ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif (original) (raw)
Abstract
ArfGAP1 promotes GTP hydrolysis in Arf1, a small G protein that interacts with lipid membranes and drives the assembly of the COPI coat in a GTP-dependent manner. The activity of ArfGAP1 increases with membrane curvature, suggesting a negative feedback loop in which COPI-induced membrane deformation determines the timing and location of GTP hydrolysis within a coated bud. Here we show that a central sequence of about 40 amino acids in ArfGAP1 acts as a lipid-packing sensor. This ALPS motif (ArfGAP1 Lipid Packing Sensor) is also found in the yeast homologue Gcs1p and is necessary for coupling ArfGAP1 activity with membrane curvature. The ALPS motif binds avidly to small liposomes and shows the same hypersensitivity on liposome radius as full-length ArfGAP1. Site-directed mutagenesis, limited proteolysis and circular dichroism experiments suggest that the ALPS motif, which is unstructured in solution, inserts bulky hydrophobic residues between loosely packed lipids and forms an amphipathic helix on highly curved membranes. This helix differs from classical amphipathic helices by the abundance of serine and threonine residues on its polar face.
Keywords: amphipathic helix, COPI, GTPase
Introduction
Protein coats are dynamic polymers that assemble at the surface of cell membranes to form transport vesicles. Most protein coats are controlled by small G proteins of the Arf and Sar families (Kirchhausen, 2000; McMahon and Mills, 2004). In the GTP conformation, Arf and Sar proteins bind avidly to lipid membranes and recruit coat complexes such as coatomer in the COPI coat, Sec23/24 in the COPII coat, and AP and GGA adaptors in clathrin coats. Coat complexes make additional links with the membrane, notably with short export motifs harboured by membrane proteins, and self-assemble laterally to form a curved lattice that induces bending of the underlying membrane. As a result, a network of interactions maintains the coat in an assembled state until the vesicle pinches off.
One unresolved issue in protein coat dynamics is the timing of GTP hydrolysis. In vitro, GTP hydrolysis in Arf or Sar is required for coat disassembly but not for assembly (Tanigawa et al, 1993; Barlowe et al, 1994; Reinhard et al, 2003). In vivo however, protein coats may lose most Arf or Sar molecules through GTP hydrolysis before disassembly. Indeed, clathrin- and COPII-coated vesicles isolated with GTP contain low amounts of Arf and Sar, respectively (Barlowe et al, 1994; Zhu et al, 1998). One possibility is that the lateral interactions between the polymerized coat proteins and their binding to other membrane components compensate for the loss of Arf-GTP or Sar-GTP. This compensation should be easier within lattice interior, where the network of lateral interactions is complete, than at the coat edge, where new coat proteins assemble. In analogy with microtubules (Desai and Mitchison, 1997), we suggested that the GTP hydrolysis reaction may be spatially organized so as to keep enough Arf/Sar-GTP at the edge of the coat (Antonny and Schekman, 2001).
Several mechanisms could cooperate to restrict the spatial distribution of Arf/Sar-GTP to the edge of a coated membrane area. In the COPII coat, the guanine nucleotide exchange factor Sec12 is excluded from the coat, thus providing new Sar-GTP molecules only at the coat periphery, whereas fast GTP hydrolysis requires Sar1 to be trapped by all COPII units (Antonny et al, 2001; Futai et al, 2004). As a result, the probability of having Sar1-GTP should decrease from the edge to the centre of the coat. In the COPI coat, for which similar mechanisms may apply, we have described an additional mechanism that should prevent GTP hydrolysis at the edge of a coated bud, while accelerating it in the interior. This mechanism lies on the remarkable sensitivity of ArfGAP1, a GTPase-activating protein for Arf1, to membrane curvature (Bigay et al, 2003).
