Essential Role of Interferon Regulatory Factor 3 in Direct Activation of RANTES Chemokine Transcription (original) (raw)
Abstract
Localized and systemic cytokine production in virus-infected cells play an important role in the outcome of viral infection and pathogenicity. Activation of the interferon regulatory factors (IRF) in turn is a critical mediator of cytokine gene transcription. Recent studies have focused on the 55-kDa IRF-3 gene product as a direct transcriptional regulator of type 1 interferon (IFN-α and IFN-β) activation in response to virus infection. Virus infection induces phosphorylation of IRF-3 on specific C-terminal serine residues and permits cytoplasmic-to-nuclear translocation of IRF-3, activation of DNA binding and transactivation potential, and association with the CBP/p300 coactivator. We previously generated constitutively active [IRF-3(5D)] and dominant-negative forms of IRF-3 that control IFN-β and IFN-α gene expression. In an effort to characterize the range of immunoregulatory genes controlled by IRF-3, we now demonstrate that endogenous human RANTES gene transcription is directly induced in tetracycline-inducible IRF-3(5D)-expressing cells or paramyxovirus-infected cells. We also show that a dominant-negative IRF-3 mutant inhibits virus-induced expression of the RANTES promoter. Specific mutagenesis of overlapping ISRE-like sites located between nucleotides −123 and −96 in the RANTES promoter reduces virus-induced and IRF-3-dependent activation. These studies broaden the range of IRF-3 immunoregulatory target genes to include at least one member of the chemokine superfamily.
Virus infection of susceptible host cells activates a set of cellular genes, including interferons (IFNs), cytokines, and chemokines, involved in antiviral defense, cell growth regulation, and immune activation (36). In part, the molecular mechanisms by which pathogens induce expression of these cellular genes involves the activation of transcription regulatory proteins, such as the well-characterized members of the NF-κB family and the IFN regulatory factors (IRFs) (3, 24, 30). IRF-1 and IRF-2 are the best-characterized members of the IRF family, originally identified by studies of the transcriptional regulation of the human IFN-β gene (7, 8, 11, 17). Their discovery preceded the recent expansion of this group of IFN-responsive proteins which now includes seven other members: IRF-3, ISGF3γ/p48, ICSBP, Pip/ICSAT/IRF-4, IRF-5, IRF-6, and IRF-7 (24). Structurally, the Myb oncoproteins also have homology with the IRF family, although their relationship to the IFN system is unclear (35). Interestingly, virally encoded forms of IRF proteins were recently recognized in the genome of the human herpesvirus 8/Kaposi’s sarcoma herpes simplex virus, which contains four open reading frames encoding proteins showing homology to the cellular IRFs (18, 31).
The IRF-3 gene encodes a 55-kDa protein which is expressed constitutively in all tissues (2). Expression of the IRF-3 gene is not stimulated by virus infection or IFN treatment but demonstrates a unique response to viral infection. Recent studies with IRF-3 demonstrate that virus- and dsRNA-inducible phosphorylation represents an important posttranslational modification, leading to cytoplasmic to nuclear translocation of phosphorylated IRF-3, stimulation of DNA binding, and transcriptional activation of the type 1 IFN and IFN-responsive genes and association with the CREB binding protein (CBP)-p300 coactivator (14, 37–39).
It is becoming clear that IRF-3 is targeted by several different classes of viruses, not all with the same biological outcome. Most of our studies have utilized the RNA-containing paramyxoviruses Sendai and Newcastle disease virus as classical viral activators of IFN production. Human cytomegalovirus (CMV), a member of the beta herpesvirus family and an important human pathogen, was recently shown to cause transcriptional activation of the interferon-stimulated gene ISG-54 by a protein synthesis and signal transducer and activator of transcription (STAT) pathway-independent mechanism (21). Characterization of the CMV-induced IFN-stimulated response element binding factor (CIF) revealed that CIF was composed of IRF-3 and the CBP coactivator, but not p300 (21). In contrast to the activation of CIF by human CMV, adenovirus infection of human fibroblasts was able to downmodulate IRF-3-mediated transcriptional activity, an effect mediated by adenovirus E1A gene product (12). IRF-3 appeared in a completely different context, identified in a yeast two-hybrid screen as an interacting partner with human papillomavirus type 16 (HPV-16) E6 protein (29). Although the E6 oncoprotein may possess numerous functions, E6 has been extensively characterized as a viral product that targets the tumor suppressor p53 for ubiquitination and degradation by forming a ternary complex with the E6AP ubiquitin protein ligase. In the context of IRF-3, HPV-16 E6 did not result in ubiquitination or degradation of IRF-3; rather, E6 inhibited the transactivation function of IRF-3 and interfered with Sendai virus-mediated induction of IFN-β expression in primary keratinocytes (29).
These biological features implicate IRF-3 as one of the most important IRFs with regard to direct induction of the antiviral, growth-regulatory and immune-modulatory functions of the IFN system. Substitution of the phosphorylated serine residues with the phosphomimetic aspartic acid created a constitutively activated IRF-3 that alone was able to stimulate IFN-β expression as strongly as virus infection (14). We reasoned that IRF-3 may also play a role in the virus-mediated induction of other cytokines and chemokines (14). We now demonstrate that endogenous human RANTES (Regulated on Activation Normal T-cell Expressed and Secreted) gene transcription is directly induced by IRF-3 in tetracycline-inducible IRF-3(5D)-expressing cells or paramyxovirus-infected cells.
MATERIALS AND METHODS
Plasmid constructions and mutagenesis.
