Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 Signalosome Subunit 5 Is Sufficient to Cause the cop/det/fus Mutant Phenotype in Arabidopsis (original) (raw)
Abstract
The COP9 signalosome (CSN) was originally identified based on the constitutively photomorphogenic/de-etiolated/fusca (cop/det/fus) mutants from Arabidopsis thaliana. CSN is evolutionary conserved, and its subunit 5 (CSN5) mediates the deconjugation of NEDD8 from the cullin subunit of E3 ubiquitin ligases (deneddylation). Here, we report on Arabidopsis mutants deficient in CSN5 function. We show that these mutants are phenotypically indistinguishable from the previously described cop/det/fus mutants of other CSN subunits. However, we also show that these mutants retain the CSN complex (lacking CSN5), and this finding is in contrast with the previously described CSN subunit mutants, which lack the CSN complex. We therefore conclude that loss of CSN5 as part of CSN is sufficient to cause the cop/det/fus mutant phenotype. Furthermore, we show that mutants defective in CSN5 as well as mutants defective in CSN are unable to deneddylate the Arabidopsis cullins AtCUL1, AtCUL3A, and AtCUL4. Because these are representative cullin subunits of the three cullin-containing E3 families present in Arabidopsis, we postulate that the cop/det/fus mutant phenotype may be the result of the defects caused by impaired CSN5-dependent deneddylation of cullin-containing E3s.
INTRODUCTION
The COP9 signalosome (CSN) is an evolutionary-conserved multiprotein complex that was originally identified in plants (Wei et al., 1994; Chamovitz et al., 1996; Kwok et al., 1996). To date, CSN complexes have been identified from all eukaryotic model organisms, and mutant studies have shown that CSN is required for many developmental and cellular processes ranging from cell cycle control in yeasts to proper embryo development in mice (reviewed in Wei and Deng, 2003; Schwechheimer, 2004). CSN is composed of eight subunits, six subunits containing the conserved PCI (proteasome, COP9 signalosome, eukaryotic initiation factor) domain and two subunits containing the conserved MPN (MOV34, PAD N-terminal) domain (Wei and Deng, 2003; Schwechheimer, 2004). In Arabidopsis thaliana, mutants of all six PCI-domain subunits display the pleiotropic constitutively photomorphogenic/de-etiolated/fusca (cop/det/fus) phenotype, which is characterized by the short hypocotyl and open cotyledons of the dark-grown seedling, by the accumulation of the plant pigment anthocyanin, and by the expression of light-induced genes in the dark (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999; Peng et al., 2001b; Serino et al., 2003). The indistinguishable phenotype of these mutants can and has been explained by the fact that all of them lack CSN, seemingly as a consequence of the destabilizing effect of the mutation in a single PCI-domain subunit. The MPN-domain subunits CSN5 and CSN6 are encoded by two genes in Arabidopsis, and their gene products have been proposed to have redundant function (Kwok et al., 1998; Peng et al., 2001a). Loss-of-function mutants for CSN5 and CSN6 have not been identified as yet in forward genetic screens.
E3 ubiquitin ligases (E3s) confer substrate specificity to the ubiquitin-proteasome system in that they specifically recognize the proteolysis target and mediate its polyubiquitylation by an associated E2 ubiquitin conjugating enzyme (E2) (Hershko and Chiechanover, 1998; Deshaies, 1999). To date, various proteins and protein complexes with E3 activity have been described, including four types of E3 complexes, which are specified by a distinct cullin subunit (Chiba and Tanaka, 2004; Schwechheimer and Villalobos, 2004). These cullin-containing E3s are referred to as SCF (SKP1, Cullin 1, F-Box protein) complexes containing cullin 1, VCB (Von-Hippel-Lindau, Elongin C, Elongin B) complexes containing cullin 2 or cullin 5, BTB/POZ (BRIC-A-BRAX, TRAMTRACK and BROAD COMPLEX/POX VIRUS and ZINC-FINGER) complexes containing cullin 3, and DCX (DAMAGED DNA BINDING PROTEIN1, CULLIN 4A, X-Box) complexes containing cullin 4 (Deshaies, 1999; Kamura et al., 1999; Higa et al., 2003; Xu et al., 2003; Wertz et al., 2004). It has been shown or proposed that all of the above-mentioned cullin-containing E3s can alter their substrate specificity by association with different degradation substrate receptor subunits, such as F-box proteins in the case of SCF, and that thereby they can acquire different substrate specificities. Based on protein complex characterization and sequence identities, it is predicted that all types of cullin-containing E3s are conserved in Arabidopsis with the exception of VCB (Gray et al., 1999; Gagne et al., 2002; Shen et al., 2002; Risseeuw et al., 2003; Wang et al., 2004; Weber et al., 2004; Wertz et al., 2004; Dieterle et al., 2005).
Several laboratories reported that CSN interacts with cullin-containing E3s (Lyapina et al., 2001; Schwechheimer et al., 2001; Liu et al., 2002; Feng et al., 2003; Groisman et al., 2003; Higa et al., 2003; Liu et al., 2003; Wang et al., 2003). Although in most cases it is clear that the CSN–E3 interaction is required for the efficient ubiquitylation and degradation of the E3 substrate, the precise role of CSN for E3 function is not understood. One hypothesis suggests that CSN is part of a proteasomal complex and that the CSN–E3 interaction may ensure rapid degradation of the E3 substrate (Peng et al., 2003). Alternatively, it has been hypothesized that CSN participates in the efficient assembly of E3 complexes or that it is required for the exchange or the stability of their substrate receptor subunits (Wolf et al., 2003; Wee et al., 2005). To date, CSN has been shown to interact with cullin 1–containing SCF-type E3s from yeasts and Arabidopsis, with a cullin 3–containing E3 from fission yeast, and with cullin 4–containing E3s from fission yeast and human (Lyapina et al., 2001; Schwechheimer et al., 2001; Zhou et al., 2001; Liu et al., 2002, 2003; Feng et al., 2003; Groisman et al., 2003; Higa et al., 2003; Pintard et al., 2003; Wang et al., 2003). Specifically in Arabidopsis, CSN interacts with SCFTIR1, SCFCOI1, and SCFUFO, E3 complexes that regulate auxin responses, jasmonate responses, and floral development, respectively (Schwechheimer et al., 2001; Feng et al., 2003; Wang et al., 2003). Furthermore, there is striking genetic evidence that Arabidopsis CSN represses photomorphogenic growth in the dark by interacting with COP1 and DET1, which may function in the context of a cullin 4–containing DCX complex (Osterlund et al., 2000; Schroeder et al., 2002; Groisman et al., 2003; Wertz et al., 2004). However, a physical interaction between Arabidopsis CSN and COP1, DET1, or a DCX-complex has not been established yet.
