular RNA was isolated from postnatal day 0 (P0) and P9 brains with the GenElute kit (Sigma-Aldrich). Aliquots of RNA were transcribed into cDNA (Superscript II; Invitrogen) and amplified with GoTaq (Promega) and primers specific for Rac1 (forward, 5′-CCAATACTCCTATCATCCTC-3′, and reverse, 5′-ACAGCAGGCATTTTCTCTTC C-3′) or Rac3 (forward, 5′-CACACACACCCATCCTTCT-3′, and reverse, 5′-GAATACAGTGCACTTCTTGCC-3′). The following amplification protocol was used: 94°C for 2 min and 30 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. Products were electrophoresed through a 1.6% agarose gel.
Northern blot analysis of total RNA from the indicated organs (15 μg/lane) was performed as previously described (26). Blots were hybridized with a 0.28-kb PCR fragment corresponding to the 3′ untranslated region of the cDNA for Rac3, with a 1-kb fragment of mouse Rac3 cDNA (the same used for in situ hybridization), with a 378-bp PCR fragment corresponding to bp 181 to 558 of the translated Rac2 cDNA (amino acids 61 to 186), or with a 1.2-kb fragment corresponding to part of the 3′ untranslated region of the Rac1 cDNA. Hybridization took place in hybridization buffer supplemented with 32P-labeled probes (1 × 106 to 2 × 106 cpm ml−1) for 15 h at 65°C. Following high-stringency washes at 65°C, X-ray films were exposed for 3 to 12 h to the hybridized filters.
Biochemical analysis.
P9 brains were extracted with lysis buffer (0,5% Triton X-100, 150 mM NaCl, 20 mM Tris-Cl, pH 7.5, and 10 μg ml−1 each of antipain, chymostatin, leupeptin, and pepstatin). Human platelets were isolated from fresh blood by separation on Ficoll gradients, washed, and lysed in lysis buffer. Mouse peritoneal macrophages were recovered after treatment for 5 days with 600 μl per mouse of 3% thioglycolate. Cell and tissue lysates were clarified by centrifugation. For each immunoprecipitation, 10 to 20 μl of immune or preimmune Rac3 antiserum was preadsorbed to 25 μl protein A-Sepharose beads (Amersham Biosciences) and added to lysates (1.5 to 3 mg protein/immunoprecipitation). After 3 h at 4°C with rotation, immunoprecipitates were washed four times with 0,75 ml of lysis buffer, and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Immunoblotting was performed on brain lysates and on immunoprecipitates. Filters were incubated with a Rac3 polyclonal antiserum diluted 1:100 (6), with a Rac1 monoclonal antibody (0.25 μg ml−1; Transduction Laboratories-Becton Dickinson) or with a tubulin monoclonal antibody (0.3 μg ml−1; Amersham Biosciences). For the detection of primary antibodies, blots were incubated with 0.2 μCi ml−1 of [125I]protein A or [125I]anti-mouse immunoglobulin (Amersham Biosciences), washed, and exposed to Amersham Hyperfilm-MP.
Histopathology.
Prior to collection of adult tissue, animals were deeply anaesthetized, and cardiac perfusion with 4% paraformaldehyde was performed. Whole brains were removed and postfixed overnight. Eyes were dissected and directly fixed in 4% paraformaldehyde. After fixation, organs were dehydrated and embedded in paraffin; 10-μm sections were stained with cresyl violet or with hematoxylin and eosin, according to standard protocols.
Immunofluorescence.
P7 mice were fixed under deep anesthesia by transcardial perfusion with 4% paraformaldehyde in phosphate-buffered saline. Brains were removed from skulls and immersed in fixative overnight at 4°C. Samples were washed with phosphate-buffered saline, cryoprotected with sucrose in phosphate-buffered saline, and frozen in O.C.T. compound (VWR International Ltd., Poole, Dorset, United Kingdom); 12-μm-thick sections were blocked in 15% serum, 0.3% Triton X-100, 450 mm NaCl, 20 mm sodium phosphate buffer, pH 7.4. Sections were first incubated overnight at 4°C with primary antibodies, followed by incubation for 1.5 h at room temperature with fluorescently labeled secondary antibodies (Molecular Probes). Primary antibodies included the G177 polyclonal antibody against synapsin I (46) and mouse monoclonal antibodies against MAP2 (HM-2 from Sigma-Aldrich), calbindin (Swant), and GAD65 and GAD67 (Chemicon). Control sections were incubated with secondary antibodies only. Sections were analyzed with a Bio-Rad MRC 1024 confocal microscope (Bio-Rad, Hercules, CA).
In situ hybridization of Rac3 mRNA.
Embryos were fixed in 4% paraformaldehyde immediately after removal, while postnatal brains were fixed by perfusion and postfixed overnight at 4°C in 4% paraformaldehyde; 10-μm sections were cut after freezing. Probes were obtained by in vitro transcription of antisense (Rac3-AS) and sense (Rac3-S) single-stranded RNA probes from a 1-kb fragment of mouse Rac3 cDNA (including the region coding for the protein flanked by a short 5′ and the 3′ nontranslated region), cloned into the plasmid BlueScript (Stratagene). The antisense Rac1 (Rac1-AS) single-stranded RNA probe was obtained from a 1.2-kb fragment of mouse Rac1 cDNA (including part of the 3′ nontranslated region). The digoxigenin-labeled antisense and sense probes were synthesized with T7 and T3 RNA polymerases and digoxigenin-11-UTP (Roche) as previously described (37). The transcripts were used as probes at a concentration of 1 μg/ml in hybridization buffer at high stringency (60°C). The hybridized probe was detected with alkaline phosphatase-coupled antibodies to digoxigenin according to the manufacturer's protocol (Roche).
Accelerating rotarod test.
For behavioral phenotyping, we used individuals obtained by crossing chimeras carrying 129/Sv ES cells targeted for the Rac3 locus with pure-bred 129/Sv individuals (Charles River). Thus, mutants are to be considered coisogenic with pure-bred 129/Sv and genetically homogeneous. Tests were performed on littermates.
Motor ability was determined using an accelerating rotarod apparatus (model 7650, Ugo-Basile SRL), as described (9, 16). Mice were tested using two trials per day (6 h rest between the two trials) on 3 consecutive days. Each trial consisted of three tests per animal, with at least 15-min rests between tests. During a trial, the rod accelerated from 4 to 40 rpm over 6 min and then remained at 40 rpm for an additional 9 min. Each trial lasted until the mouse fell from the rod or for a maximum of 15 min. If a mouse was observed to rotate passively with the bar for a full rotation, the test was stopped. Statistical significance was assessed by Student's t test. Differences were considered significant at P < 0.05.
Footprint analysis.
After the coating of their hind feet with a nontoxic paint, animals were allowed to walk through an illuminated, narrow, 7-cm-wide, 110-cm-long, 10-cm-high alley. The footprint patterns made on the paper lining the floor of the alley were scored for four parameters. The left and right strides (average distance of forward movement between alternate steps) and the base (average lateral distance between opposite left and right steps) were calculated by measuring the average values from five clearly visible consecutive footprints. Average values were normalized by the length of each animal. Statistical significance was assessed by Student's t test. Differences were considered significant at P < 0.05.