The wild-type (wt) and mutated forms of IRF-3-expressing plasmids were described previously (14). CMVt-IRF-3(5D) was constructed by cloning of IRF-3(5D) cDNA downstream of the doxycycline (Dox)-responsive promoter CMVt at the _Bam_HI site of the neo CMVt BL vector (25). The RANTES promoter was amplified by PCR with Vent DNA polymerase from human 293 genomic DNA. The gene-specific primers used for PCR were 5′-CCTTCCATGGATGAGGGAAAG-3′ and 5′-CATGGTACCTGTGGGAGAGGC-3′. This 483-bp PCR product was digested with _Pst_I and cloned into pCAT-Basic vector (Promega) by using _Hin_dIII (filled in with Klenow enzyme) and _Pst_I sites. Mutated forms of the RANTES promoter were generated by overlap PCR mutagenesis using Vent DNA polymerase with two internal primers as follows: 5′-CTATTTCAGTAAACTAAACCGTTTTGTG-3′ and 5′-CACAAAACGGTTTAGTTTACTGAAATAG-3′ (mutated nucleotides are underlined) for mutAB; 5′-CTATTTCAGTAAACTTTTCCGTTTTGTG-3′ and 5′-CACAAAACGGAAAAGTTTACTGAAATAG-3′ for mutA; 5′-CTATTTCAGTTTTCTAAACCGTTTTGTG-3′ and 5′-CACAAAACGGTTTAGAAAACTGAAATAG-3′ for mutB. The κB site-mutated RANTES promoter, RANTES-mutKB/CAT, was generated by PCR with primers 5′-CCTTCCATGGATGAGGGAAAG-3′ and 5′-TCCTCTGCAGCTCAGGCTGGCCCTTTAT AGGGCCAGTTGAGGTTCAAGGCCTAAGGCCTGTTAGCAAAATAGC AACCAAGC-3′ (27). Mutations were confirmed by sequencing. The IRF-7-expressing plasmid was a kind gift of Dimitris Thanos (Columbia University).
Generation of IRF-3 and IRF-3(5D) cell lines.
Plasmid CMVt-rtTA (25) was introduced into human 293 cells by the calcium phosphate method. Cells were selected beginning at 48 h after transfection for about 1 week in the alpha modification of Eagle’s medium (αMEM) (GIBCO-BRL) containing 10% heat-inactivated fetal bovine serum, glutamine, antibiotics, and 2.5 ng of puromycin (Sigma)/μl. Resistant cells carrying the CMVt-rtTA plasmid (rtTA-293 cells) were then transfected with the CMVt-IRF-3 and CMVt-IRF-3(5D) plasmids. Cells were selected beginning at 48 h for a period of approximately 2 weeks in αMEM containing 10% heat-inactivated calf serum, glutamine, antibiotics, 2.5 ng of puromycin/μl, and 400 μg of G418 (Life Technologies, Inc.)/ml. For generation of IRF-3 dominant-negative (ΔN) cells, IRF-3 (ΔN)/pEGFPC1 plasmid was introduced into human 293 cells by the calcium phosphate method, and cells were selected with G418 as described above.
Cell culture and transfections.
All transfections for a chloramphenicol acetyltransferase (CAT) assay were carried out in human embryonic kidney 293 or Jurkat T cells grown in αMEM (293) or RPMI 1640 (Jurkat) medium (GIBCO-BRL) supplemented with 10% fetal bovine serum, glutamine, and antibiotics. Subconfluent 293 cells were transfected with 5 μg of CsCl purified CAT reporter and expression plasmids by the calcium phosphate coprecipitation method. Jurkat cells were transfected by electroporation. In some experiments, transiently transfected IRF-3-expressing cells were infected with Sendai virus (80 hemagglutinating units [HAU]/ml). For individual transfections, 5 μg (293) or 50 μg (Jurkat) of total protein extract was assayed for 2 h at 37°C. The CAT activity was normalized with cotransfected β-galactosidase, and all transfections were performed three to six times.
Western blot analysis of IRF-3.
To screen and characterize the kinetics of IRF-3 and IRF-3(5D) induction, the expressing cells were cultured in the presence of 1 μg of Dox (Sigma)/ml for various periods of time. Cells were then washed with phosphate-buffered saline (PBS) and lysed in 10 mM Tris-Cl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% Nonidet P-40 (NP-40), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 5 μg of leupeptin/ml, 5 μg of pepstatin/ml, and 5 μg of aprotinin/ml. To examine subcellular localization of the IRF-3(5D) protein, nuclear and cytoplasmic extracts were prepared from the IRF-3(5D)-expressing cells after induction with Dox for different periods of time. The cells were washed in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF) and were resuspended in buffer A containing 0.1% NP-40. The cells were then chilled on ice for 10 min before centrifugation at 10,000 × g. This procedure was performed twice to remove cytoplasmic contaminants in the nuclear extracts. After centrifugation, supernatants were kept as cytoplasmic extracts. The pellets were then resuspended in buffer B (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 5 μg of leupeptin/ml, 5 μg of pepstatin/ml, 5 μg of aprotinin/ml, 5 μg of spermine/ml, 5 μg of spermidine/ml). Samples were incubated on ice for 15 min before being centrifuged at 10,000 × g. Nuclear extract supernatants were diluted with buffer C (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 50 mM KCl, 0.5 mM DTT, 0.5 mM PMSF). Equivalent amounts of nuclear, cytoplasmic, or whole-cell extract (10 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% polyacrylamide gel. After electrophoresis, the proteins were transferred to Hybond transfer membrane (Amersham) in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol for 1 h. The membrane was blocked by incubation in PBS containing 5% dried milk for 1 h and then probed with IRF-3 antibody in 5% milk-PBS, at a dilution of 1:3,000. These incubations were done at 4°C overnight or at room temperature for 1 to 3 h. After four 10-min washes with PBS, membranes were reacted with a peroxidase-conjugated secondary goat anti-rabbit antibody (Amersham) at a dilution of 1:2,500. The reaction was then visualized with the enhanced chemiluminescence (ECL) detection system as recommended by the manufacturer (Amersham).
EMSA.