The cullin subunits of all cullin-containing E3s described above are reversibly modified by the ubiquitin-like protein NEDD8/RUB1 (NEDD8) in a process referred to as neddylation and deneddylation (Hori et al., 1999; Pan et al., 2004). Although it has been observed that the neddylation status of cullins affects the stability of E3 complex adaptor subunits, the precise role of neddylation and deneddylation for cullin or E3 function is not understood. It is however established that both neddylation and deneddylation are essential for proper development in higher eukaryotes and that both processes function together to mediate proteolysis (reviewed in Chiba and Tanaka, 2004; Pan et al., 2004; Wee et al., 2005). There is strong evidence that cullin deneddylation is a biochemical activity of CSN5, thus providing a biochemical link between CSN and cullin-containing E3s (Komari et al., 1996; Zhou et al., 2001; Cope et al., 2002; Wee et al., 2002; Liu et al., 2003; Pintard et al., 2003; Tran et al., 2003). CSN5's deneddylation activity resides within a specific motif that is conserved in the MPN-domain of CSN5 but not in the MPN-domain of CSN6. Consequently, specific mutations in the MPN-domain of CSN5 impair deneddylation in yeast and Arabidopsis (Cope et al., 2002; Tran et al., 2003; Gusmaroli et al., 2004).
In Arabidopsis, studies on CSN5 function have so far been restricted to plants that are defective in one of the two Arabidopsis CSN5 genes or to plants that express mutant deneddylation-deficient CSN5 proteins (Peng et al., 2001a; Schwechheimer et al., 2001, 2002; Gusmaroli et al., 2004). In this study, we characterize CSN5 loss-of-function mutants from Arabidopsis. We show that these CSN5 mutants mimic the pleiotropic cop/det/fus mutant phenotype, and we find that these mutants retain a CSN complex that lacks CSN5. Because CSN5 mutants are indistinguishable from the previously described cop/det/fus mutants that lack the entire CSN complex, we propose that loss of CSN5 function in the CSN complex is sufficient to cause the pleiotropic cop/det/fus mutant phenotype. Furthermore, we show that CSN5 loss-of-function mutants as well as mutants that lack the entire CSN are deficient in deneddylation of the cullins AtCUL1, AtCUL3A, and AtCUL4. Because these proteins are representative cullin subunits of the three families of cullin-containing E3s present in Arabidopsis, we propose that the cop/det/fus mutant phenotype may be the collective effect of impaired E3 function caused by a block in cullin deneddylation.
RESULTS
CSN5 Loss-of-Function Mutants Display the Severe Phenotype of the cop/det/fus Mutants
A total of 10 genes encode the eight CSN subunits in Arabidopsis (Wei et al., 1994; Staub et al., 1996; Kwok et al., 1998; Karniol et al., 1999; Serino et al., 1999, 2003; Peng et al., 2001a, 2001b). Loss-of-function mutants have been described for the six PCI-domain subunits, namely CSN1 through CSN4, CSN7, and CSN8, and all of these mutants are phenotypically indistinguishable in that they display the pleiotropic cop/det/fus mutant phenotype, which is characterized by a short hypocotyl and open cotyledons in the dark-grown seedling, the accumulation of the anthocyanin pigment, and the expression of light-induced genes in the dark (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999, 2003; Peng et al., 2001b). CSN5A (AJH1, At1g22920) and CSN5B (AJH2, At1g71230) are the two genes that encode CSN5 in Arabidopsis (Kwok et al., 1998). CSN5 loss-of-function mutants have not been reported yet. To obtain a CSN5 loss-of-function mutant, we isolated mutants that carry T-DNA insertions in CSN5A and CSN5B, which we designated _csn5a_-1, _csn5a_-2, and _csn5b_-1 (Figures 1A and 1B). We found that both csn5a mutant alleles have identical phenotypes in that they display reduced growth at the seedling and adult stage, reduced lateral root formation, reduced root hair formation, and reduced flower size (Figures 1D, 1E, 1H, 1I, 1O, and 1S). Despite these severe growth defects, both csn5a mutant alleles are fertile and can be propagated as homozygous mutants. By contrast, the csn5b mutation gives rise to comparably subtle phenotypes, such as reduced and delayed root hair development, but more phenotypes became apparent during subsequent quantitative analyses (Figures 1F, 1K, 1P, 1T, and studies described below). Next, we generated _csn5a-_1 _csn5b-_1 and _csn5a_-2 _csn5b_-1 double mutants. Both csn5 double mutant combinations result in seedlings with identical dramatic phenotypes, including a short hypocotyl and open cotyledons in seedlings grown in any light condition and an apparent accumulation of anthocyanin (Figure 1L; see below). Thus, the csn5 double mutants mimic the phenotype of the previously described cop/det/fus mutants, such as the CSN8 subunit mutant cop9/fus8-S253, suggesting that the same developmental pathways are affected in both types of mutants (Figures 1L and 1M).
Figure 1.
Phenotypic Characterization of Mutant Alleles of the Two CSN5 Genes from Arabidopsis.
(A) Schematic representation of the CSN5A and CSN5B genes. Exons are shown as black boxes and introns and upstream sequences as lines. Boxes and lines are drawn to scale. Positions of the T-DNA insertions of the three available mutants are indicated.
(B) Homozygous mutants of all three single mutants as well as the csn5a-2 csn5b-1 double mutant were identified by PCR genotyping using the primer combinations described in Methods. Results of PCR-based genotyping of the csn5 single and double mutants are shown. Presence or absence of a band indicates presence or absence of the respective wild-type or mutant loci.
(C) to (F) Phenotype of 8-d-old seedlings of the wild type and the csn5 mutants as indicated.
(G) to (K) Phenotype of 4-week-old seedlings of the wild type and the csn5 mutants as indicated. Bar = 5 cm in (G) and (H) and 1 cm in (H) and (I).