Hot plate test.
Hot plate nociception is a reflex that requires higher brain centers (11, 32). The mouse is placed in a Plexiglass cylinder (10.2-cm diameter by 30.5 cm high) on the surface of a hot plate which is maintained at 55°C. The time that passes before the mouse raises and licks its paw or jumps up is recorded. The mouse is then immediately removed from the hot plate. If the mouse had not responded within 30 s, it was removed from the hot plate to prevent tissue damage. Hot plate test data were analyzed by the Student t test, with P < 0.05 considered significant.
RESULTS AND DISCUSSION
Generation of Rac3-deficient mice.
The translated region of the mouse Rac3 gene includes six exons spanning about 2 kb on mouse chromosome 11 E2. We isolated and partially sequenced genomic clones that contain exons 1 to 6 (Fig. 1A). The full sequence of this region of mouse chromosome 11 has since been confirmed by NCBI (MGI:2180784) and the Mouse Genome Sequencing Consortium (49).
To determine the function of Rac3 in vivo, we used homologous recombination in ES cells to disrupt the Rac3 gene. We removed the 2-kb region of the gene encoding the full-length Rac3 polypeptide (exons 2 to 5 and part of exons 1 and 6; Fig. 1A). The targeting vector was designed to replace this region with the bacterial Rac3 gene and the neomycin resistance gene. The deleted region (2 kb) encodes the full-length Rac3 polypeptide. Independent ES cell clones with homologous recombination confirmed by Southern blotting were injected into blastocysts to generate chimeras. Male chimeras from two different ES cell clones were crossed with C57BL/6 females to establish strains with a mixed genetic background (129-C57) heterozygous for the mutated allele. Alternatively, male chimeras were crossed with 129/Sv females to establish strains with the 129/Sv genetic background. Homologous recombination in offspring was confirmed by both PCR (Fig. 1B) and Southern blot analysis (Fig. 1C).
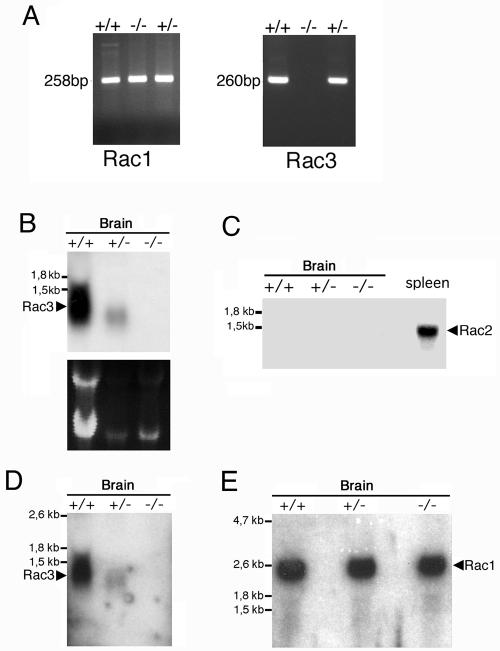
The absence of Rac3 mRNA in the knockout mice was confirmed by reverse transcription PCR with cDNA generated from cerebral RNA from P9 animals. No Rac3 product was amplified from knockout tissue, while a band of the expected size was amplified from wild-type and heterozygous tissue (Fig. 2A). Primers specific for Rac1 were used to ensure that expression of these genes was not dysregulated. An amplification product of the expected size was also obtained with the Rac1 primers from knockout tissue. The absence of Rac3 mRNA from knockout animals was confirmed by Northern blotting using two different probes on P0 mRNA from the brain (Fig. 2B and D). Rac2 is not expressed in the brain (15), and Northern blot analysis on knockout animals showed no evident upregulation of either Rac2 (Fig. 2C) or Rac1 (Fig. 2E) expression in brain. As a positive control for Rac2, this gene was clearly expressed in the control spleen, as expected (Fig. 2C).
FIG. 2.
Analysis of the expression of the transcripts of Rac proteins in mutant mice. (A) Total RNA prepared from P9 brains of wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice was reverse transcribed and then used for PCR amplification with primers specific for Rac1 and Rac3. No amplification product for Rac3 was observed in the knockout mice. Total RNA (15 μg/lane) from P0 (B and D) or P9 (C and E) brains of wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice was used for Northern blot analysis. Filters were incubated at high stringency with a 0.28-kb Rac3-specific probe (B), a 0.38-kb Rac2-specific probe (C), the 1-kb Rac3-specific probe also used for in situ hybridization (D), or a 1.2-kb Rac1-specific probe (E). A control lane with 15 μg of total RNA from adult spleen was included in panel C. No RNA for Rac3 was detectable in the knockout mice (B and D).
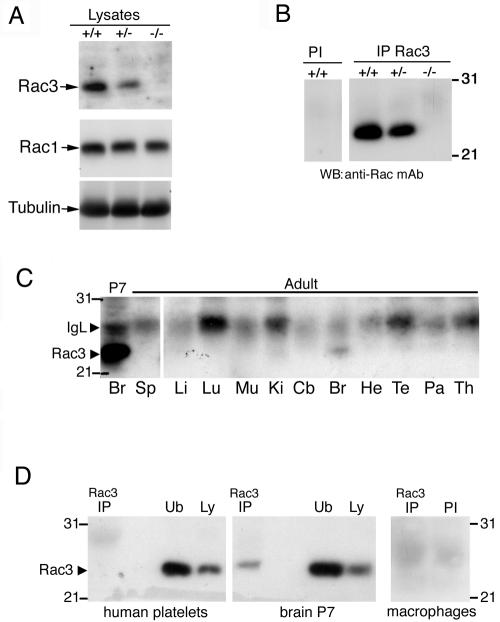
The Rac3 protein shows specific expression in the mouse brain, with a peak of expression around P7 (6). By using a Rac3-specific antibody, we found that the Rac3 protein was absent from knockout animals, by both immunoblotting (Fig. 3A) and immunoprecipitation from P9 brain lysates (Fig. 3B). As a control, immunoblotting with an anti-Rac1 antibody showed no differences in the protein levels in wild-type and knockout mouse brains (Fig. 3A). Altogether these data confirm the absence of Rac3 in the knockout mice and show no evident upregulation of expression of the other two Rac proteins in the brains of these animals.
FIG. 3.
Expression of the Rac3 protein. (A) Immunoblotting on P9 brain lysates. Filters were incubated with the anti-Rac3 (upper), anti-Rac1 (middle), or antitubulin (lower) antibodies. No Rac3 protein was detected in the brain lysate from knockout mice. (B) P9 brain lysates were incubated with protein A-Sepharose beads conjugated to preimmune (PI) or immune anti-Rac3 serum (IP Rac3). After blotting, the filters were incubated with the anti-Rac monoclonal antibody. No Rac3 protein was immunoprecipitated from the brain lysate of knockout mice. (C) Aliquots of 2 mg protein of P7 brain lysate and of 3 mg protein of adult tissue lysates were immunoprecipitated with anti-Rac3 antibody. After blotting, the filters were incubated with the anti-Rac monoclonal antibody recognizing both Rac3 and Rac1. Br, brain; Sp, spleen; Li, liver; Lu, lung; Mu, skeletal muscle; Ki, kidney; Cb, cerebellum; He, heart; Te, testis; Pa, pancreas; Th, thymus. (D) Aliquots of lysates from human platelets (1.12 mg protein), P7 brain (2 mg protein), and mouse peritoneal macrophages (1.2 mg protein) were immunoprecipitated with anti-Rac3 (IP) or with preimmune serum (PI) and blotted with the anti-Rac monoclonal antibody. No Rac3 could be detected in platelets and macrophages. Ub, unbound material after immunoprecipitation (250 μg/lane); Ly, lysate (100 μg/lane).