ArfGAP1 is a Golgi-localized protein that controls the dynamics of the COPI coat (Cukierman et al, 1995; Liu et al, 2005). ArfGAP1 is the founder member of the ArfGAP family (Donaldson, 2000; Randazzo and Hirsch, 2004). These proteins are characterized by a Zn-finger domain of about 120 amino acids (aa), which promotes GTP hydrolysis in Arf (Goldberg, 1999; Mandiyan et al, 1999). Since Arf-GTP is attached to lipid membranes, the targeting of ArfGAPs to membranes is crucial for their function. Some ArfGAP proteins contain structural domains (e.g. PH domains) that bind to specific lipids such as phosphoinositides (Donaldson, 2000; Randazzo and Hirsch, 2004). In ArfGAP1 however, the Zn-finger GAP domain (aa 10–130) is the only recognizable domain in the full 415 aa sequence. Moreover, although the binding of ArfGAP1 to model lipid membranes and its activity towards Arf1-GTP are remarkably sensitive to lipid composition, no specific ArfGAP1–lipid interaction seems to be involved (Antonny et al, 1997b). Instead, it seems that the key parameter governing the adsorption of ArfGAP1 to membranes is lipid packing, a physical parameter that itself depends on the shape of lipid molecules and on the curvature of the membrane. At constant liposome radius, the activity of ArfGAP1 increases when conical lipids such as dioleolylglycerol are introduced at the expense of cylindrical lipids (e.g. phosphatidylcholine (PC)) (Antonny et al, 1997b). Conversely, when the liposome composition is kept constant, the activity of ArfGAP1 increases with liposome curvature (Bigay et al, 2003). We proposed that ArfGAP1 recognizes the defects in lipid packing that appear when the actual curvature of the membrane exceeds its spontaneous curvature (Bigay et al, 2003). Functionally, the hypersensitivity of ArfGAP1 to lipid packing could determine the timing and location of GTP hydrolysis within a coated membrane area. When the membrane is curved by the mechanical force imposed by the COPI coat, ArfGAP1 would gradually eliminate Arf1-GTP molecules from the coat except at the edge where membrane curvature is negative (Bigay et al, 2003).
In this study, we show that a central region of ArfGAP1, which is conserved from yeast to mammals, acts as a lipid-packing sensor and helps anchor ArfGAP1 at the surface of highly curved membrane, thus allowing GTP hydrolysis on Arf1. This region, which is not structured in solution, adopts an amphipathic helical structure on curved membranes through insertion of bulky hydrophobic residues between loosely packed lipids.
Results
Gcs1p, the yeast homologue of ArfGAP1, is sensitive to lipid membrane curvature
We previously reported that whereas ArfGAP1 is sensitive to lipid membrane curvature, a construct derived from ASAP1, another GTPase-activating protein for Arf, is not (Bigay et al, 2003). Except for the presence of the Zn-finger GAP domain, ASAP1 and ArfGAP1 have no sequence homology. We therefore postulated that ArfGAP1 contains outside the Zn-finger GAP domain a specific region that binds to highly curved membranes. Yeast Saccharomyces cerevisiae contains six ArfGAP proteins, of which Gcs1p is the closest homologue of ArfGAP1 (Poon et al, 1996). In contrast to other yeast ArfGAPs, Gcs1p shares homology with ArfGAP1 not only for the N-terminal Zn-finger domain but also for a central region of about 60 aa (Figure 1A). This prompted us to examine whether Gcs1p is sensitive to membrane curvature. For this, we used a spectroscopic assay in which the GTP to GDP transition of Arf1, bound to liposomes with a Golgi-like composition, is followed by tryptophan fluorescence. As shown in Figure 1B and C, the time course of Arf1 inactivation initiated by the addition of Gcs1p was very sensitive to liposome radius. As previously observed for rat ArfGAP1 (Bigay et al, 2003), a 100-fold increase in the apparent activity of Gcs1p was observed when the liposome radius was reduced from 140 to 30 nm, with a very sharp effect at radii below 50 nm. Gcs1p had weaker activity than ArfGAP1, an effect that we attributed to the use of mammalian Arf1 as a substrate (Figure 1C). Interestingly, the sensitivity of Gcs1p to liposome radius was shifted to higher radii with liposomes containing unsaturated lipids (C18:1–C18:1) as compared to liposomes made from natural sources (Figure 1C). Yeast cells are known to contain more unsaturated lipids than mammalian cells and Gcs1p could be adapted to this feature.
Figure 1.