The GST-IRF-3(133) fusion protein was expressed and purified as described previously (32). Whole-cell extracts were prepared from 293 cells with or without infection with Sendai virus (80 HAU/ml). In some experiments, extracts were prepared from 293 cells transfected with pFLAG/CMV-2 vector, IRF-3/pFLAG, or IRF-3 (5D)/pFLAG (infected with Sendai virus or left untreated as indicated in individual experiments). Cells were washed in PBS and lysed in 10 mM Tris-Cl(pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, 0.5 mM PMSF, 10 μg of leupeptin/ml, 10 μg of pepstatin/ml, 10 μg of aprotinin/ml, 0.5 ng of chymostatin/μl, and 0.25 μM microcystin. Recombinant IRF-3 protein or whole-cell extracts were subjected to an electrophoretic mobility shift assay (EMSA) by using a 32P-labelled probe corresponding to the ISRE region of the RANTES promoter (wt, 5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′; mutAB, 5′-CTATTTCAGTAAACTAAACCGTTTTGTG-3′; mutA, 5′-CTATTTCAGTAAACTTTTCCGTTTTGTG-3′; and mutB, 5′-CTATTTCAGTTTTCTAAACCGTTTTGTG-3′) or the ISRE region of the ISG-15 promoter (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′). The binding mixture (20 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM DTT, 5% glycerol, and 0.5% NP-40; 62.5 μg of poly(dI-dC)/ml was added to reduce nonspecific binding. After a 20-min incubation with probe, the resulting protein-DNA complexes were resolved on a 5% polyacrylamide gel and exposed to X-ray film. To demonstrate the specificity of protein-DNA complex formation, a 200-fold molar excess of unlabelled oligonucleotide was added to the cell extract before adding labelled probe or preincubated with anti-CBP antibody A-22 (Santa Cruz) and anti-FLAG antibody M2 (Sigma).
RPA and ELISAs.
For a ribonuclease protection assay (RPA), cells were induced with Dox for 48 h or for different lengths of time as indicated and were either left untreated, infected with Sendai virus (80 HAU/ml) for 16 h, treated with IFN-α/β, or treated with neutralizing antibody for type I IFN (Sigma). Total RNA was prepared from the cell pellets by using the Qiagen RNeasy Kit. Total RNA (5 μg) was subjected to an RPA using the hCK-5 chemokine template of RiboQuant multiprobe RPA kit in accordance with the manufacturer’s instructions (Pharmingen, San Diego, Calif.). Culture supernatants were collected and clarified by centrifugation at 5,000 × g for 10 min, and the supernatants were stored at −80°C until used for the enzyme-linked immunosorbent assay (ELISA). The concentration of secreted RANTES protein was determined by using the Human RANTES ELISA kit, following the manufacturer’s instructions (BioSource International, Camarillo, Calif.).
RESULTS
Tet-inducible IRF-3-expressing cells.
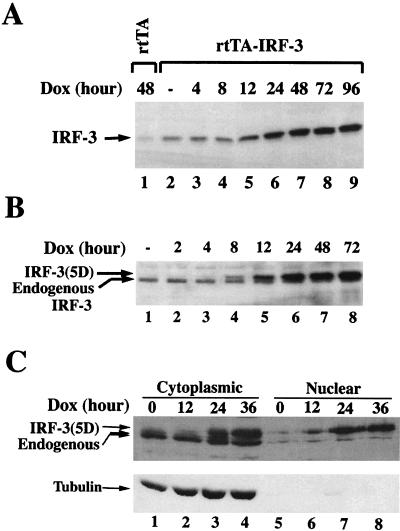
To begin to identify other cytokine-chemokine genes that may be regulated by the IRF-3 transcription factor, human embryonic kidney (293) cells inducibly expressing IRF-3 and IRF-3(5D) were created by using the reverse tTA activator (rtTA) (10, 25), which permits Dox-inducible expression of IRF-3 and IRF-3(5D). Individual clones of double transformants (20 from each transfection) were expanded and screened for protein expression by immunoblot analysis. Two individual clones from rtTA-IRF-3 or rtTA-IRF-3(5D) cells that possessed minimal uninduced protein levels and high Dox-inducible expression were chosen for further study. Figure 1 illustrates the kinetics of Dox-inducible transgene induction from one of each of these clones. Beginning at 12 h after induction in the wt IRF-3-expressing cell line (Fig. 1A, lane 5) and at 8 h in the IRF-3(5D)-expressing cell line (Fig. 1B, lane 4), Dox-induced transgene expression was detected by immunoblot analysis. Induced IRF-3(5D) migrated more slowly than endogenous IRF-3 at the position of phosphorylated IRF-3. Previously, IRF-3(5D) was shown to localize to both the cytoplasm and the nucleus in unstimulated cells (14). Using biochemical fractionation of cytoplasmic and nuclear extracts, Dox-induced IRF-3(5D)—detectable at 12 h after induction (Fig. 1C, lanes 2 and 6)—also partitioned to both the nucleus and cytoplasm (Fig. 1C, lanes 2 to 4 and 6 to 8), whereas the endogenous IRF-3 was almost exclusively cytoplasmic (Fig. 1C, lanes 1 to 4) as was the tubulin control. Thus, IRF-3(5D) stably expressed as a Tet-inducible protein in 293 cells also localized to both the nucleus and cytoplasm, as previously demonstrated by green fluorescent protein analysis in transiently transfected cells (14).
FIG. 1.
Inducible expression of IRF-3 and IRF-3(5D) in 293 cells. Whole-cell extracts (10 μg) prepared from rtTA, IRF-3 (A), and IRF-3(5D) (B) cells induced with Dox for 0 to 96 h were subjected to SDS-PAGE and transferred to nitrocellulose membrane. (C) To examine IRF-3(5D) protein subcellular localization, cytoplasmic and nuclear extracts (20 μg) from IRF-3(5D) cells induced with Dox for 0 to 36 h were prepared, subjected to SDS-PAGE, and transferred to nitrocellulose membrane. IRF-3 and IRF-3(5D) protein levels were detected by using a polyclonal IRF-3 antibody. IRF-3(5D) protein migrated slower than endogenous IRF-3 protein, at the position previously identified for C-terminal phosphorylated IRF-3 (14). To verify the purity of the cytoplasmic and nuclear extracts, extracts were probed with an α-tubulin monoclonal antibody (ICN Biomedicals).