We have previously shown that the Rac3 protein is detectable only in brain among a number of organs obtained from P7 mice (6), while it is undetectable in other tissues, including the thymus. Here, we tested the expression of Rac3 in different adult organs and detected some Rac3 protein only in the brain (Fig. 3C). We also checked for the expression of Rac3 in human platelets and mouse peritoneal macrophages, and we were unable to detect any protein by immunoprecipitation (Fig. 3D). This finding is in agreement with previous findings showing that Rac3 is not expressed in mouse bone marrow-derived macrophages (50). In general, we have so far been unable to detect the endogenous Rac3 protein in cells or tissues other than the nervous system, although low levels of RNA can often be detected by reverse transcription-PCR (data not shown).
To disrupt the Rac3 gene, we have replaced the portion corresponding to Rac3 with the bacterial lacZ gene (Fig. 1A), with the aim of studying the expression of β-galactosidase in knockout animals. Unfortunately, we have been unable to uti-lize the expression of β-galactosidase to detect the localization of the transcript of the mutant gene in knockout animals, probably due to inefficient expression of the mutant gene. In fact, while a weak band of the expected size was detected by reverse transcription-PCR with oligonucleotides amplifying the 3′ untranslated region of Rac3 or a region of the bacterial lacZ gene, no signal was detectable by Northern blotting or by lacZ staining on sections or whole-mount preparations of knockout embryos (data not shown).
Genotype distributions at birth matched expected Mendelian ratios as a result of intercrossing heterozygotes, indicating that disruption of Rac3 did not result in embryonic lethality. Rac3-null mice were indistinguishable from their wild-type and heterozygous littermates in size, weight, and external appearance.
To investigate the effect of Rac3 on both male and female fertility, a number of matings between knockout animals were performed. Thirty-six litters from five Rac3−/− females mated with knockout males revealed that both knockout males and females were fertile and that litter size and average number of pups were not significantly different from those observed in wild-type-only matings. The time interval between the addition of a knockout male and the first litter was comparable between wild-type and knockout females, suggesting normal mating behavior.
Analysis of Rac3 expression by in situ hybridization.
Previous findings have shown that expression of avian and mouse Rac3 is developmentally regulated in the nervous system of chicken embryos and mouse brain, respectively (6, 29). We have previously shown staining with the Rac3 antibody in the cerebellum of P7 mice that was specific according to a number of controls (6). Unfortunately, we found that this antibody still gave a signal in the cerebellum of knockout animals (data not shown). Therefore, although the specificity of our antibody was confirmed by the biochemical analysis shown in this study (see Fig. 3), we have to hypothesize that a cross-reacting antigen is recognized by the antibody under the conditions utilized for immunofluorescence on sections in mouse brain.
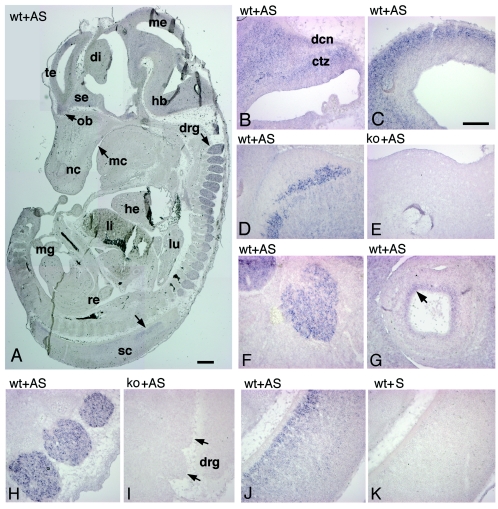
Here, we used in situ hybridization with an antisense RNA probe specific for mouse Rac3 to analyze in more detail the distribution of the Rac3 transcript in E13 and P7 mice. Parasagittal sections of E13 mouse embryos hybridized with antisense digoxigenin-labeled cRNAs corresponding to Rac3 were examined for signal detection. Rac3 mRNA was specifically and widely expressed in the developing nervous system of E13 mouse embryos (Fig. 4A). Rac3 was evident in several areas of the brain, including developing cerebellum (Fig. 4B), mesencephalon and pretectum (Fig. 4C), medulla oblungata (Fig. 4D), and the inner layer of the developing retina (Fig. 4G). A prominent signal was also detected at this stage in trigeminal ganglia, in dorsal root ganglia, and in the ventral part of the spinal cord (Fig. 4F, H, and J, respectively). Inspection of other organs revealed no evident signal for Rac3 at this stage. The specificity of the signal was confirmed by the absence of a signal using the corresponding Rac3 sense probe on sections of wild-type animals (Fig. 4K) and by the lack of the signal in Rac3-null embryos incubated with the antisense probe (Fig. 4E and I).
FIG. 4.
Expression of Rac3 mRNA in E13 mice. Parasagittal sections of E12.5 to E13 wild-type (wt) and knockout (ko) mouse embryos were hybridized with antisense (AS) or sense (S) digoxigenin-labeled cRNAs for Rac3 and examined for signal detection. (A) Rac3 is specifically and widely expressed in the developing nervous system. (B) Cerebellum; (C) mesencephalon; (D and E) medulla oblongata; (F) trigeminal ganglion; (G) eye; (H and I) dorsal root ganglia; (J and K) spinal cord. Abbreviations: ctz, cortical transitory zone; dcn, deep cerebellar nuclei; di, diencephalon; drg, dorsal root ganglia; hb, hindbrain; he, heart; li, liver; lu, lung; mc, mouth cavity; me, mesencephalon; mg, midgut; nc, nasal cavity; ob, olfactory bulb; re, rectum; sc, spinal cord; se, septum; te, telencephalic vesicle. Bars: 500 μm (A); 200 μm (B to K).