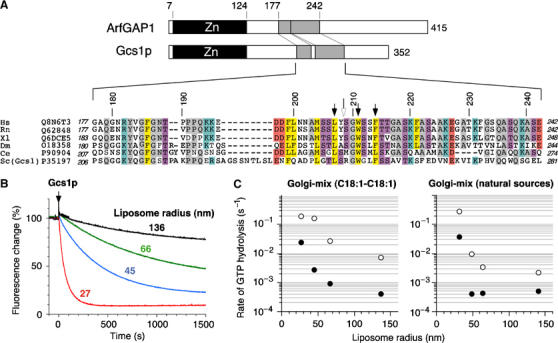
Gcs1p is sensitive to membrane curvature. (A) Sequence alignment of the central region of ArfGAP1 orthologues, including yeast Gcs1p. The abbreviations used are as follows: H.s. Homo sapiens, R.n. Rattus norvegicus, X.l. Xenopus laevis, D.m. Drosophila melanogaster, C.e. Caenorhabditis elegans, S.c. Saccharomyces cerevisiae. The Uniprot accession numbers are indicated. Black arrows indicate point mutations introduced in rat ArfGAP1 and the white arrow indicates a cleavage site by chymotrypsin. Conserved residues are highlighted with the following colour code: yellow, hydrophobic; purple, serine and threonine; blue, basic; red, acidic; grey, other residues. (B) Time course of GTP hydrolysis in Arf1 (0.5 μM) on Golgi-mix (C18:1–C18) liposomes of defined size upon the addition of Gcs1p (50 nM). The reaction was followed by tryptophan fluorescence. The hydrodynamic radius of the liposomes was determined by dynamic light scattering (DLS) and is indicated. (C) Rate (1/_t_1/2) of GTP hydrolysis in Arf1 as a function of the liposome radius in the presence of 50 nM Gcs1p (black circles) or 50 nM ArfGAP1 (white circles). Golgi-mix liposomes were made from natural lipids or enriched in C18:1–C18:1 lipids (see Materials and methods).
ArfGAP1 and Gcs1p bind preferentially to small liposomes
To quantify in a direct manner the avidity of ArfGAP1 and Gcs1p for curved lipid membranes, we used a flotation assay, in which liposomes and associated proteins were recovered by centrifugation at the top of dense sucrose cushions (Figure 2). We found this method more reliable than sedimentation, as the efficiency of liposome recovery by sedimentation diminished as the size of the liposomes decreased. Proteins were incubated with liposomes of defined radii at a protein:lipid molar ratio of about 1:1000. A control experiment was performed in the absence of liposomes. The suspension was adjusted to 30% w/v with sucrose, overlaid with two cushions of lower density and then centrifuged in a swing rotor for 1 h at 240 000 g. The liposomes contained 0.2 mol% of the fluorescent lipid nitrobenzoxadiazoldihexadecanoyl-phosphatidylethanolamine (NBD-PE) and could be visualized in the tubes before and after centrifugation using a fluorescence imaging system (Figure 2A). Whatever their size, the liposomes moved almost completely from the high-sucrose cushion to the sucrose-free cushion as a result of centrifugation. We typically collected 80–90% of the lipids in the top 100 μl. The protein content of the various fractions was determined by SDS–PAGE. As shown in Figure 2B, the binding of ArfGAP1 to liposomes strongly increased with membrane curvature. Thus, if only 14% of ArfGAP1 was found associated with large liposomes (Rh=88 nm), 98% was found associated with small liposomes (Rh=31 nm). Gcs1p bound also in a radius-dependent manner to C18:1–C18:1 Golgi-mix liposomes (Figure 2C). Altogether, the results of the GTPase and flotation assays suggested that ArfGAP1 and Gcs1p contain a region that binds avidly to highly curved lipid membranes.
Figure 2.