Activation of RANTES transcription by IRF-3 and virus.
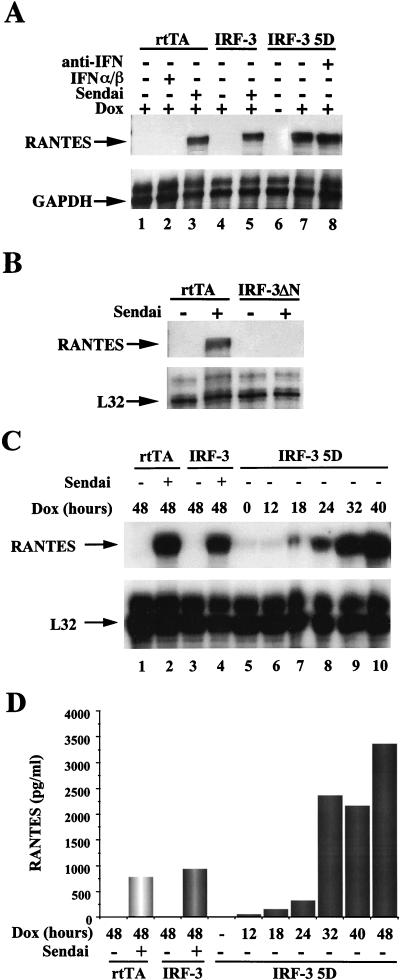
The IRF-3-inducible cells were next used to determine whether other cytokine-chemokine genes may be regulated by IRF-3; an RPA with multiple human cytokine-chemokine probes (Pharmingen) was used to examine RNA derived from rtTA-IRF-3 or rtTA-IRF-3(5D) cells. Strikingly, the RANTES gene was highly expressed in the IRF-3(5D)-inducible cells, as well as in virus-infected cells (Fig. 2A, lanes 3, 5, and 7) but not in uninfected rtTA- or wt IRF-3-expressing cells (Fig. 2A, lanes 1 and 4). Since IRF-3(5D) was a strong transactivator of the IFN-β promoter in transient transfection assays, the possibility of an autoregulatory effect of IFN-α/β expression on transcription of RANTES promoter via JAK-STAT activation was considered. Activation of RANTES did not occur secondary to the production of IFN-α/β, since RANTES mRNA was not detected in control rtTA-expressing cells treated directly with IFN-α/β (Fig. 2A, lane 2); furthermore, addition of neutralizing antibody directed against type I IFN did not block the stimulation of RANTES gene expression by IRF-3(5D) (Fig. 2A, lane 8). Other experiments also demonstrated that IRF-3 itself was not activated by IFN treatment (13a). Inducible expression of RANTES in cells stably expressing a dominant-negative form of IRF-3 which lacks the N-terminal amino acids 9 to 133 and does not bind to DNA was also examined. As shown in Fig. 2B, RANTES gene transcription was induced by Sendai virus in control (rtTA) cells (Fig. 2B) but not in IRF-3(ΔN)-expressing cells (Fig. 2B). This experiment indicates that a non-DNA binding, dominant-negative mutant of IRF-3 is able to block completely virus-induced activation of RANTES transcription.
FIG. 2.
IRF-3 inducible expression of RANTES gene. (A) Stimulation of RANTES gene transcription in virus-infected and IRF-3(5D)-expressing cells. The rtTA, IRF-3, and IRF-3(5D) cells were cultured in the presence or absence of Dox as indicated. After 30 h, cells were either left untreated, infected with Sendai virus (80 HAU/ml) for 16 h, or treated with IFN-α/β (100 IU/ml). The neutralizing antibody for type I IFN (Sigma) was added at the time of Dox addition. Total RNA was isolated from each sample and analyzed by RPA using the hCK5 kit (Pharmingen) as described in Materials and Methods. (B) Repression of virus-induced RANTES gene transcription by a dominant-negative form of IRF-3. The rtTA- and IRF-3(ΔN)-expressing cells were either left untreated or infected with Sendai virus (80 HAU/ml) for 16 h. Total RNA was isolated from each sample and analyzed by RPA. (C) The kinetics of RANTES expression induced by IRF-3(5D). Total RNA from IRF-3(5D)-expressing cells was isolated from each sample after Dox addition and analyzed by RPA. (D) Cell culture supernatants were analyzed for the presence of RANTES protein by an ELISA performed as specified by the manufacturer (Biosource International).
The kinetics of IRF-3 transgene induction and RANTES mRNA expression were characterized at various times following Dox induction. IRF-3(5D) was detected at 8 to 12 h with peak levels at 24 h following Dox addition (Fig. 1B). RANTES mRNA was first detectable at 18 h after Dox induction with peak levels at 40 h (Fig. 2C, lanes 5 to 10). Induction of RANTES protein expression as detected by ELISA (Fig. 2D) was first observed at 12 h after Dox induction of IRF-3, in good agreement with the mRNA levels, and accumulated thereafter with a dramatic increase between 24 and 32 h after stimulation, also in agreement with mRNA levels. The possibility that IRF-3(5D) may be directly activating another transcription factor, such as NF-κB, which in turn would stimulate RANTES transcription, was also considered. No evidence for IRF-3(5D)-mediated activation of NF-κB DNA binding activity was observed; similarly, IRF-3(5D) expression did not activate the human immunodeficiency virus (HIV)-long terminal repeat, a complex promoter controlled by NF-κB and other transcription factors (data not shown).