(L) and (M) The _csn5a-_2 _csn5b_-1 double mutant (L) is phenotypically indistinguishable from the previously described CSN subunit mutants, such as cop9/fus8-S253 (M). Bar = 1 mm.
(N) to (Q) Root hair development in the wild type and the csn5 mutants as indicated.
(R) to (T) Floral phenotype of the wild type and the csn5 mutants as indicated. All pictures belonging to one series were taken at the same scale unless otherwise noted.
Mutant studies from CSN5 function in mice invite the hypothesis that the mouse CSN5 gene is haploinsufficient. Homozygous mutant mice arrest growth during embryogenesis, whereas mice carrying one functional copy of CSN5 are smaller than their wild-type littermates, a difference that has been attributed to a reduction in cell number (Tomoda et al., 2004). During the process of establishing homozygous csn5a single mutant lines from hemizygous parents, we repeatedly noticed Arabidopsis plants that displayed semidwarfed growth, stem fasciation, and reduced apical dominance, hence phenotypes that were distinct from the wild type and also distinct from the more severe phenotypes observed in the homozygous csn5a mutants (data not shown). All of these plants were found to be hemizygous for the CSN5A T-DNA insertions. It can therefore be envisioned that the phenotypes observed in the hemizygous CSN5A plants may also be the result of CSN5A haploinsufficiency in Arabidopsis.
Light Responses in csn5 Mutants
The cop/det/fus mutants mimic the phenotype of light-grown seedlings when grown in the dark (Kwok et al., 1996). HY5 and HY5-HOMOLOG (HYH) are positive regulators of photomorphogenesis that regulate the expression of light-induced genes (Osterlund et al., 2000; Holm et al., 2002). Mutants of CSN subunit genes fail to degrade HY5 and HYH in the dark, and they consequently misexpress light-induced genes (Kwok et al., 1996; Osterlund et al., 2000; Holm et al., 2002). To study the photomorphogenesis phenotype of the csn5 mutants, we grew the csn5 single and double mutants in different light conditions. In all conditions examined, the csn5 double mutant displayed the constitutive photomorphogenic phenotype of the cop/det/fus mutants, such as _cop9/fus8_-S253 (Figures 2A and 2B). Furthermore, increased photomorphogenic responses were observed in all light conditions with both csn5a mutant alleles or in selected light conditions with the _csn5b_-1 mutant allele (Figures 2A and 2B). In any light condition, the relative contribution of CSN5A to the respective phenotype was stronger than that of CSN5B. Taken together, these data indicate that the two CSN5 genes have redundant function in repressing photomorphogenic responses and that the combined csn5 mutations give rise to the constitutive photomorphogenic cop/det/fus mutant phenotype that had previously been reported for mutants defective in other CSN subunits.
Figure 2.
The Two CSN5 Genes Contribute Differentially to the Constitutive Photomorphogenic Phenotype Observed in the _csn5a-_2 _csn5b_-1 Double Mutant.
(A) Phenotypes of wild-type seedlings, the csn5 mutant seedlings, and _cop9/fus8-_S253 mutant seedlings grown for 4 d in the light as indicated. D, dark; W, white light; FR, far-red light; R, red light; B, blue light.
(B) Quantitative analysis of the csn5 mutant seedlings shown in (A) (n ≥ 10). Black bar, the wild type; dark-gray bar, _csn5a_-2; light-gray bar, _csn5b-_1; white bar, _csn5a-_2 _csn5b-_1.
(C) Semiquantitative RT-PCR analysis of light- and dark-grown wild type, _csn5a_-2 _csn5b_-1, and _cop9/fus8-_S253 mutant seedlings as indicated. Thirty-five PCR cycles were used to amplify the PSBA, RBCS, and ACTIN genes as indicated.
Next, we used RT-PCR to compare light-induced gene expression in dark- and light-grown wild-type, csn5, and cop9/_fus8_-S253 seedlings (Figure 2C). We analyzed the expression of the light-induced genes PSBA and RBCS1, which encode the 32-kD protein of photosystem II and the small subunit of ribulose-1,5-biphosphate carboxylase, respectively. Although we detected strong PSBA and RBCS expression in all light-grown samples, we found their expression to be essentially absent in the dark-grown wild-type controls but strongly upregulated in the csn5 double mutants and in the cop9/_fus8_-S253 mutant (Figure 2C). These data show that the csn5 loss-of-function mutants also display the molecular aspects of the constitutive photomorphogenesis phenotype described for the cop/det/fus mutants that carry mutations in other CSN subunit genes.
CSN5 Mutants Have Defects in Auxin Response Pathways
Defects in photomorphogenesis are the most obvious phenotypes discernable in the Arabidopsis cop/det/fus mutants. However, biochemical studies and genetic studies have linked CSN to the activity of SCFTIR1, an E3 required for proper auxin response (Gray et al., 2001; Schwechheimer et al., 2001). The phytohormone auxin controls numerous developmental processes, including primary and lateral root growth as well as root hair formation. At the molecular level, auxin response can be characterized by analysis of auxin-induced gene expression, which is under the control of the unstable AUX/IAA repressors, the degradation targets of SCFTIR1 (Gray et al., 2001). We reasoned that mutants deficient in CSN5 may have impaired auxin responses similar to the previously characterized cop/det/fus mutants (Schwechheimer et al., 2001, 2002). In fact, our analyses of the csn5 single and double mutants revealed that several auxin-dependent growth processes, including root growth, lateral root formation, and root hair formation, are defective in these mutants (Figures 1C to 1F, 1N to1Q, 3A, and 3B). Furthermore, we also determined that the csn5 single mutants are insensitive to the root growth inhibiting effects of exogenously applied auxin (Figure 3C). In line with the previous phenotypic analyses, the relative severity of these phenotypes was more pronounced in the csn5a mutants than in the csn5b mutant. Based on these morphological criteria, we conclude that csn5 single and double mutants have reduced auxin responses.
Figure 3.
Auxin Responses in the csn5 Mutants.
(A) and (B) Graph showing total root length and lateral root number of 10-d-old Arabidopsis seedlings of the genotypes as indicated. _csn5a_-2 _csn5b_-1 double mutants do not form lateral roots.
(C) Graph showing the relative length of the wild type (black bars), _csn5a_-2 (gray bars), and _csn5b_-1 (white bars) mutant roots grown on different concentrations of the synthetic auxin 2,4-D. The _csn5a_-2 _csn5b_-1 double mutants are not included because they arrest growth at earlier stages of development.