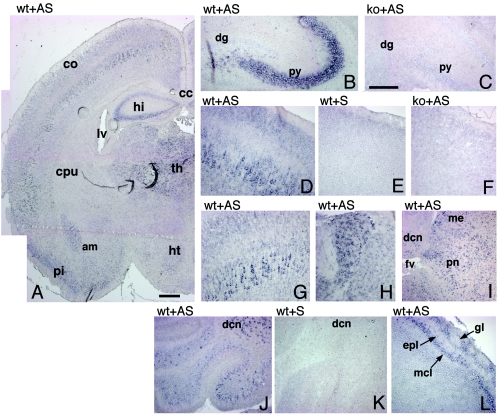
Previously, we detected the highest levels of Rac3 protein by immunoprecipitation from P7 mouse brain lysates (6). Here, mRNA in situ hybridization analysis revealed high levels of Rac3 mRNA in several regions of P7 mouse brain (Fig. 5). Rac3 was particularly evident in the hippocampus, with a strong signal in the CA1-CA3 region and a much weaker signal in the gyrus dentatus (Fig. 5B), while no signal could be detected in the hippocampus of Rac3-deficient mice (Fig. 5C). In the cerebral cortex, the signal was strongest in the pyramidal neurons of the developing fifth layer, but a weaker signal was also observed in the second and third layers (Fig. 5D to G). Rac3 mRNA was evident in several nuclei of the thalamus, in the amygdala (Fig. 5A and H), and in the pons (Fig. 5I), mesencephalon, and colliculi (data not shown). In the cerebellum, the strongest staining was observed in the deep cerebellar nuclei and in scattered cells in the internal granular layer, probably corresponding to Golgi type II cells, while a weaker signal could be detected in the Purkinje cell layer (Fig. 5J). Virtually all territories in which the gene is expressed contain projection neurons involved in long and complex neuronal networks. Notably, no white matter region ever showed a signal for Rac3, suggesting that the gene is not significantly expressed in oligodendrocytes. In the olfactory bulb, Rac3 was present in the mitral cell layer and in the glomerular layer (Fig. 5L).
FIG. 5.
Expression of Rac3 mRNA in P7 mouse brain. In situ hybridization on P7 mouse brain coronal (A, C, F to H) and parasagittal (B, D, E, I to L) sections from wild-type (wt) and knockout (ko) mice, using a Rac3 antisense (AS) or sense (S) probe. (A) Rac3 is specifically expressed in several areas of the P7 brain. (B and C) hippocampus; (D to G) cerebral cortex; (H) thalamus; (I) pons; (J and K) cerebellum; (L) olfactory bulb. Abbreviations: am, amygdala; cc, corpus callosum; co, cortex; cpu, caudatus-putamen; dcn, deep cerebellar nuclei; dg, dentate gyrus; epl, external plexiform layer; fv, fourth ventricle; gl, glomerular layer of olfactory bulb; hi, hippocampus; ht, hypothalamus; lv, lateral ventricle; mcl, mitral cell layer; me, mesencephalon; pi, piriform cortex; pn, pons; py, pyramidal cell layer of the hippocampus; th, thalamus. Bars: 500 μm (A); 200 μm (B to L).
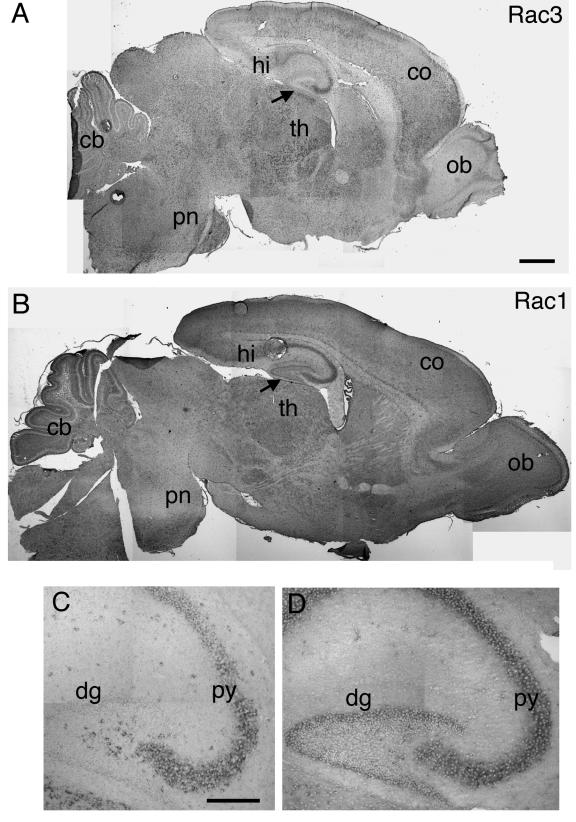
Previous studies have shown that Rac1 is ubiquitously expressed in the adult rat brain and in the developing mouse brain (6, 44). Moreover, in situ analysis has shown that the Rac1 transcript is present in both CA1-CA3 and dentate gyrus of the P8 rat hippocampus (34). Here we have shown by in situ analysis that Rac1 was quite homogenously expressed in P7 mouse brain (Fig. 6B). We also confirmed the expression of Rac1 in both CA1-CA3 and the dentate gyrus of the hippocampus (Fig. 6B and D), in contrast to Rac3, which showed the highest expression specifically in the CA1-CA3 region but was absent from the dentate gyrus (Fig. 6A and C).
FIG. 6.
Expression of Rac1 and Rac3 mRNA in P7 mouse brain. In situ hybridization on P7 mouse brain parasagittal sections from wild-type mice, using Rac3 (A and C) or Rac1 (B and D) antisense probes, as detailed in the text. Abbreviations: cb, cerebellum; co, cortex; dg, dentate gyrus; hi, hippocampus; ob, olfactory bulb; pn, pons; py, pyramidal cell layer of the hippocampus; th, thalamus. Arrows in A and B indicate an evident difference in labeling in the dentate gyrus between Rac3 and Rac1. Bars: 500 μm (A and B); 200 μm (C and D).
Histopathological and immunohistochemical analysis of Rac3-deficient mice.
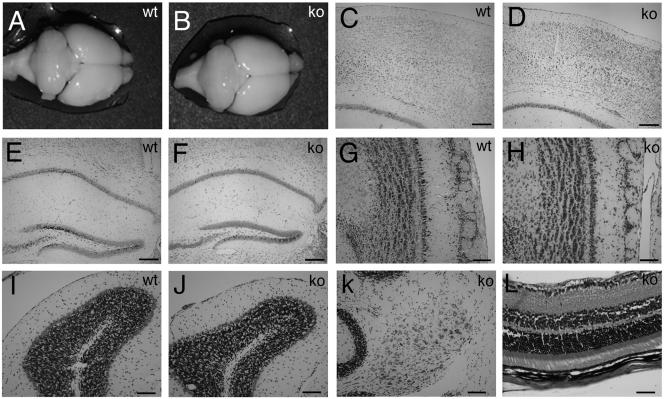
The gross morphology of adult brains from Rac3 knockout animals did not differ from that of wild-type brains (Fig. 7A and B). A closer histological inspection of distinct regions of the brain both at P7 (not shown) and in the adult (Fig. 7) revealed no major abnormalities. From the analysis of cresyl violet-stained sections, the structure and layering of the cortex were indistinguishable between wild-type and _Rac3_-null mice (Fig. 7C and D). No evident differences could be detected in the organization of the hippocampus, which showed the characteristic cytoarchitecture with a fully shaped dentate gyrus (Fig. 7E and F). The olfactory bulb of adult Rac3 knockout mice appeared normal, with no noticeable changes in the different layers (Fig. 7G and H). Foliation of the cerebellum was normal, and the molecular, Purkinje cell, and granule cell layer as well as the deep cerebellar nuclei were present (Fig. 7I to K). The eye and the stratification of the retina were normal (Fig. 7L). Taken together, these results show that Rac3 is dispensable for the normal macroscopic development of the brain.
FIG. 7.