IRF-3 binds to overlapping ISRE-like elements in the RANTES promoter.
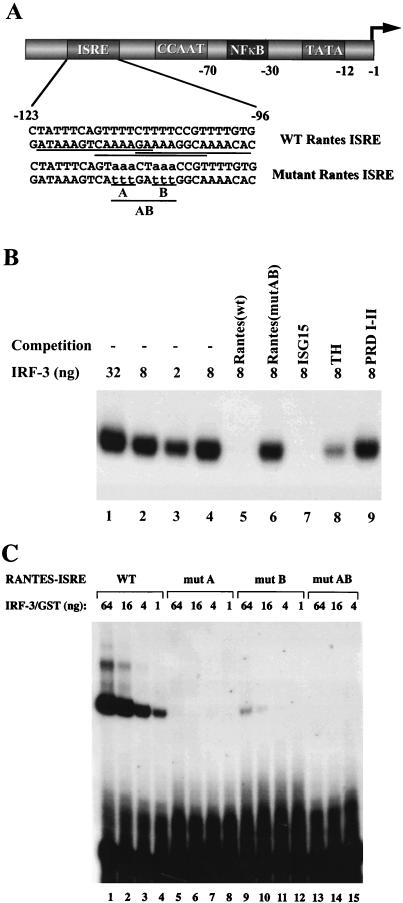
Several potential transcription factor binding sites were found within the region immediately upstream of human RANTES promoter (22), including three overlapping ISRE sites located between nucleotides −123 and −96 in the minus strand that share 11 of 14, 10 of 14, and 10 of 14 nucleotides with the consensus ISRE site (Fig. 3A). The binding of recombinant N-terminal IRF-3 to this region was examined by EMSA. IRF-3 bound strongly to the wt ISRE probe (Fig. 3B, lanes 1 to 3). This protein-DNA complex was specific since IRF-3 binding was efficiently competed with a 200-fold excess of unlabelled wt ISRE from the RANTES promoter and with the ISRE element from the ISG15 promoter and was partially competed with the synthetic tetrahexamer-TH (AAGTGA4) oligonucleotide (Fig. 3B, lanes 5, 7, and 8) but was unaffected by a 200-fold excess of mutated RANTES ISRE or the PRDI-PRDII element of the IFN-β promoter (Fig. 3B, lanes 7 and 9). Furthermore, IRF-3 failed to bind to oligonucleotides which contained mutations blocking all three ISRE sites (mutAB) or two upstream ISRE sites (mutA) (Fig. 3C, lanes 5 to 8 and 13 to 15) and bound only weakly to the oligonucleotide containing mutations in two downstream ISRE sites (Fig. 3C, lanes 9 to 12).
FIG. 3.
Binding of IRF-3 to ISRE-like sites in RANTES promoter. (A) The upstream sequence of RANTES gene contains putative ISRE sites. Numbers indicate the nucleotide positions relative to the transcriptional start site (22). The wild-type (wt) and mutated ISRE probes used for the EMSA are also shown. Three overlap putative ISRE sites are underlined. (B) Recombinant N-terminal IRF-3 binds to the RANTES ISRE probe. EMSA was performed with the indicated amounts of recombinant protein and 32P-labelled wt RANTES ISRE (as shown in panel A). For competition, 200-fold excess of unlabelled oligonucleotide was added as indicated. (C) EMSA was performed with the indicated amounts of recombinant protein and 32P-labelled wt RANTES and mutated ISRE probes (as shown in panel A).
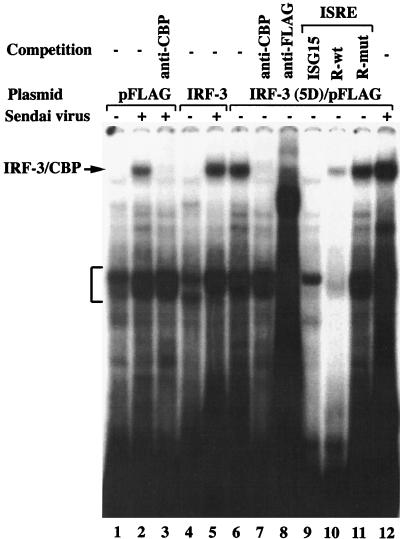
IRF-3 and IRF-3(5D) DNA binding activity was also examined by an EMSA using whole-cell extracts (10 μg) derived from 293 cells transfected with pFLAG-CMV2 vector, IRF-3/pFLAG, or IRF-3(5D)/pFLAG and either left untreated or infected with Sendai virus (Fig. 4). In virus-induced control or IRF-3/pFLAG-expressing cells, a new protein-DNA complex was identified by EMSA (Fig. 4, lanes 2 and 5). This protein-DNA complex, which was previously characterized in detail (14, 38, 39), contained CBP coactivator as confirmed by supershift analysis with the A22 antibody (Fig. 4, lane 3). Strikingly, the same complex was present in untreated IRF-3(5D)/pFLAG-expressing cells (Fig. 4, lane 6), further demonstrating the constitutive DNA binding of the activated form of IRF-3. Supershift analysis demonstrated that this protein-DNA complex also contained IRF-3 and CBP (Fig. 4, lanes 7 and 8). Competition with excess ISRE elements from the RANTES or ISG-15 promoters successfully depleted the binding of the complex (Fig. 4, lanes 9 and 10), whereas mutated RANTES ISRE failed to compete for IRF-3(5D) binding activity (Fig. 4, lane 11).
FIG. 4.
Binding of IRF-3 to ISRE-like sites in RANTES promoter. An EMSA was performed using whole-cell extracts (20 μg) derived from control 293 cells transfected with pFLAG-CMV2, IRF-3/pFLAG, or IRF-3(5D)/pFLAG which was either left untreated or infected with Sendai virus. The 32P-labelled probe corresponds to the ISRE of the ISG-15 gene (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′). Anti-CBP antibody A22 and anti-FLAG antibody M2 were added as indicated to demonstrate the presence of CBP and IRF-3 in the high-molecular-weight protein-DNA complex, indicated by the arrow. The bracket indicates the position of IRF-2 binding to the probe. For oligonucleotide competition, a 200-fold excess of unlabelled oligonucleotide was added as indicated above the lanes.