(D) Schematic representation of the auxin-responsive GH3-2:luciferase (LUC) construct and its GH3-2mut:LUC mutant version used for quantitative auxin induction assays.
(E) Graph showing a typical auxin-induction experiment (5 μM 2,4-D) with the GH3-2:LUC reporter construct in the presence and absence of the 26S proteasome inhibitor MG132 (100 μM).
(F) Scatter blot showing the auxin induction rates (increase in relative light units/minute between 30 and 110 min) obtained with GH3-2:LUC in the wild type, csn5, _cop9/fus8_-S253, and _cop13/fus11_-U203 mutant backgrounds as well as with the GH3-2mut:LUC lines. Please note the logarithmic scale (n ≥ 5).
Auxin Induction of the Auxin-Responsive GH3-2:LUC Reporter Is Impaired in CSN5 and Other CSN Subunit Mutants
Auxin responses are controlled by the AUX/IAA transcriptional repressors (Gray et al., 2001). In the absence of auxin, AUX/IAAs repress the activity of AUXIN REPONSE FACTORS (ARFs), transcriptional activators that recognize the auxin-responsive promoter element (AuxRE) TGTCTC (Ulmasov et al., 1997a, 1997b). In the presence of auxin, AUX/IAAs are degraded by the ubiquitin-proteasome system, thus allowing the induction of auxin-induced genes by the ARF activators (Gray et al., 2001). To obtain a quantifiable assay system for the AUX/IAA and ARF-controlled gene expression system, we generated transgenic lines that express the luciferase reporter under control of an 800-bp promoter fragment of the auxin-induced GH3-2 gene At4g37390 (Figure 3D) (Tian and Reed, 1999). In these GH3-2:LUC lines, luciferase activity was not detected in the absence of auxin but was induced as early as 30 min after auxin application (Figure 3E). Luciferase expression generally reached its maximum at 90 to 120 min followed by a more or less pronounced decline in activity.
The selected promoter fragment contains a single AuxRE with the canonical TGTCTC sequence. We found that auxin-induced gene expression was strongly reduced in transgenic Arabidopsis lines that contained an equivalent mutant construct GH3-2mut:LUC, where the AuxRE TGTCTC sequence had been mutated to TG_G_CTC, a mutation known to prevent ARF binding (Figures 3D and 3F) (Ulmasov et al., 1997a). Similarly, concomittant application of the 26S proteasome inhibitor MG132 partially blocked auxin induction, suggesting that auxin induction is dependent on the degradation of an unstable repressor (Figure 3E). In summary, we suggest that GH3-2:LUC expression is controlled by ARF transcription factors and that their activition is repressed by inhibiting AUX/IAA degradation.
To examine GH3-2:LUC expression in the different mutant backgrounds, we selected the reporter line GH3-2:LUC#3-8, which has a single locus transgene insertion, and crossed the transgene to the CSN5 mutants as well as to the CSN subunit mutants _cop9/fus8_-S253 (CSN8) and _cop13/fus11-_U203 (CSN3). We isolated lines homozygous for CSN subunit mutations based on the cop/det/fus mutant phenotype and genotype, and we subsequently examined auxin induction of the GH3-2:LUC reporter construct. In our analysis, we found that the csn5 single mutants are partially blocked and that the csn5 double mutants as well as the other CSN subunit mutants are fully blocked in auxin-induced gene expression of the GH3-2:LUC reporter (Figure 3F). Based on these molecular phenotypes, we conclude that the csn5 mutants as well as the other previously described CSN subunit mutants are insensitive to the gene expression–inducing effects of auxin, and we reason that this is the result of the inefficient degradation of AUX/IAA repressors.
CSN5 Mutants Fail to Express CSN5 but Retain CSN
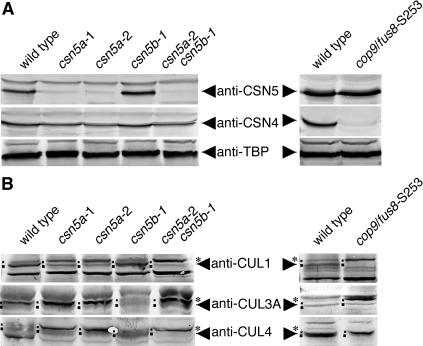
Based on the mutant phenotypes and based on the position of the T-DNA insertions in the CSN5 genes, we predicted that the csn5a and csn5b single mutants may fail to express the respective CSN5 proteins. We therefore analyzed CSN5 protein levels in the different mutants using an antibody that recognizes CSN5A as well as CSN5B (Kwok et al., 1998). Consistent with the comparatively weak phenotype of the _csn5b_-1 mutant allele, CSN5 levels were not significantly altered in the _csn5b-_1 mutant when compared with the wild type (Figure 4A). However, consistent with the strong developmental defects observed in the csn5a mutants, we found CSN5 protein levels to be significantly reduced in both csn5a mutant alleles as well as in the csn5 double mutant (Figure 4A). Thus, we observed a close correlation between the severity of the developmental defects and the reduction in CSN5 protein levels in the csn5 mutants.
Figure 4.
Quantitative Analysis of CSN Subunit Protein Levels in Light-Grown Seedlings and Correlation with the Deneddylation Activity.
(A) Immunoblot of identical protein extracts derived from wild-type and csn5 mutant seedlings as indicated as well as wild-type and _cop9/fus8-_S253 mutant seedlings probed with anti-CSN4 and anti-CSN5 as well as anti-TATA box binding protein (anti-TBP) (loading control).
(B) Immunoblot of protein extracts shown in (A) probed with anti-CUL1, anti-CUL3A, and anti-CUL4 antibodies. Larger points mark the position of the unneddylation cullins (CUL1, calculated molecular mass 81 kD; CUL3A, 80 kD; CUL4, 87 kD), and smaller points and asterisks mark the positions of the neddylated cullins (CUL1, calculated molecular mass 90 kD; CUL3A, 89 kD; CUL4, 96 kD). The apparent differences in cullin protein abundance in the individual samples may at least in part be attributed to the different morphologies of the individual genotypes.