Histology of wild-type and knockout adult brains. Macroscopic analysis of brains from 3-month-old wild-type (A) and knockout (B) mice revealed no major abnormalities. Coronal sections stained with cresyl violet of brains through cerebral cortex (C and D), hippocampus (E and F), olfactory bulbs (G and H), cerebellar cortex (I and J), and deep cerebellar nuclei (K) showed normal cell layering and an unaltered cytoarchitecture in both wild-type and knockout mice. (L) Section through knockout adult retina stained with hematoxylin and eosin, showing normal layering. Bars: 200 μm (C to F), 100 μm (G to K), 50 μm (L).
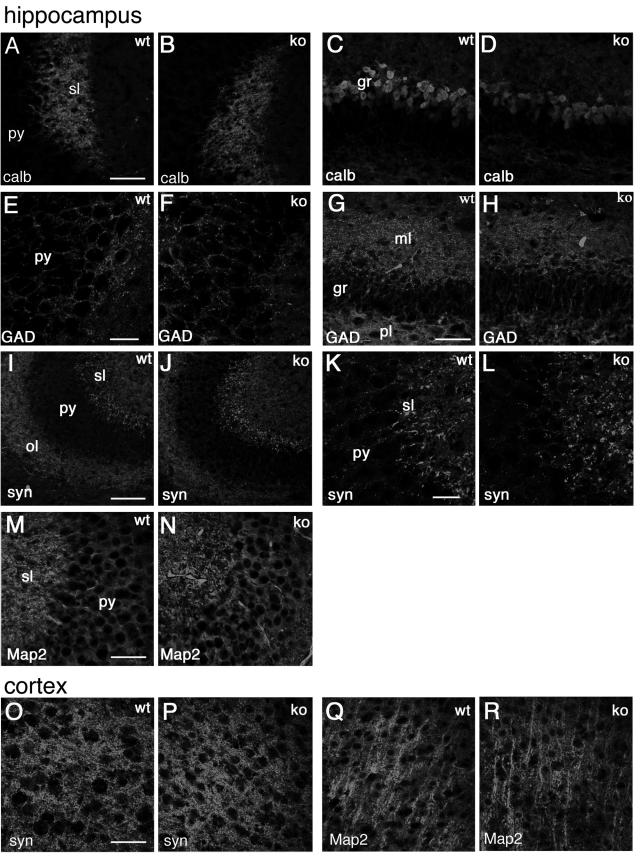
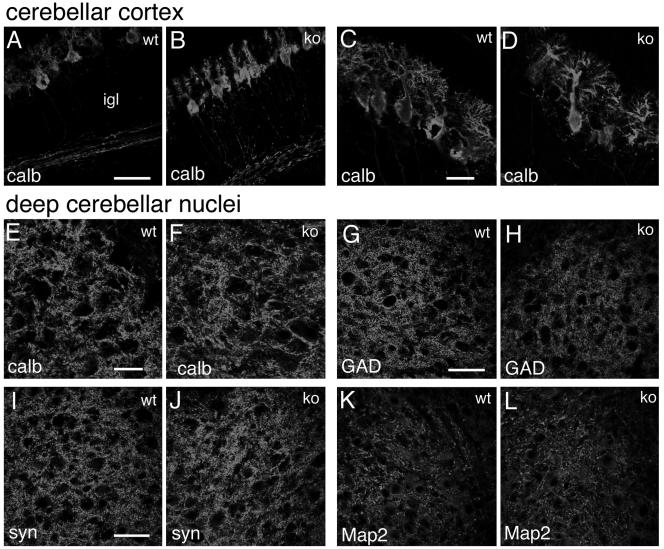
To look more carefully into possible alterations in the organization of the brain of Rac3 knockout animals, we performed a careful immunohistochemical analysis using a number of neuronal markers specific for axons and dendrites, such as calbindin and Map2 (22, 30) or, for synaptic sites, synapsin I and GAD65/67 (43, 46). Representative data from this analysis are shown, including some of the regions with high levels of Rac3 transcript expression (Fig. 4 to 6), such as the hippocampus (Fig. 8A to N), the cerebral cortex (Fig. 8O to R), and the cerebellum with deep cerebellar nuclei (Fig. 9). The general conclusion is that no detectable differences could be observed at this level of analysis in the general organization of the neurites and of synapses.
FIG. 8.
Distribution of neuronal markers in the hippocampus and in the cortex of P7 mice. Immunofluorescence on P7 brain coronal sections from wild-type (wt) and knockout (ko) mice, incubated with the indicated primary antibodies, and Alexa-488-labeled secondary antibodies. (A to N) Hippocampus; (O to R) fourth to sixth layers of the cerebral cortex. Abbreviations: gr, granular layer of dentate gyrus; ml, molecular layer of dentate gyrus; ol, oriens layer of the CA3 region; sl, stratum lucidem; pl, polymorph layer of dentate gyrus; py, pyramidal cell layer of the CA3 region; calb, calbindin; syn, synapsin I; GAD, GAD65 and GAD67 staining. Bars: 100 μm (I and J); 50 μm (A to D, G, H, M, and N); 25 μm (E, F, K, and L).
FIG. 9.
Distribution of neuronal markers in the cerebellum of P7 mice. Immunofluorescence on P7 cerebellar parasagittal sections from wild-type (wt) and knockout (ko) mice, incubated with the indicated primary antibodies and Alexa-488-labeled secondary antibodies. (A to D) Cerebellar cortex; (E to L) deep cerebellar nuclei. Abbreviations: P, Purkinje cells; igl, inner granular layer. Bars: 50 μm (A, B, and G to L); 25 μm (C to F).
Behavioral differences between _Rac3_-null and wild-type animals.
Our data show that Rac3 is dispensable for normal development. Rac3 shows a broad distribution throughout the nervous system during development. We performed a number of behavioral tests on 129/Sv _Rac3_-null mice to look for effects of the Rac3 mutation that may be caused by so far undetected alterations in the nervous system. Sensory abilities and motor functions were analyzed by a number of specific tests, including observation of home cage behaviors, footprint pathway, visual cliff, pain threshold (hot plate task), and rotarod motor coordination (11). We obtained some significant differences in the footprint pathway and in the hot plate test and quite striking differences with the rotarod test.
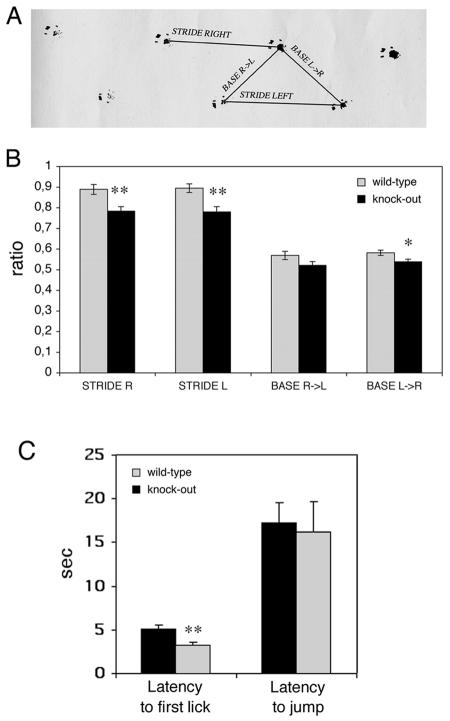
Gait abnormalities were assessed using footprint pattern analysis (8). Footprint analysis is a useful task to study slight locomotor impairments, which are generally undetectable in terms of motor properties (35). We detected a slight but significant difference between the hind-paw footprint pathways of the mutant (four females and five males) and wild-type (four females and five males) 24- to 28-week-old animals. The difference between wild-type and knockout mice could be detected for left and right strides (Fig. 10A). Both strides were decreased by 12% (P < 0,05) in Rac3-deficient mice compared to wild-type animals (Fig. 10B). The parameters obtained by footprint analysis provide an anatomical and locomotor index to parameters such as the functional efficiency of the sciatic nerve. In our case, analysis of the sciatic nerve from wild-type and knockout animals did not reveal any detectable abnormality (data not shown).