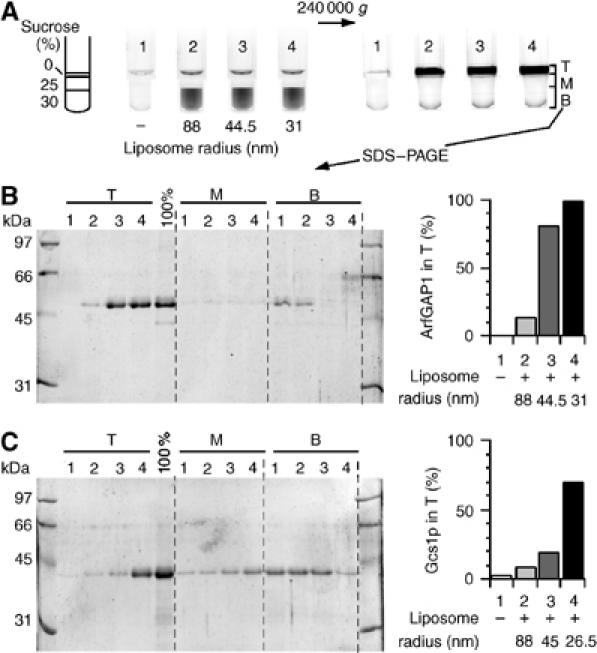
The binding ArfGAP1 and Gcs1p to liposomes increases with membrane curvature. (A) Flotation assay. ArfGAP1 (0.75 μM) was incubated with Golgi-mix liposomes (0.75 mM lipids) of decreasing size (tubes 2–4) or with no liposome (tube 1). The suspension was adjusted to 30% w/v sucrose and overlaid with two cushions of decreasing sucrose density. The NBD fluorescence of the liposomes was visualized in the tubes before and after centrifugation using a fluorescence imaging system. NBD fluorescence appears as black. (B) After centrifugation, the top (T), middle (M) and bottom (B) fractions were collected and analysed by SDS–PAGE. Proteins were stained with sypro-orange. The 100% lane was used to determine the ratio of ArfGAP1 present in the top fraction (right panel). (C) Same analysis with Gcs1p using C18:1–C18:1 Golgi-mix liposomes.
The central region of ArfGAP1 is necessary and sufficient for binding to curved lipid membranes
We constructed several truncated forms of ArfGAP1 in which the central homology region with Gcs1p has been either conserved or deleted (Figure 3A). The [1–257], [1–196], [1–148] and [137–257] constructs contained an N-terminal polyhistidine tag and were purified from bacteria inclusion bodies by nickel chromatography under denaturing conditions. After renaturation by dialysis, proteins were further purified by anion exchange or gel filtration chromatography (Figure 3B). As determined by analytical gel filtration on a Superose-12 column, these constructs displayed an apparent molecular weight within once and twice the calculated molecular weight, suggesting no significant residual aggregation (Figure 3C). In addition, we constructed two protein fusions in which aa 192–257 or 192–231 of ArfGAP1 were added to GST (Figure 3A and B). The sensitivity of the various constructs to membrane curvature was assessed by the flotation assay and, for those containing the GAP domain, by the fluorescence GAP assay.
Figure 3.
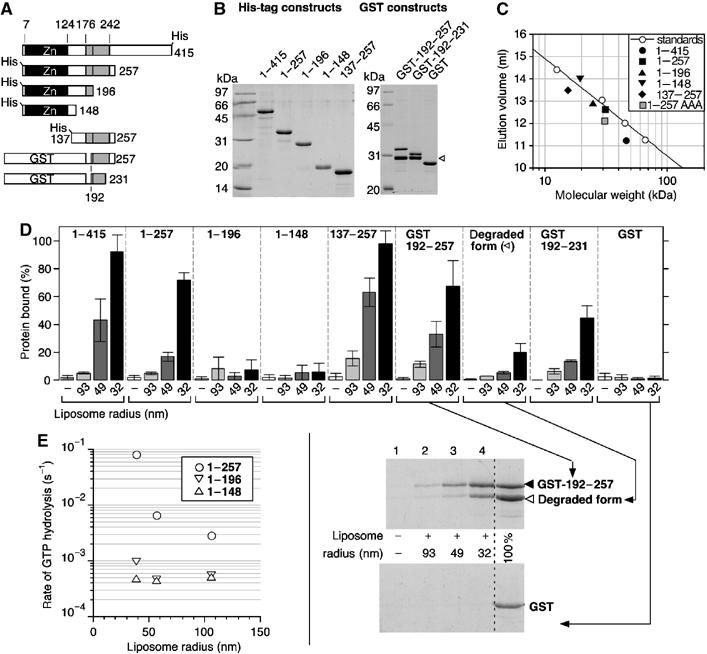
The central region of ArfGAP1 is necessary and sufficient to sense membrane curvature. (A) Diagram of the various constructs. (B) SDS–PAGE of the purified proteins visualized with sypro-orange. Note the presence of a degradation product in the GST constructs (white arrow). This form probably corresponds to a GST-192–214 fusion form as assessed by HPLC and mass spectrometry analysis (see Materials and methods). (C) Size-exclusion chromatography of the constructs with a polyhistidine tag on a superose-12 column. The straight line represents the best fit for the elution of molecular weight standards. The grey square (1–257AAA) represents the elution peak of [1–257]ArfGAP1 carrying the triple L207A–W211A–F214A mutation. (D) Avidity of the various constructs for Golgi-mix liposomes of decreasing size. For all constructs, flotation experiments similar to that described in Figure 2 were conducted except that the protein concentration was 1 μM and the lipid concentration 0.5 mM. The error bars show the variation observed between two independent experiments using different batches of liposomes. A detail view of the SDS–PAGE is shown for some GST constructs. (E) GAP assay using Arf-GTP bound to Golgi-mix liposomes of defined size. ArfGAP1[1–148] and ArfGAP1[1–196] were used at 100 nM. ArfGAP1[1–257] was used at 50 nM.