Seemingly as a result of the destabilizing effect of the loss of one CSN subunit, Arabidopsis mutants of the six PCI-domain containing CSN subunits lack the CSN complex and the corresponding CSN subunit monomers (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999, 2003; Peng et al., 2001b). At the same time, these CSN subunit mutants retain a monomeric form of CSN5. CSN5 monomers have been reported in any organism where CSN has been studied, and it has been suggested that the CSN5 monomer plays a biological role, which in turn may or may not be connected to the CSN complex or its function (Kwok et al., 1998; Seeger et al., 1998; Oron et al., 2002). When we analyzed the effect of loss of CSN5 protein on the abundance of other CSN subunits by examining CSN4 protein levels, we found that CSN4 protein levels are unaltered in the csn5 double mutants that lack CSN5 (Figure 4A). This observation is in contrast with all other previously described CSN subunit mutants from Arabidopsis and gave rise to the two alternative hypotheses that either CSN complex integrity is not affected in the csn5 double mutants or that CSN4 monomers accumulate in the csn5 double mutants.
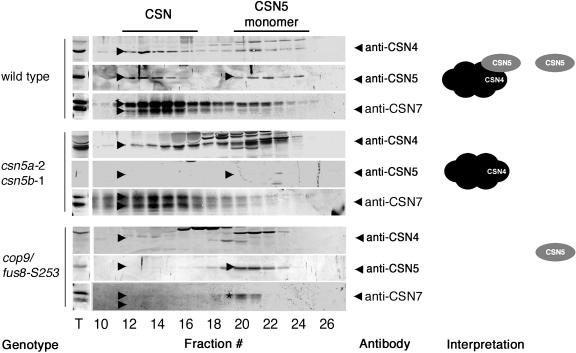
To examine this point in more detail, we turned to gel filtration analyses followed by immunoblotting to analyze CSN complex integrity in the wild type, the CSN5 subunit mutants, and the CSN8 subunit mutant _cop9/fus8-_S253 (Figure 5). As expected and consistent with previous reports, the examination of wild-type protein extracts revealed the presence of CSN4, CSN5, and CSN7 as subunits of CSN as well as the presence of a CSN5 monomer (Figure 5) (Kwok et al., 1998; Karniol et al., 1999; Serino et al., 1999). The same elution profile was detected in the comparably weak _csn5b_-1 mutant as well as for the comparably strong csn5a mutants, the latter requiring longer exposure times to reveal the presence of the less abundant CSN5B protein (data not shown). Also consistent with previous reports, CSN5 and CSN7 monomer but no CSN complex was detectable in the CSN8 subunit mutant cop9/fus8-S253 as demonstrated by the absence of CSN4, CSN5, and CSN7 in the CSN complex fractions (Figure 5). Most interestingly, however, we still detected a CSN complex lacking the CSN5 protein in the csn5 double mutant as demonstrated by the presence of CSN4 and CSN7 in the same size fractions that elute the CSN complex in the wild type (Figure 5). Based on these observations, we conclude that CSN complex stability is not affected by the loss of CSN5. Furthermore, we conclude that loss of CSN5 monomer in the csn5 double mutant does not give rise to a phenotype that distinguishes the csn5 mutants from the other CSN subunit mutants of the cop/det/fus mutant series. It may therefore be that the CSN5 monomer does not play a role in development that is discernable at the morphological level.
Figure 5.
Gel Filtration Analyses Reveal That csn5 Loss-of-Function Mutants Retain CSN Complex.
Protein extracts of the genotypes as indicated were subjected to gel filtration analyses and probed with anti-CSN4, anti-CSN5, and anti-CSN7 antibodies. Arrowheads indicate the position of the respective CSN subunits in the different analyses. CSN7 elutes as a CSN7 monomer in the _cop9/fus8_-S253 mutant and is indicated by an asterisk. The interpretation of the gel filtration experiments with respect to CSN and CSN5 are shown in the panels at the right. The gel filtration fractions 1 to 9 are not shown to improve clarity of the presentation.
Arabidopsis csn5 Mutants Are Deneddylation Deficient
The cullin subunits of E3s are subject to NEDD8 conjugation and deconjugation (Hori et al., 1999; Pan et al., 2004). Five cullin proteins that contain the conserved motif for NEDD8 modification are expressed in Arabidopsis (Risseeuw et al., 2003). Based on biochemical analyses and in analogy to E3s described in other eukaryotes, these cullins are thought to be engaged in SCF complexes (AtCUL1 and AtCUL2), BTB/POZ complexes (AtCUL3A and AtCUL3B), and DCX complexes (AtCUL4), respectively (Gray et al., 1999; Gagne et al., 2002; Shen et al., 2002; Risseeuw et al., 2003; Wang et al., 2004; Weber et al., 2004; Wertz et al., 2004; Dieterle et al., 2005). In Arabidopsis, CSN has been shown to interact with SCF-type E3s, and loss of CSN function has been shown to block deneddylation of the AtCUL1 subunit of SCF E3s (Schwechheimer et al., 2001). Subsequent studies revealed a specific role for CSN5 in cullin 1 deneddylation (Cope et al., 2002; Gusmaroli et al., 2004). Because all Arabidopsis cullins mentioned above contain the conserved sequence motif for NEDD8 modification, it can be proposed that all of them are neddylated and that all of them are subject to CSN5-mediated deneddylation. To examine this point, we investigated the role of CSN5 and CSN on deneddylation of the cullins AtCUL1, AtCUL3A, and AtCUL4, representative cullin subunits of the three types of cullin-containing E3 complexes that have been proposed to exist in Arabidopsis. Although we detected unneddylated/deneddylated as well as neddylated forms of all three cullins in wild-type extracts, we found exclusively neddylated cullins in the csn5 double mutant as well as the CSN8 mutant _cop9/fus8-_S253 (Figure 4B). Furthermore, consistent with the observed reduction in CSN5 protein levels and their relatively strong phenotype, we found that both csn5a single mutants contain significantly increased levels of the neddylated cullins when compared with the csn5b single mutant and the wild type (Figure 4B). Also consistent with previous reports, we observe that the CSN5 monomer as present in the CSN8 mutant _cop9/fus8-_S253 is not sufficient to deneddylate the Arabidopsis cullins (Figure 4B). Because mutants lacking CSN5 and mutants lacking CSN have the same molecular defects with regard to cullin deneddylation, and because both types of mutants lack CSN5 in the CSN complex, we propose that CSN5 as subunit of CSN is essential for the deneddylation of all cullins present in Arabidopsis. At the same time, we also propose that these deneddylation defects block E3 ubiquitin ligase-mediated biological processes and that the combination of the resulting defects may be the molecular cause for the pleiotropic phenotype observed in the cop/det/fus mutants.