FIG. 10.
Behavioral characterization of wild-type and Rac3 knockout mice: analysis of footprint patterns and hot plate test. (A) Experimental set-up used for footprint analysis. Stride right, stride left, base R>L (right to left) and base L>R (left to right) indicate the four parameters considered. (B) Footprint patterns of 24- to 28-week-old wild-type (n = 9, grey bars) and knockout (n = 9, black bars) mice were quantitatively assessed for stride R, stride L, base R>L, and base L>R, as detailed in Materials and Methods. The mean values (± standard error of the mean) normalized by the body length are shown. Rac3 knockout animals had slightly decreased strides. (C) Ten wild-type and 10 knockout mice were placed on a hot plate at 55°C. Latency to first paw lick and latency to jump were recorded. Each bar shows the mean ± standard error of the mean. *, P < 0.05; **, P < 0.005.
In the hot plate test, 10 wild-type animals (five females and five males) and 10 knockout animals (five females and five males) were analyzed. We found a significant difference in the latency to first paw lick after placing the mice on the hot plate, with an average 60% increase in the latency measured in knockout animals compared to control wild-type mice (Fig. 10C). On the other hand, no significant difference was observed in the latency to jump between knockout and wild-type animals (Fig. 10C).
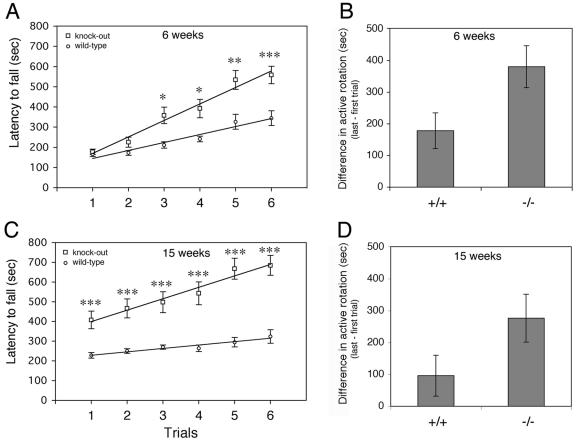
A more striking difference was observed with the accelerating rotarod test. The rotarod test is used to screen locomotor performance measuring motor coordination and balance control (16). Two groups of 10 wild-type animals (five females and five males) and knockout animals (five females and five males) about 6 weeks old were tested on the accelerating rotarod (Fig. 11A). Two trials, each consisting of three consecutive tests, were performed for 3 consecutive days, with 6 h of rest between the two trials on the same day. While the animals performed similarly during the first two trials (day 1), a significant difference was observed from the second day. Although both groups of animals showed improvement over the 3 days, Rac3 knockout animals performed significantly better from the third trial (day 2, P < 0,005). The difference between the wild-type and knockout mice became more evident on day 3 (P < 0,001).
FIG. 11.
Rotarod learning is improved in Rac3 knockout mice. (A) Time that mice remained on the rotarod before falling as a function of training session of 6-week-old animals. Open circles represent wild-type Rac3+/+ (n = 10) and open squares represent _Rac3_−/− mice (n = 10). (C) Performance on the accelerating Rotarod was tested on the same groups of mice at 15 weeks. Each point represents the mean of three tests. Lines of best fit had a correlation coefficient of 0.92 or greater. Error bars indicate standard error of the mean. Statistical significance in panels A and C: *, P < 0.005; **, P < 0.001; ***, P < 0,0005. (B) Motor learning for animals at 6 weeks and (D) 15 weeks of age. Bars represent differences in active rotation behavior (trial 6 − trial 1) for wild-type (+/+) and knockout (−/−) animals (P < 0.05 in panels B and D).
While performance on the first day gives information on the performance of “naive” mice, the time before the mouse falls in the final test is a measure of motor learning. Comparison of the differences between the last and the first trials in the two groups of animals showed a clear improvement in the learning ability of the Rac3 knockout mice (Fig. 11B). Interestingly, when the test was repeated with the same animals at 15 weeks of age, the Rac3-deficient mice showed a better performance even at the first trial (Fig. 11C, P < 0,001). Moreover, as for the first set of trials (Fig. 11A), the performance ability of Rac3-null mice increased more compared to that of wild-type animals, resulting in better motor learning (Fig. 11D). These findings suggest that Rac3 gene inactivation improves motor learning and memory retention of motor behaviors involving coordinated movements.
Typically, mice with structural abnormalities in the cerebellum or with disruptions in genes richly expressed in the cerebellum exhibit performance deficits on the rotarod (25, 41). On the other hand, the ability to perform this task cannot easily be attributed to a single brain region (10, 27). Recently, a few cases of improved rotarod performance after gene inactivation in mice have been described. Mice deficient for the histidine decarboxylase gene have histamine deficiency and show improved rotarod performance (14). These mice have altered acetylcholine concentrations in the frontal cortex and the neostriatum, which may be related to the observed behavioral changes. In the case of the Rac3 knockout, given the high expression of Rac3 in neurons at the time of synaptogenesis, one could postulate that alteration of the circuitry in regions important for the learning and memory of motor behaviors may be responsible for the observed altered behavior on the rotarod.
Interestingly, retinal degeneration may also play a role in performance on the rotarod, since mouse strains without retinal degeneration have a much shorter active rotation time (31), suggesting that vision influences performance on the rotarod, although the exact mechanism is unknown. Rac3 is expressed in the inner layer of the developing retina containing the retinal ganglion cells. Although histological inspection of the retina of knockout animals showed no evident alterations and the visual cliff utilized to test vision and depth perception in Rac3 knockout mice gave negative results, more sophisticated methods to measure visual ability will have to be tried.
Conclusions.
Most studies dealing with neuronal development in mammals have focused primarily on Rac1 (28). On the other hand, our in situ hybridization analysis indicates that Rac3 is specifically expressed in several populations of neural cells of the developing mouse. Intriguingly, deletion of Rac3 appears to have no major effects on morphogenesis and histogenesis in regions of the brain where it is normally expressed. While it is possible that Rac1 is able to functionally compensate for the lack of Rac3 during neuronal development, no evidence was observed for upregulation of Rac1 and Rac2 in Rac3-null mice. From the data presented here on Rac3-deficient mice, it is clear that in contrast to Rac1, Rac3 is not strictly required for normal development, and knockout mice survive embryogenesis.