Regulation of RANTES transcription by ISRE and NF-κB sites.
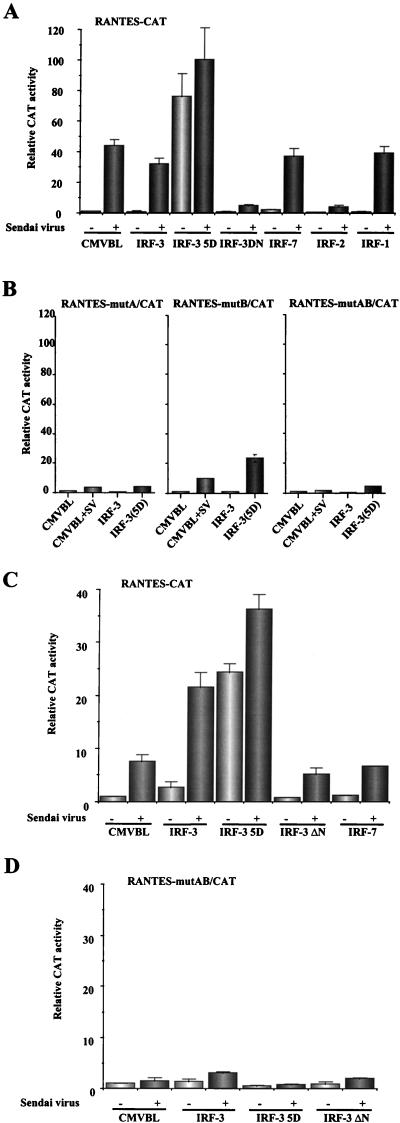
To further explore IRF-3 regulation of RANTES promoter activity, a 432-bp (nucleotides −425 to +7) sequence of the wt and mutated human RANTES gene promoter (22) was cloned into the pCAT-reporter vector (Promega), and uninduced and induced reporter gene activity after transient cotransfection into 293, Jurkat, and U937 cell lines was tested. In uninfected cells, both wt and mutated forms of the RANTES promoter had low basal activity (Fig. 5A). Virus infection resulted in a 40-fold induction of RANTES activity in 293 cells (Fig. 5A) and an eightfold induction in Jurkat cells (Fig. 5C). In contrast, the ISRE-mutated RANTES promoters were not activated upon Sendai virus infection (Fig. 5B and D). Cotransfection with the IRF-3(5D)-expressing plasmid stimulated expression of wt RANTES promoter up to 80-fold in 293 cells without virus infection (Fig. 5A); additional Sendai virus induction stimulated promoter activity in the IRF-3(5D)-expressing cells only slightly (Fig. 5A). Both the A and B mutations of the ISRE-like sites also blocked RANTES activation by virus and IRF-3(5D), again reflecting the requirement for an intact ISRE element (Fig. 5B). Interestingly, inducible expression of the RANTES promoter by Sendai virus infection was inhibited by cotransfection with a dominant-negative mutant of IRF-3 which lacks the DNA binding domain (14) or with the IRF-2 repressor (Fig. 5A). Two other IRF family members—IRF-1 and IRF-7—had no effect on expression of the RANTES promoter (Fig. 5A). A difference between the two cell types was observed with wt IRF-3 coexpression; in 293 cells, additional IRF-3 did not further stimulate Sendai virus activation of RANTES (Fig. 5A) whereas in Jurkat cells, IRF-3 overexpression more than doubled virus-induced activation of the RANTES promoter (Fig. 5C), possibly reflecting quantitative differences in the levels of IRF-3 in the two cell types.
FIG. 5.
Transactivation of the RANTES promoter by IRF-3. 293 (A and B) and Jurkat (C and D) cells were transfected with wt (A and C) or mutated (B and D) RANTES promoter-CAT reporter plasmids and various expression plasmids as indicated below each bar graph. At 24 h posttransfection, cells were infected with Sendai virus for 16 h or were left uninfected as indicated. CAT activity was analyzed at 48 h posttransfection with 5 μg (293) or 50 μg (Jurkat) of total protein extract for 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of CMV-B1 vector alone after normalization with cotransfected β-galactosidase activity). The values represent the averages of three to six experiments with standard deviations shown in the error bars.
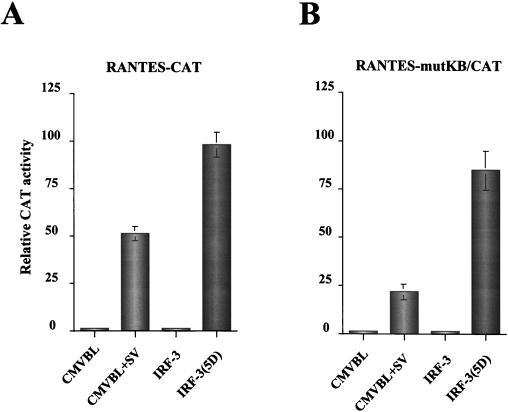
Two NF-κB sites contribute to RANTES transcriptional activation by stimuli such as phorbol myristate acetate-ionomycin, proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and interleukin 1 [IL-1]) or anti-CD3 and anti-CD28 antibodies (19, 27). The possible utilization of these two NF-κB sites in virus- and IRF-3-mediated activation of RANTES gene transcription was examined by analyzing the effects of NF-κB site mutations on RANTES induction. Mutation of the NF-κB sites reduced virus-activated RANTES promoter activity from 50- to 25-fold—an overall twofold reduction—indicating a small but significant role for the NF-κB sites in virus-induced RANTES expression (Fig. 6). However, mutation of the NF-κB sites had essentially no effect on the inducible expression of RANTES promoter by IRF-3(5D) (Fig. 6).