DISCUSSION
In this article, we report on the physiological and molecular characterization of CSN5 loss-of-function mutants from Arabidopsis. We show that plants deficient in CSN5 function mimic the phenotypes of the previously described cop/det/fus mutants that carry mutations in one of the six PCI-domain containing CSN subunits (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999, 2003; Peng et al., 2001b). These phenotypes include constitutively photomorphogenic growth, accumulation of anthocyanins, expression of light-induced genes in the dark, insensitivity to the phytohormone auxin, as well as growth arrest at the seedling stage. In comparison with the CSN5 double mutant, single mutants of the two CSN5 genes are only partially impaired in the different responses examined, suggesting that the two CSN5 genes have redundant function. Taken together, our findings suggest that CSN5 is essential for normal plant growth and development and that loss of CSN5 function causes phenotypic defects that are equivalent to those reported for the cop/det/fus mutants defective in any other CSN subunit.
In contrast with all previous reports on CSN subunit mutations, our biochemical analyses reveal that CSN5 loss-of-function mutants retain a CSN complex. Interestingly, a CSN5 subunit mutant from Drosophila was also found to lack CSN5 but to retain CSN (Oron et al., 2002). Therefore, unlike any of the six PCI-domain CSN subunits, the MPN-domain subunit CSN5 is seemingly not required for CSN complex stability (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999, 2003; Peng et al., 2001b). Furthermore, because Arabidopsis mutants that lack CSN5 show the same dramatic phenotypes as those mutants that lack CSN, including CSN5, the molecular cause of the mutant phenotype may now be attributed to loss of CSN5 function in CSN.
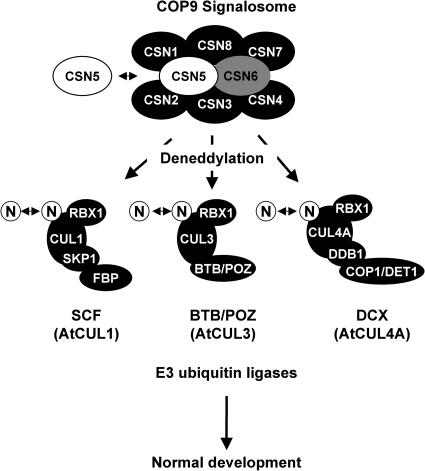
CSN5 as subunit of CSN but not as a CSN5 monomer is the proposed cullin deneddylating enzyme (Cope et al., 2002; Tran et al., 2003). To date, Arabidopsis CSN5 as part of CSN has only been implicated in the deneddylation of Arabidopsis cullin 1 (Schwechheimer et al., 2001). Our results show that CSN5 as part of the CSN complex mediates the deneddylation of Arabidopsis AtCUL1, AtCUL3A, and AtCUL4. These cullins are representatives of the five member cullin protein family, and they are thought to engage in SCF-type (AtCUL1 and AtCUL2), BTB/POZ-type (AtCUL3A and AtCUL3B), and DCX-type E3s (AtCUL4), respectively. Therefore, our results show that cullins from all proposed cullin-containing E3 complexes of Arabidopsis require CSN5-mediated deneddylation. Because deneddylation is essential for the activity of cullin-containing E3s and because hundreds of such E3 complexes seemingly exist in Arabidopsis, the mutant phenotype observed in the CSN5 and other CSN subunit mutants may be the result of the impaired function of a host of these E3-dependent developmental processes (Figure 6).
Figure 6.
The CSN Acts on Three Different Types of E3 Ubiquitin Ligases in Arabidopsis.
Schematic summary of the results obtained in this study where CSN, and more specifically CSN5, deneddylates cullins that belong to at least three different E3 ubiquitin ligase families from Arabidopsis. N, NEDD8; FBP, F-box protein.
With regard to CSN5, it needs to be noted that mutants of CSN subunits other than CSN5 lack CSN but still retain a CSN5 monomer. The CSN5 loss-of-function mutants described in this study lack also the CSN5 monomer. It was previously noted, and our studies confirm these observations, that the presence of the CSN5 monomer is not sufficient for cullin deneddylation and that cullin deneddylation is an activity of CSN5 as subunit of CSN (Lyapina et al., 2001; Schwechheimer et al., 2001; Cope et al., 2002). We observe that both types of mutants, those that contain and those that lack the CSN5 monomer, have identical phenotypes. Therefore, the CSN5 monomer does not have a function that leads to morphologically discernable phenotypes that are different from loss of CSN function alone. Based on our observation that CSN5 is only an associated rather than an integral CSN subunit, it may now also be postulated that the CSN5 monomer is a CSN-dissociated form of CSN5 that does not have a CSN-independent biochemical or biological activity (Figure 6).
In summary, we show that mutants deficient in CSN5 function have the cop/det/fus mutant phenotype, which is the phenotype that characterizes mutants of other CSN subunit genes. In contrast with these other CSN subunit mutants, however, loss of CSN5 function does not lead to destabilization of the CSN complex. The cop/det/fus mutant phenotype may therefore be caused by the loss of CSN5 function in the CSN complex. We furthermore show that loss of CSN5 or loss of CSN causes a deficiency to deneddylate multiple cullins; therefore, the dramatic phenotype of the cop/det/fus mutants may be the result of the combined impairment of these E3 ubiquitin ligase activities.
METHODS
Biological Material
Arabidopsis thaliana mutants were grown on soil or on growth medium in standard growth conditions under continuous light. The CSN5 T-DNA insertion lines SALK_063436 (_csn5a_-1), SALK_027705 (_csn5a_-2), and SALK_007134 (_csn5b_-1) were identified in the SIGNAL database (Alonso et al., 2003). The CSN5A T-DNA insertion lines were PCR genotyped using the oligonucleotides AJH1FW2 5′-GTTTTGGATTAGCATTAGTCCCCAAATC-3′ and AJH1RV2 5′-TTCAAACATAAATGTGAAAAACAACAT-3′ to test for the presence of the wild-type CSN5A gene, and AJH1FW2 and LBb1 5′-CAGCGTGGACCGCTTGCTGCAACTCTCTCA-3′ to test for the presence of the T-DNA insertions. The CSN5B T-DNA insertion lines were PCR genotyped using AJH2FW 5′-AAGATCTCAGCGCTCGCTCTTCTTAAG-3′ and AJH2RV 5′-ATGGCACAACTCCTCCAAAGCGAGAC-3′ to test for the presence of the wild-type gene, and AJH2RV and LBb1 to test for the presence of the T-DNA insertion. The cop9 mutant allele _fus8_-S253 carries a mutation in CSN8 and was used as a CSN mutant control for all experiments (Miséra et al., 1994; Wei et al., 1994). The _fus11_-U203 mutant allele used in luciferase assays is a loss-of-function mutant of CSN3 (Peng et al., 2001b).