Although no obvious anatomical abnormalities were detected in these mice, it is possible that subtler defects are present that may have important consequences for behavior and may explain the behavioral abnormalities described here. One interesting aspect is the elevated expression of Rac3 in projection neurons. Future studies will focus on the identification of morphological and functional differences in the fine organization of the neuronal networks. Moreover, in vitro studies with neuronal cultures will be required to establish possible deficits in neuronal development under more stringent environmental conditions. Given the interesting findings obtained with some of the behavioral tests described here, we will continue the examination of behavioral traits correlating with the expression patterns observed.
Acknowledgments
We thank Andras Nagy for the R1 ES cells and Lorenza Ronfani and the Core Facility for Conditional Mutagenesis at the San Raffaele Scientific Institute for helping with the screening of ES cells and the production of chimeric mice. We thank the following people for helpful discussion and assistance with this work: Ivana Benzoni, Annalisa Bolis, Patrizia D'Adamo, Laura Feltri, Cinzia Ferri, Alessandro Nodari, and Larry Wrabetz.
This work is supported by Telethon-Italy (grant GGP02190 to I.D.C.) and by the Italian Ministry of University and Research (MIUR) within the framework of project FIRB (FIRB RBNE01WY7P). Part of this work was carried out in Alembic, an advanced microscopy laboratory established by the San Raffaele Scientific Institute.
REFERENCES
- 1.Abdel-Latif, D., M. Steward, D. L. Macdonald, G. A. Francis, M. C. Dinauer, and P. Lacy. 2004. Rac2 is critical for neutrophil primary granule exocytosis. Blood 104**:**832-839. [DOI] [PubMed] [Google Scholar]
- 2.Abo, A., E. Pick, A. Hall, N. Totty, C. G. Teahan, and A. W. Segal. 1991. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature 353**:**668-670. [DOI] [PubMed] [Google Scholar]
- 3.Albertinazzi, C., D. Gilardelli, S. Paris, R. Longhi, and I. de Curtis. 1998. Overexpression of a neural-specific Rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons. J. Cell Biol. 142**:**815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ando, S., K. Kaibuchi, T. Sasaki, K. Hiraoka, T. Nishiyama, T. Mizuno, M. Asada, H. Nunoi, I. Matsuda, Y. Matsuura, P. Polakis, F. McCormick, and Y. Takai. 1992. Post-translational processing of Rac p21s is important both for their interaction with the GDP/GTP exchange proteins and for their activation of NADPH oxidase. J. Biol. Chem. 267**:**25709-25713. [PubMed] [Google Scholar]
- 5.Benvenuti, F., S. Hugues, M. Walmsley, S. Ruf, L. Fetler, M. Popoff, V. L. Tybulewicz, and S. Amigorena. 2004. Requirement of Rac1 and Rac2 expression by mature dendritic cells for T cell priming. Science 305**:**1150-1153. [DOI] [PubMed] [Google Scholar]
- 6.Bolis, A., S. Corbetta, A. Cioce, and I. de Curtis. 2003. Differential distribution of Rac1 and Rac3 GTPases in the developing mouse brain: implications for a role of Rac3 in Purkinje cell differentiation. Eur. J. Neurosci. 18**:**2417-2424. [DOI] [PubMed] [Google Scholar]
- 7.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116**:**167-179. [DOI] [PubMed] [Google Scholar]
- 8.Carter, R. J., L. A. Lione, T. Humby, L. Mangiarini, A. Mahal, G. P. Bates, S. B. Dunnett, and A. J. Morton. 1999. Characterization of progressive motor deficits in mice transgenic for the human Huntington's Disease mutation. J. Neurosci. 15**:**3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, H. B., E. N. Burright, W. S. Yunis, S. Larson, C. Wilcox, B. Hartman, A. Matilla, H. Y. Zoghbi, and H. T. Orr. 1997. Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by a progressive cerebellar dysfunction and histological alterations. J. Neurosci. 17**:**7385-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa, R. M., D. Cohen, and M. A. Nicodelis. 2004. Differential corticostriatal plasticity during fast and slow motor skill learning in mice. Curr. Biol. 14**:**1124-1134. [DOI] [PubMed] [Google Scholar]
- 11.Crawley, J. N. 1999. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 835**:**18-26. [DOI] [PubMed] [Google Scholar]
- 12.Croker, B. A., E. Handman, J. D. Hayball, T. M. Baldwin, V. Voigt, L. A. Cluse, F. C. Yang, D. A. Williams, and A. W. Roberts. 2002. Rac2-deficient mice display perturbed T-cell distribution and chemotaxis, but only minor abnormalities in T(H)1 responses. Immunol. Cell Biol. 80**:**231-240. [DOI] [PubMed] [Google Scholar]
- 13.Croker, B. A., D. M. Tarlinton, L. A. Cluse, A. J. Tuxen, A. Light, F. C. Yang, D. A. Williams, and A. W. Roberts. 2002. The Rac2 guanosine triphosphatase regulates B lymphocyte antigen receptor responses and chemotaxis and is required for establishment of B-1a and marginal zone B lymphocytes. J. Immunol. 168**:**3376-3386. [DOI] [PubMed] [Google Scholar]
- 14.Dere, E., M. A. De Souza-Silva, R. E. Spieler, J. S. Lin, H. Ohtsu, H. L. Haas, and J. P. Huston. 2004. Changes in motoric, exploratory and emotional behaviours and neuronal acetylcholine content and 5-HT turnover in histidine decarboxylase-KO mice. Eur. J. Neurosci. 20**:**1051-1058. [DOI] [PubMed] [Google Scholar]
- 15.Didsbury, J., R. F. Weber, G. M. Bokoch, T. Evans, and R. Snyderman. 1989. Rac, a novel ras-related family of proteins that are botulinum toxin substrates. J. Biol. Chem. 264**:**16378-16382. [PubMed] [Google Scholar]
- 16.Dunham, N. W., and T. S. Miya. 1957. A note on a simple apparatus for detecting neurological deficit in rats and mice. J. Am. Pharm. Assoc. 46**:**208-209. [DOI] [PubMed] [Google Scholar]
- 17.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420**:**629-635. [DOI] [PubMed] [Google Scholar]
- 18.Gu, Y., M. C. Byrne, N. C. Paranavitana, B. Aronow, J. E. Siefring, M. D'Souza, H. F. Horton, L. A. Quilliam, and D. A. Williams. 2002. Rac2, a hematopoiesis-specific Rho GTPase, specifically regulates mast cell protease gene expression in bone marrow-derived mast cells. Mol. Cell. Biol. 22**:**7645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, Y., M. D. Filippi, J. A. Cancelas, J. E. Siefring, E. P. Williams, A. C. Jasti, C. E. Harris, A. W. Lee, R. Prabhakar, S. J. Atkinson, D. J. Kwiatkowski, and D. A. Williams. 2003. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science 302**:**445-449. [DOI] [PubMed] [Google Scholar]
- 20.Haataja, L., J. Groffen, and N. Heisterkamp. 1997. Characterization of RAC3, a novel member of the Rho family. J. Biol. Chem. 272**:**20384-20388. [DOI] [PubMed] [Google Scholar]
