ARTS: a web-based tool for the set-up of high-throughput genome-wide mapping panels for the SNP genotyping of mouse mutants (original) (raw)
Abstract
Genome-wide mapping in the identification of novel candidate genes has always been the standard method in genetics and genomics to correlate a clinically interesting phenotypic trait with a genotype. However, the performance of a mapping experiment using classical microsatellite approaches can be very time consuming. The high-throughput analysis of single-nucleotide polymorphisms (SNPs) has the potential of being the successor of microsatellite analysis routinely used for these mapping approaches, where one of the major obstacles is the design of the appropriate SNP marker set itself. Here we report on ARTS, an advanced retrieval tool for SNPs, which allows researchers to comb freely the public mouse dbSNP database for multiple reference and test strains. Several filters can be applied in order to improve the sensitivity and the specificity of the search results. By employing the panel generator function of this program, it is possible to abbreviate the extraction of reliable sequence data for a large marker panel including several different mouse strains from days to minutes. The concept of ARTS is easily adaptable to other species for which SNP databases are available, making it a versatile tool for the use of SNPs as markers for genotyping. The web interface is accessible at http://andromeda.gsf.de/arts.
INTRODUCTION
The identification and understanding of genes and their respective function is the general aim of both modern genetics and genomics. The most common approach for the identification of aberrations on the genomic level is the genome-wide mapping of affected individuals and their non-affected relatives in order to identify the causative gene defect.
In particular, the large-scale _N_-ethyl-_N_-nitrosourea (ENU) mutagenesis screens set-up in the late 1990s produced a stock of several hundred mouse mutant lines showing an interesting clinical phenotype (1–3). However, the identification of possibly mutated candidate genes of such lines lags behind the production of new mutants if the genome-wide genotyping of individuals is not performed in a high-throughput manner. Usually, a set of microsatellite marker sequences is amplified by PCR and analyzed by agarose or PAGE. Although there have been some advances in terms of throughput and automation in the recent years (4), technologies designed for high-throughput single-nucleotide polymorphism (SNP) analysis, e.g. MALDI-TOF MS and Taqman genotyping (5,6), are promising as replacements of microsatellite marker analysis in the genome-wide genotyping of several hundred animals per month.
However, the generation of assays for mapping panels consisting of one or several hundred SNP markers and their flanking sequences is not an easy task when using the established public or proprietary search engines for mouse SNPs. Their input masks allow the search for SNPs that are either reported for one strain (EnsMart: http://www.ensembl.org/Multi/martview), or that are reported as polymorphic between two strains (Roche MouseSNP database: http://mousesnp.roche.com/cgi-bin/msnp.pl; and Celera MyScience: https://myscience.appliedbiosystems.com) or that are polymorphic between one reference strain and several outcross strains (NCBI mouse dbSNP: http://www.ncbi.nlm.nih.gov/SNP/MouseSNP.cgi). Although these tools deliver valuable results, they lack some flexibility necessary for an efficient set-up of advanced SNP mapping panels like a free definition of the number of strains evaluated for polymorphisms, or some additional filters to increase the confidence of the query results. Moreover, they do not offer an automated approach for the generation of mapping panels, and the output format of their search hits usually cannot be inserted into spreadsheets or primer design software without post-processing. Nevertheless, all these demands are necessary for a reliable design of SNP marker panels in silico with a reduced amount of validation in vitro and redesign of assays that failed due to the lack of polymorphisms or inconsistency of the flanking sequences.
ARTS (Advanced Retrieval Tool for SNPs) has been designed to fulfill all of these demands. It offers the retrieval of SNPs that are polymorphic between several different mouse reference and outcross strains as well as the automatic generation of marker panels consisting of nucleotide sequences and clearly assignable SNP alleles. Employing several filter parameters facilitates the improved chances of identifying candidate SNP markers in order to validate their polymorphic state. The program is designed to be most comprehensive for the researcher working in the field of genetics and genomics.
THE GENERATION OF SNP ASSAYS WITH ARTS
Generating and exporting results from ARTS requires filling out two forms.