Physiological Experiments
For hypocotyl elongation experiments, seedlings were grown for 6 d in the dark and in white (121 μmol), far-red (0.2 μmol), red (40 μmol), and blue (6.8 μmol) light. Hypocotyl length was measured using the NIH Image software (National Institutes of Health, Bethesda, MD). The number of lateral roots was determined from 10-d-old light-grown seedlings. The effect of auxin on root elongation was measured from 10-d-old seedlings 5 d after transfer from auxin-free medium to a medium containing the synthetic auxin 2,4-D as previously described (Schwechheimer et al., 2001, 2002).
GH3-2:LUC Experiments
An 800-bp GH3-2 (At4g37390) promoter fragment was amplified by PCR using the oligonucleotides GH3-2 FW 5′-AGATCTGTCGACATGCTATAGATTGA-3′ and GH3-2 RV 5′-CCATGGTTGTTTTTTTTTCTAAAAGAAAAACAT-3′ and cloned as a _Bgl_II/_Nco_I fragment into the plant transformation vector LUCTRAP-1, which will be described elsewhere. The mutant GH3-2mut:LUC construct was generated by overlap extension PCR with the oligonucleotides pGH3FW2w 5′-ACATAAGCTTATTGTCGACTGTACCTTTTGTCCCCCGTCTCG-3′ and pGH3RV2m 5′-CAGTCGACAATAAGCTAATGTTAGTTAATGGAGCCATAAGGG-3′ in combination with the GH3-2 FW and GH3-2 RV primers (Ho et al., 1989). GH3-2:LUC#3-8, a GH3-2:LUC line with a single locus transgene insertion, was selected and crossed into mutant backgrounds. Lines homozygous for the mutation and the GH3-2:LUC transgene were selected for further analysis. For luciferase reporter analyses, 5-d-old seedlings were incubated with growth medium containing 250 μM D-luciferin (PJK, Kleinblittersdorf, Germany), and luminescence was quantified in regular intervals using a Mithras LB940 luminometer (Berthold Technologies, Bad Wildbad, Germany).
Gene Expression Analyses
The expression of light-regulated genes was determined by RT-PCR from 7-d-old dark- and light-grown seedlings. RNA was extracted using the RNeasy Plant Mini kit (Qiagen, Valencia, CA), and RT-PCR was performed using the OneStep RT-PCR kit (Qiagen) with 0.5 μg of total RNA as starting material. For PCR amplification, the following oligonucleotides were used: PsbA-FW 5′-ATGACTGCAATTTTAGAGAGACG-3′ and PsbA-RV 5′-CCTCAACAGCAGCTAGGTCTAG-3′ for the 32-kD protein of photosystem II (PsbA); RBCS1A-FW 5′-TCAGTCACACAAAGAGTAAAG-3′ and RBCS1A-RV 5′-TCAACACTTGAGCGGAGTCGGTG-3′ for the small subunit of ribulose-1,5-biphosphate carboxylase (RbcS); and Actin-FW 5′-ATTCAGATGCCCAGAAGTCTTGTTC-5′ and Actin-RV 5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′ for Actin.
Immunoblots and Size Exclusion Chromatography
For immunoblots and gel filtration analyses, protein extracts were prepared from 7- to 10-d-old Arabidopsis seedlings in protein extraction buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 10% glycerol, 1 mM PMSF, and 1 mM β-mercaptoethanol). Forty micrograms of total protein were loaded in each lane in immunoblots. The anti-CUL1 and anti-TBP antibodies were previously described (Schwechheimer et al., 2002). Anti-CUL3A and anti-CUL4 were a gift from P. Genschik (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) (Weber et al., 2004). Anti-CSN4 and anti-CSN5 antibodies were previously described and purchased from BioMol (Hamburg, Germany) (Kwok et al., 1998; Serino et al., 1999). The anti-CSN7 antibody was previously described and is a gift from D. Chamovitz (Tel Aviv University, Israel) (Karniol et al., 1999). Size exclusion chromatography of 300 μg of protein extract was performed using a Superose 6 HR column (Amersham Pharmacia, Freiburg, Germany) as previously described (Schwechheimer and Deng, 2002).
Acknowledgments
We thank Luz Irina A. Calderon-Villalobos and Melina Zourelidou (Centre for Plant Molecular Biology, Tübingen, Germany) for critically reading the manuscript. We thank the Salk Institute Genomic Analysis Laboratory for generating the sequence-indexed Arabidopsis T-DNA insertion mutants and the Nottingham Arabidopsis Stock Centre (Nottingham, UK) for providing seeds of these lines. We also thank Pascal Genschik (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) and Danny Chamovitz (Tel Aviv University, Israel) for providing antibodies. This work is supported by a grant from the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 446; Teilprojekt A21) to C.S. E.M.N.D. is recipient of a fellowship from the Landesgraduiertenförderung Baden-Württemberg.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Claus Schwechheimer (claus.schwechheimer@zmbp.uni-tuebingen.de).