- 21.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279**:**509-514. [DOI] [PubMed] [Google Scholar]
- 22.Jande, S. S., L. Maler, and D. E. Lawson. 1981. Immunohistochemical mapping of vitamin D-dependent calcium-binding protein in brain. Nature 294**:**765-767. [DOI] [PubMed] [Google Scholar]
- 23.Kinsella, B. T., R. A. Erdman, and W. A. Maltese. 1991. Carboxyl-terminal isoprenylation of ras-related GTP-binding proteins encoded by rac1, rac2, and ralA. J. Biol. Chem. 266**:**9786-9794. [PubMed] [Google Scholar]
- 24.Knaus, U. G., P. G. Heyworth, T. Evans, J. T. Curnutte, and J. M. Bokoch. 1991. Regulation of phagocyte oxygen radical production by the GTP-binding protein Rac 2. Science 254**:**1512-1515. [DOI] [PubMed] [Google Scholar]
- 25.Lalonde, R., A. N. Bensoula, and M. Filali. 1995. Rotorod sensorimotor learning in cerebellar mutant mice. Neurosci. Res. 42**:**3-6. [DOI] [PubMed] [Google Scholar]
- 26.Lehrach, H., D. Diamond, J. M. Wozney, and H. Boedtker. 1977. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 16**:**4743-4751. [DOI] [PubMed] [Google Scholar]
- 27.Llinas, R., and J. P. Welsh. 1993. On the cerebellum and motor learning. Curr. Opin. Neurobiol. 9**:**58-65. [DOI] [PubMed] [Google Scholar]
- 28.Luo, L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1**:**173-180. [DOI] [PubMed] [Google Scholar]
- 29.Malosio, M. L., D. Gilardelli, S. Paris, C. Albertinazzi, and I. de Curtis. 1997. Differential expression of distinct members of Rho family GTP-binding proteins during neuronal development: identification of Rac1B, a new neural-specific member of the family. J. Neurosci. 17**:**6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matus, A. 1990. Microtubule-associated proteins and the determination of neuronal form. J. Physiol. (Paris) 84**:**134-137. [PubMed] [Google Scholar]
- 31.McFadyen, M. P., G. Kusek, V. J. Bolivar, and L. Flaherty. 2003. Differences among eight inbred strains of mice in motor ability and motor learning on a rotorod. Genes Brain Behav. 2**:**214-219. [DOI] [PubMed] [Google Scholar]
- 32.McIntosh, J. M., G. O. Corpuz, R. T. Layer, J. E. Garrett, J. D. Wagstaff, G. Bulaj, A. Vyazovkina, D. Yoshikami, L. J. Cruz, and B. M. Olivera. 2000. Isolation and characterization of a novel conus peptide with apparent antinociceptive activity. J. Biol. Chem. 275**:**32391-32397. [DOI] [PubMed] [Google Scholar]
- 33.Moll, J., G. Sansig, E. Fattori, and H. van der Putten. 1991. The murine rac1 gene: cDNA cloning, tissue distribution and regulated expression of rac1 mRNA by disassembly of actin microfilaments. Oncogene 6**:**863-866. [PubMed] [Google Scholar]
- 34.Nakayama, A. Y., M. B. Harms, and L. Luo. 2000. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20**:**5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norreel, J. C., M. Jamon, G. Riviere, E. Passage, M. Fontes, and F. Clarac. 2001. Behavioral profiling of a murine Charcot-Marie-Tooth disease type 1A model. Eur. J. Neurosci. 13**:**1625-1634. [DOI] [PubMed] [Google Scholar]
- 36.Olson, M. F., A. Ashworth, and A. Hall. 1995. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 269**:**1270-1272. [DOI] [PubMed] [Google Scholar]
- 37.Pringle, N. P., W.-P. Yu, M. Howell1, J. S. Colvin, D. M. Ornitz, and W. D. Richardson. 2003. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development 130**:**93-102. [DOI] [PubMed] [Google Scholar]
- 38.Ridley, A. J., H. F. Paterson, C. L. Johnston, D. Diekmann, and A. Hall. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70**:**401-410. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, A. W., C. Kim, L. Zhen, J. B. Lowe, R. Kapur, B. Petryniak, A. Spaetti, J. D. Pollock, J. B. Borneo, G. B. Bradford, S. J. Atkinson, M. C. Dinauer, and D. A. Williams. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 10**:**183-196. [DOI] [PubMed] [Google Scholar]
- 40.Shirsat, N. V., R. J. Pignolo, B. L. Kreider, and G. Rovera. 1990. A member of the ras gene superfamily is expressed specifically in T, B and myeloid hemopoietic cells. Oncogene 5**:**769-772. [PubMed] [Google Scholar]
- 41.Storm, D. R., C. Hansel, B. Hacker, A. Parent, and D. J. Linden. 1998. Impaired cerebellar long-term potentiation in type I adenylyl cyclase mutant mice. Neuron 1**:**199-210. [DOI] [PubMed] [Google Scholar]
- 42.Sugihara, K., N. Nakatsuji, K. Nakamura, K. Nakao, R. Hashimoto, H. Otani, H. Sakagami, H. Kondo, S. Nozawa, A. Aiba, and M. Katsuki. 1998. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 17**:**3427-3433. [DOI] [PubMed] [Google Scholar]
- 43.Takayama, C., and Y. Inoue. 2004. Morphological development and maturation of the GABAergic synapses in the mouse cerebellar granular layer. Brain Res. Dev. Brain Res. 150**:**177-190. [DOI] [PubMed] [Google Scholar]
- 44.Tanabe, K., T. Tachibana, T. Yamashita, Y. H. Che, Y. Yoneda, T. Ochi, M. Tohyama, H. Yoshikawa, and H. Kiyama. 2000. The small GTP-binding protein TC10 promotes nerve elongation in neuronal cells, and its expression is induced during nerve regeneration in rats. J. Neurosci. 20**:**4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valtorta, F., F. Benfenati, and H. Basudev. 1996. Role of protein kinases in nerve terminal maturation and function. Biochem. Soc. Trans. 24**:**645-653. [DOI] [PubMed] [Google Scholar]
- 46.Valtorta, F., R. Jahn, R. Fesce, P. Greengard, and B. Ceccarelli. 1988. Synaptophysin (p38) at the frog neuromuscular junction: its incorporation into the plasma membrane and recycling after intense quantal secretion. J. Cell Biol. 107**:**2719-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11**:**2295-2322. [DOI] [PubMed] [Google Scholar]
- 48.Walmsley, M. J., S. K. Ooi, L. F. Reynolds, S. H. Smith, S. Ruf, A. Mathiot, L. Vanes, D. A. Williams, M. P. Cancro, and V. L. Tybulewicz. 2003. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science 302**:**459-462. [DOI] [PubMed] [Google Scholar]
- 49.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:**520-562. [DOI] [PubMed] [Google Scholar]
- 50.Wells, C. M., M. Walmsley, S. Ooi, V. Tybulewicz, and A. J. Ridley. 2004. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 117**:**1259-1268. [DOI] [PubMed] [Google Scholar]
- 51.Yamauchi, A., C. Kim, S. Li, C. C. Marchal, J. Towe, S. J. Atkinson, and M. C. Dinauer. 2004. Rac2-deficient murine macrophages have selective defects in superoxide production and phagocytosis of opsonized particles. J. Immunol. 173**:**5971-5979. [DOI] [PubMed] [Google Scholar]