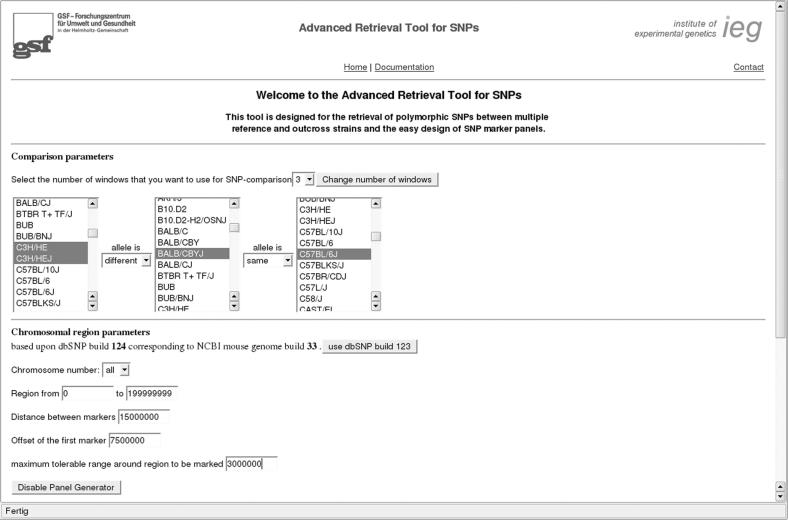
In the first form, the researcher can open up to six selection windows needed for strain comparisons and define their polymorphic relationship by using drop down menus. For more flexibility, it is also allowed to select several strains per window (Figure 1).
Figure 1.
The main input screen of the ARTS search engine.
The target chromosome and region are set afterwards. If the ‘Panel Generator’ feature is enabled, the distance between each single marker and the offset of the first marker relative to the starting point as well as the tolerable radius around each marker region can be defined.
Then, the additional filter parameters are set. If desired, indels, SNPs with no reported genotype, SNPs with more than two genotypes or SNPs with an unclear genotype association (e.g. C/G or A/T reported on both the plus and the minus strands) can be filtered. Further filter options are the minimum number of genotyped animals per SNP and the minimum number of submissions reporting that SNP. The next filter option defines the maximum percentage of sequencing errors and masked sequences within the 5′ and 3′ flanking regions 20 bp of the SNP.
In the second form, the ARTS displays the retrieved SNPs along with a preview of their flanking sequences (Figure 2). The first nucleotide within the brackets always represents the allele of the reference strains whereas the second one represents the allele of the outcross strains. This facilitates the assignment of alleles to their appropriate strains.
Figure 2.
The preview output screen of the ARTS search engine.
The researcher can then choose the data that will be included in the final output screen. It has to be noted that ARTS strictly relies on the original sequence data found in the FASTA files on the NCBI's FTP server (ftp.ncbi.nlm.nih.gov/snp/mouse/rs_fasta); therefore, the sequence length might be shorter than requested if they have not been originally submitted.
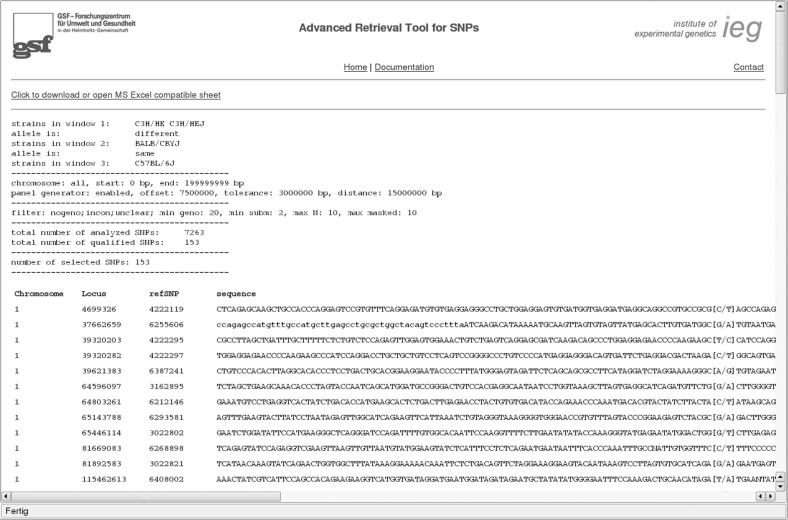
The data from the final output screen can be copied and pasted into the appropriate target application. In addition, a file is provided that can be imported into a spreadsheet (Figure 3).
Figure 3.
The final output screen of the ARTS search engine.
The program is written completely in PERL (www.perl.org) and makes use of the additional DBI, XML and CGI modules that can be obtained from www.cpan.org. It consists of a parser script building a MySQL database (www.mysql.com) out of the genotype XML files on the NCBI's FTP server (ftp.ncbi.nlm.nih.gov/snp/mouse/genotype) together with the FASTA files mentioned above, and a web interface for database access. The interface is accessible at andromeda.gsf.de/arts. A copy of the scripts can be obtained upon request.