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301**,** 633–657. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.-W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86**,** 115–121. [DOI] [PubMed] [Google Scholar]
- Chiba, T., and Tanaka, K. (2004). Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr. Protein Pept. Sci. 5**,** 177–184. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., Suh, G.S.B., Aravind, L., Schwarz, S.E., Zipursky, S.L., Koonin, E.V., and Deshaies, R.J. (2002). Role for predicted metalloprotease motif of JAB1/Csn5 in cleavage of NEDD8 from CUL1. Science 298**,** 608–611. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15**,** 435–467. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Thomann, A., Renou, J.-P., Parmentier, Y., Cognat, V., Lemonnier, G., Müller, R., Shen, W.-H., Kretsch, T., and Genschik, P. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41**,** 386–399. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.-W. (2003). The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15**,** 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99**,** 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13**,** 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414**,** 271–276. [DOI] [PubMed] [Google Scholar]
- Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J.-i., Siaijo, M., Tanaka, K., and Nakatani, Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113**,** 357–367. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Feng, S., and Deng, X. (2004). The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16**,** 2984–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Chiechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67**,** 425–479. [DOI] [PubMed] [Google Scholar]
- Higa, L.A.A., Mihaylov, I.S., Banks, D.P., Zhneg, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5**,** 1008–1015. [DOI] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Fuller, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77**,** 51–59. [DOI] [PubMed] [Google Scholar]
- Holm, M., Ma, L., Qu, L.-J., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16**,** 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Osaka, F., Chiba, T., Miyamoto, C., Okabayashi, K., Shimbara, N., Kato, S., and Tanaka, K. (1999). Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18**,** 6829–6834. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin, W.G.J., Elledge, S.J., Conaway, R.C., Harper, J.W., and Conaway, J.W. (1999). RBX1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284**,** 657–661. [DOI] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11**,** 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komari, T., Hiei, Y., Saito, Y., Murai, N., and Kumashiro, T. (1996). Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10**,** 165–174. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Miséra, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110**,** 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.-W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10**,** 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Powell, K.A., Mundt, K., Wu, L., Carr, A.M., and Caspari, T. (2003). Cop9/signalosome subunits and Pcu4 regulate ribonucletide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17**,** 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-C., Schiff, M., Serino, G., Deng, X.-W., and Dinesh-Kumar, S.P. (2002). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14**,** 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). COP9 signalosome promotes cleavage of NEDD8-CUL1 conjugates. Science 292**,** 1382–1385. [DOI] [PubMed] [Google Scholar]
- Miséra, S., Müller, A.J., Weiland-Heidecker, U., and Jürgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244**,** 242–252. [DOI] [PubMed] [Google Scholar]
- Oron, E., Mannervik, M., Rencus, S., Harari-Steinberg, O., Neumann-Silberberg, S., Segal, D., and Chamovitz, D. (2002). COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129**,** 4399–4409. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.-W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405**,** 462–466. [DOI] [PubMed] [Google Scholar]
- Pan, Z.Q., Kentsis, A., Dias, D.C., Yamoah, K., and Wu, K. (2004). Nedd8 on cullin: Building an expressway to protein destruction. Oncogene 23**,** 1985–1997. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.-W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted development processes in Arabidopsis. Plant Cell 13**,** 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.-W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128**,** 4277–4288. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Shen, Y., Feng, S., Wang, X., Chittetei, B.N., Vierstra, R.D., and Deng, X.W. (2003). Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr. Biol. 13**,** R504–R505. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Kurz, T., Glaser, S., Willis, J.H., Peter, M., and Bowerman, B. (2003). Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13**,** 911–921. [DOI] [PubMed] [Google Scholar]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34**,** 753–767. [DOI] [PubMed] [Google Scholar]
- Schroeder, D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2002). De-Etiolated 1 and Damaged DNA Binding Protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12**,** 1462–1472. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C. (2004). The COP9 signalosome (CSN): An evolutionary conserved proteolysis regulator in eukaryotic development. Biochim. Biophys. Acta 1695**,** 45–54. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X. (2002). FPLC gel filtration. In Arabidopsis: A Laboratory Manual, D. Weigel and J. Glazebrook, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 226–228.
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292**,** 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.-W. (2002). Multiple ubiquitin-ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14**,** 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., and Villalobos, L.I. (2004). Cullin-containing E3 ubiquitin ligases in plant development. Curr. Opin. Plant Biol. 7**,** 677–686. [DOI] [PubMed] [Google Scholar]
- Seeger, M., Kraft, R., Ferrell, K., Bech-Otschir, D., Dumdy, R., Schade, R., Gordon, C., Naumann, M., and Dubiel, W. (1998). A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 12**,** 469–478. [PubMed] [Google Scholar]
- Serino, G., Su, H., Peng, Z., Tsuge, T., Wei, N., Gu, H., and Deng, X.W. (2003). Characterization of the last subunit of the Arabidopsis COP9 signalosome: Implications for the overall structure and origin of the complex. Plant Cell 15**,** 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.-W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11**,** 1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.-H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13**,** 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., Wei, N., and Deng, X.-W. (1996). Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8**,** 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126**,** 711–721. [DOI] [PubMed] [Google Scholar]
- Tomoda, K., Yoneda-Kato, N., Fukumoto, A., Yamanaka, S., and Kato, J.-y. (2004). Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J. Biol. Chem. 279**,** 43013–43018. [DOI] [PubMed] [Google Scholar]
- Tran, H.J.T.T., Allen, M.D., Löwe, J., and Bycroft, M. (2003). Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42**,** 11460–11465. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276**,** 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9**,** 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.L.-C., Yoshida, H., Lurin, C., and Ecker, J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428**,** 945–950. [DOI] [PubMed] [Google Scholar]
- Wang, X., Feng, S., Nakayama, N., Crosby, W.L., Irish, V.F., Deng, X.W., and Wei, N. (2003). The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell 15**,** 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., Genschik, P., and Hellmann, H. (2004). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137**,** 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee, S., Geyer, R.K., Toda, T., and Wolf, D.A. (2005). CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter stability. Nat. Cell Biol. 7**,** 387–391. [DOI] [PubMed] [Google Scholar]
- Wee, S., Hetfeld, B., Dubiel, W., and Wolf, D.A. (2002). Conservation of the COP9/signalosome in budding yeast. BMC Genet. 3**,** 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.-W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78**,** 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (2003). The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19**,** 261–286. [DOI] [PubMed] [Google Scholar]
- Wertz, I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. (2004). Human De-Etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303**,** 1371–1374. [DOI] [PubMed] [Google Scholar]
- Wolf, D.A., Zhou, C., and Wee, S. (2003). The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5**,** 1029–1033. [DOI] [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.-H., Vidal, M., Elledge, S.J., and Harper, J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing Cul-3. Nature 425**,** 316–321. [DOI] [PubMed] [Google Scholar]
- Zhou, C., Seibert, V., Geyer, R., Rhee, E., Lyapina, S., Cope, G., Deshaies, R.J., and Wolf, D.A. (2001). The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2**,** 7. [DOI] [PMC free article] [PubMed] [Google Scholar]