FIG. 6.
The effect of NF-κB site mutations on RANTES activation. 293 cells were transfected with wt (A) or NF-κB site-mutated (B) RANTES promoter-CAT reporter plasmids and various expression plasmids as indicated below each bar graph. At 24 h posttransfection, cells were infected with Sendai virus for 16 h or were left uninfected as indicated. CAT activity was analyzed at 48 h posttransfection with 5 μg of 293 protein extract for 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of CMV-B1 vector alone after normalization with cotransfected β-galactosidase activity). The values represent the averages of three experiments with standard deviations shown in the error bars.
DISCUSSION
In the present study, we demonstrate that infection with Sendai virus or expression of the constitutively activated phosphomimetic form of IRF-3 directly binds to and stimulates transcription of the human RANTES promoter via the overlapping ISRE elements. Mutation of the ISRE sites blocked the IRF-3(5D)-induced or virus-mediated expression from this promoter. Together with the repression of virus-induced expression of the RANTES gene by the dominant-negative IRF-3 mutant, these results demonstrate that IRF-3 plays a primary role in the virus-inducible activation of RANTES gene. Thus at least one member of the chemokine superfamily is also directly targeted by the virus-dependent, posttranslational activation of IRF-3.
Other members of the IRF family of transcription factors are potentially able to stimulate transcription from promoters containing ISRE-like elements, including IRF-1, IRF-7, and ISGF3γ/p48. ISGF3γ/p48 recognizes and binds to various ISREs but exerts its transcriptional activity exclusively in association with the STAT1 and STAT2 proteins, as part of the ISGF-3 complex (24, 37). Based on the present data, ISGF3 does not appear to regulate direct induction of RANTES. Transcription was not activated by type I IFN (Fig. 2A, lane 2) and in transient cotransfection assays, treatment with IFN-α/β or IFN-γ had little or no effect on the activity of the RANTES-CAT reporter (data not shown), indicating the JAK-STAT pathway and the ISGF3 complex do not contribute to RANTES activation in 293 cells. Similarly, in coexpression studies, IRF-1 and IRF-7 failed to play a major role in the activation of the RANTES promoter (Fig. 5A).
RANTES, an important inflammatory chemoattractant for monocytes, T cells and eosinophils (13, 33), is a member of CC-chemokine family (26, 28) and is expressed after cellular activation in fibroblasts, T cells, monocytes, endothelial cells, and certain epithelial cells (15). RANTES and CC-chemokines MIP-1α and MIP-1β were identified as the major HIV-suppressive factors released by cytotoxic-suppressor CD8+ T-lymphocytes (4), acting via the inhibition of macrophage-tropic but not T cell-tropic HIV-1 strains (6). It is now well recognized that the chemokine receptor CCR5—a seven-transmembrane, G-protein coupled receptor found on T cells and macrophages—functions as an HIV-1 fusion cofactor and binds RANTES, MIP-1α, and MIP-1β. Expression of CCR5 in the presence of CD4 confers permissiveness to membrane fusion by M-tropic virus strains (6). It has been recently demonstrated that RANTES also inhibits infection by certain T-tropic HIV-1 viruses (34).
RANTES is expressed relatively late after activation of peripheral blood T cells by antigen or mitogens but is rapidly induced in normal fibroblasts and epithelial cells by TNF-α and IL-1β, suggesting that different control mechanisms may regulate RANTES transcriptional activation (22). Among the multiple regulatory domains identified in the RANTES upstream promoter are two binding sites for the NF-κB transcription factors, located between positions −78 to −42 upstream from the transcription initiation site (19, 23, 27). Induction of NF-κB plays an important role in RANTES gene activation by stimuli such as PMA-ionomycin, proinflammatory cytokines (TNF-α and IL-1), or anti-CD3 and anti-CD28 antibodies (19); furthermore, NF-AT-like and a CD28RE-like motifs also serve as NF-κB binding sites (19). In contrast, the NF-κB sites in the murine RANTES promoter failed to play a significant role in virus-mediated activation of this gene (15). Sequence analysis of the murine RANTES promoter revealed that a number of regulatory motifs in human RANTES promoter are also present in murine RANTES promoter, including NF-κB and an IRF-1 response element (5). However, this study demonstrates for the first time the involvement of an IRF family member in the regulation of RANTES gene expression.
Viruses are known to acquire and modify genes of cellular origin to gain a survival advantage against host defenses. This evolutionary subversion by viruses also provides important clues concerning defense strategies that are critical to the generation of a successful immune response to particular viral pathogens. For example, several viral chemokine receptors have been discovered (reviewed in reference 20). The first functional viral chemokine receptor identified and characterized was ECRF3 of herpesvirus saimiri, the receptor for the CXC chemokines IL-8, GROα, and NAP-2 (1). A viral chemokine receptor encoded by the human CMV US28 gene binds the CC-chemokines RANTES, MIP-1α, and MIP-1β (9) and interferes with chemokine function. It has been suggested that herpesvirus saimiri and human CMV may use chemokine receptors to control viral replication by regulating cell cycle progression of the host cell or by inhibiting cellular apoptosis (20). In contrast, several poxvirus cytokine receptor homologues have been discovered that are secreted into the extracellular milieu (16). Poxviruses may use these receptors as secreted cytokine antagonists to scavenge host cytokines and block the generation of an antiviral defense cascade, mediated in part by cytokine receptor signalling.