ARTS IN PRACTICE: GENERATION OF A WHOLE-GENOME MAPPING PANEL/PROOF OF PRINCIPLE
The standard strain used in our laboratory for ENU injection is the C3HeB/FeJ strain. Owing to the lack of data for this strain in the dbSNP, we have decided to evaluate the datasets of its closest strain family members C3H/HeJ and C3H/He instead. Outcrossing and backcrossing is mostly performed on the C57BL/6J strain, and for lines showing low penetrance on the C57BL/6J background, the BALB/cByJ strain, which is closely related to the C3HeB/FeJ strain, is used.
Genome-wide mapping is performed with a set of markers placed every 15 Mb on all chromosomes with the first marker placed at a distance of 7.5 Mb from the centromere. The maximum tolerable range around each marker region is set to 3 Mb resulting in a 6 Mb region around each marker location where most probable SNPs are retrieved.
Entering all these data into the ARTS advanced search engine with the standard filter options results in a preselection of 153 SNPs that can be exported by the researcher. By decreasing the level of confidence the number of retrieved SNPs can be increased dramatically. For instance, if the minimum number of submissions is decreased to one, 558 SNPs are found in this example and if the number of genotyped individuals is additionally decreased to one, 887 SNPs are found in this example.
By using the concept of ARTS, we have currently expanded our genome-wide SNP mapping panel by 49 markers that have been proved polymorphic among C3HeB/FeJ, C57BL6/J and BALB/cByJ strains.
Altogether five lines have been mapped so far using this panel, which have passed the validation of the identified locus by complementing microsatellite marker analysis.
DISCUSSION
With the physical genome maps becoming more and more precise with each assembly and the recently initiated large-scale SNP discovery efforts in mice (7,8), the employment of SNPs as markers in conjunction with a high-throughput SNP genotyping technology is an alternative to the common mapping approaches based on microsatellite analysis.
One of the major advantages is the rapid identification of a candidate region by a whole-genome mapping approach with a relatively dense SNP marker panel. Depending on the technology for SNP genotyping and the amount of manpower, SNP markers and genotyped individuals, it is possible to identify candidate regions of several different backcrossed lines within a week.
However, the retrieval of SNPs in species other than human for a genome-wide mapping approach is a time-consuming task that requires some experience in data mining. ARTS provides more concise and easier building of SNP panels, and improves the chance of identifying SNPs that pass in vitro validation.
From our experience, a manual or semi-automatic identification of SNPs from the dbSNP database without applying the data filters of ARTS, results in a validation failure rate of ∼10–20% of all SNP assays because the reported polymorphisms could not be shown. One possible reason for this is the fact that the dbSNP contains large SNP data with an unclear validation status. An automatic sorting and prefiltering of the dbSNP entries in silico is a necessary step for building large marker panels of SNPs that will eventually pass the in vitro validation process.
By adapting the import filter of ARTS, it is possible to incorporate the concept of high-throughput SNP mapping to other scientifically relevant species, such as Rattus norvegicus, Danio rerio, Drosophila melanogaster and any other kind of research inbred species.
Acknowledgments
We thank Thomas Lisse for critically reading and commenting on this manuscript. Funding to pay the Open Access publication charges for this article was provided by GSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hrabe de Angelis M., Flaswinkel H., Fuchs H., Rathkolb B., Soewarto D., Marschall S., Heffner S., Pargent W., Wuensch K., Jung M., et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nature Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 2.Nolan P.M., Peters J., Strivens M., Rogers D., Hagan J., Spurr N., Gray I.C., Vizor L., Brooker D., Whitehill E., et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nature Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 3.Justice M.J. Capitalizing on large-scale mouse mutagenesis screens. Nature Rev. Genet. 2000;1:109–115. doi: 10.1038/35038549. [DOI] [PubMed] [Google Scholar]
- 4.Iakoubova O.A., Olsson C.L., Dains K.M., Choi J., Kalcheva I., Bentley L.G., Cunanan M., Hillman D., Louie J., Machrus M., et al. Microsatellite marker panels for use in high-throughput genotyping of mouse crosses. Physiol. Genomics. 2000;3:145–148. doi: 10.1152/physiolgenomics.2000.3.3.145. [DOI] [PubMed] [Google Scholar]
- 5.Jurinke C., Oeth P., van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol. Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 6.Shi M.M. Enabling large-scale pharmacogenetic studies by high-throughput mutation detection and genotyping technologies. Clin. Chem. 2001;47:164–172. [PubMed] [Google Scholar]
- 7.Wade C.M., Kulbokas E.J., III, Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 8.Wiltshire T., Pletcher M.T., Batalov S., Barnes S.W., Tarantino L.M., Cooke M.P., Wu H., Smylie K., Santrosyan A., Copeland N.G. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl Acad. Sci. USA. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]