In many respects, IRF-3 has a mode of activation similar to that of the NF-κB factors. In unstimulated cells, NF-κB heterodimers are retained in the cytoplasm by the inhibitory IκB proteins. Upon stimulation by many inducers, including cytokines, viruses, and dsRNA, IκBα is rapidly phosphorylated and degraded, resulting in the release of NF-κB (3). Thus, both IRF-3 and NF-κB are present in an inactive form localized to the cytoplasm in unstimulated cells. Virus-induced phosphorylation of IRF-3 at the C-terminal Ser-Thr cluster between amino acids 396 to 405 appears to alter the conformation of IRF-3 to permit nuclear translocation, DNA binding, transactivation, and interaction with the CBP/p300 coactivator (14, 21, 37–39). Subsequent degradation of IRF-3 by a phosphorylation-dependent, proteasome-dependent pathway (14) is also reminiscent of IκBα phosphorylation and degradation, which leads to the induction of NF-κB DNA binding activity (3). In this study, we demonstrated that IRF-3 plays an essential role in the virus-inducible activation of RANTES gene expression and the adjacent NF-κB sites also contribute, indicating that following virus infection, activated forms of IRF-3 and NF-κB bind to the _cis_-elements of the RANTES promoter and synergistically stimulate human RANTES transcription. The possibility of the involvement of common signalling pathways controlling IRF-3 and NF-κB phosphorylation is currently under investigation.
ACKNOWLEDGMENTS
We thank Dimitris Thanos and Illka Julkunen for reagents used in this study.
This research was supported by grants from the Medical Research Council of Canada, the National Cancer Institute and the National Institutes of Health (P.M.P.). R.L. was supported in part by a Fraser Monat McPherson Fellowship from McGill University, C.H. by a FRSQ-FCAR studentship, P.G. by an ARC Fellowship, and J.H. by a MRC Senior Scientist award.
REFERENCES
- 1.Ahuja S K, Murphy P M. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. [PubMed] [Google Scholar]
- 2.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 5.Danoff T, Lally P, Chang Y, Heeger P, Neilson E G. Cloning, genomic organization, and chromosomal localization of the Scya5 gene encoding the murine chemokine RANTES. J Immunol. 1994;152:1182–1189. [PubMed] [Google Scholar]
- 6.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 7.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 8.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-β gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J L, Murphy P M. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 10.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 11.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 12.Juang Y T, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor-3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kameyoshi Y, Dorschner A, Mallet A I, Christophers E, Schroder J M. Cytokine RANTES released by thrombin-stimulated platelets is a potent attractant for human eosinophils. J Exp Med. 1992;176:587–592. doi: 10.1084/jem.176.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Lin, R. Unpublished data.
- 14.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lokuta M A, Maher J, Noe K H, Pitha P M, Shin M L, Shin H S. Mechanisms of murine RANTES chemokine gene induction by Newcastle disease virus. J Biol Chem. 1996;271:13731–13738. doi: 10.1074/jbc.271.23.13731. [DOI] [PubMed] [Google Scholar]
- 16.McFadden G, Graham K, Ellison K, Barry M, Macen J, Schreiber M, Mossman K, Nash P, Lalani A, Everett H. Interruption of cytokine networks by poxviruses: lessons from myxoma virus. J Leukoc Biol. 1995;57:731–738. doi: 10.1002/jlb.57.5.731. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to the IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 18.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 19.Moriuchi H, Moriuchi M, Fauci A S. Nuclear factor-κB potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J Immunol. 1997;158:3483–3491. [PubMed] [Google Scholar]
- 20.Murphy P M. Chemokine receptors: structure, function and role in microbial pathogenesis. Cytokine Growth Factor Rev. 1996;7:47–64. doi: 10.1016/1359-6101(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 21.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson P J, Kim H T, Manning W C, Goralski T J, Krensky A M. Genomic organization and transcriptional regulation of the RANTES chemokine gene. J Immunol. 1993;151:2601–2612. [PubMed] [Google Scholar]
- 23.Nelson P J, Ortiz B D, Pattison J M, Krensky A M. Identification of a novel regulatory region critical for expression of the RANTES chemokine in activated T lymphocytes. J Immunol. 1996;157:1139–1148. [PubMed] [Google Scholar]
- 24.Nguyen H, Hiscott J, Pitha P M. The growing family of IRF transcription factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen H, Lin R, Hiscott J. Activation of multiple growth regulatory genes following inducible expression of IRF-1 or IRF/RelA fusion proteins. Oncogene. 1997;15:1425–1435. doi: 10.1038/sj.onc.1201318. [DOI] [PubMed] [Google Scholar]
- 26.Ortiz B D, Nelson P J, Krensky A M. Switching gears during T-cell maturation: RANTES and late transcription. Immunol Today. 1997;18:468–471. doi: 10.1016/s0167-5699(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 27.Ray P, Yang L, Zhang D, Ghosh S K, Ray A. Selective up-regulation of cytokine-induced RANTES gene expression in lung epithelial cells by overexpression of IκBR. J Biol Chem. 1997;272:20191–20197. doi: 10.1074/jbc.272.32.20191. [DOI] [PubMed] [Google Scholar]
- 28.Rollins B J. Chemokines. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 29.Ronco L, Karpova A, Vidal M, Howley P. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roulston A, Lin R, Beauparlant P, Wainberg M A, Hiscott J. Regulation of HIV-1 and cytokine gene expression in myeloid cells by NF-κB/Rel transcription factors. Microbiol Rev. 1995;59:481–505. doi: 10.1128/mr.59.3.481-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P. Nucleotide sequence of the kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type 1 interferon gene expression by interferon regulatory factor 3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 33.Schall T J, Bacon K, Toy K, Goddel D V. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347:669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 34.Schols D, Proost P, Damme J V, Clercq E. RANTES and MCP-3 inhibit the replication of T-cell-tropic human immunodeficiency virus type 1 strains (SF-2, MN, and HE) J Virol. 1997;71:7300–7304. doi: 10.1128/jvi.71.10.7300-7304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 37.Wathelet M G, Lin C H, Parakh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 38.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama M, Suhara W, Fukuhara Y, Fukada M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]