The largest construct, [1–257]ArfGAP1, which contains the homology region with Gcs1p, showed strong sensitivity to liposome radius. [1–257]ArfGAP1 bound avidly to small liposomes but not to large liposomes (Figure 3D) and its activity on liposome-bound Arf1-GTP strongly increased with membrane curvature (Figure 3E). Therefore, [1–257]ArfGAP1 behaved essentially as full-length ArfGAP1 with regard to its interaction with liposomes. In marked contrast, shorter constructs ending at aa 196 or 148 did not associate to liposomes, even at low liposome radius, and were almost insensitive to liposome radius in the GTPase assay (Figure 3D and E). Because all constructs displayed comparable background GAP activity in solution on a soluble form of Arf1 ([△17]Arf1-GTP; data not shown), we concluded that the central region of ArfGAP1 (within aa 196 and 257) is necessary for coupling GTP hydrolysis in the Arf1–ArfGAP1 complex with the curvature of the supporting lipid membrane.
The central region of ArfGAP1 is not only necessary but also sufficient for interacting with highly curved lipid membrane. This was demonstrated by the fate of three constructs in the flotation assay. [137–257]ArfGAP1 as well as GST-[192–257]ArfGAP1 and GST-[192–231]ArfGAP1 bound in a radius-dependent manner to Golgi-mix liposomes, whereas pure GST did not bind at all (Figure 3D). The largest construct, [137–257]ArfGAP1, bound more avidly than the GST constructs but the recovery of GST-[192–231]ArfGAP1 with small liposomes remained substantial. Interestingly, the two GST fusions contained a degraded form that displayed residual avidity for small liposomes. As assessed by mass spectroscopy analysis, this form would correspond to the fusion of GST with the 192–214 sequence of ArfGAP1.
In conclusion, the mapping experiments presented in Figure 3 demonstrate that the region of ArfGAP1 that binds to highly curved lipid membranes is a motif of about 40 aa (between aa 192 and 231). This motif will be referred here as ALPS (for ArfGAP1 Lipid Packing Sensor). The ALPS motif is highly hydrophobic, rich in serine and threonine residues and overlaps the homology region with Gcs1p (see Figure 1A).
The ALPS motif recognizes highly curved membranes through hydrophobic interactions
We previously suggested that ArfGAP1 recognized packing defects between lipid molecules due to the mismatch between the actual curvature of the lipid membrane and the molecular shape of the various lipids (Antonny et al, 1997b; Bigay et al, 2003). This hypothesis was based on the observation that ArfGAP1 is sensitive not only to the liposome radius but also to the ratio between conical and cylindrical lipids. At constant liposome radius, the recruitment of ArfGAP1 and Gcs1p increased when lipids with small polar heads and/or bulky acyl chains (e.g. dioleoyl-glycerol) were introduced at the expense of cylindrical lipids (e.g. dipalmitoyl-PC) (Antonny et al, 1997b). Therefore, a straightforward model would be that ArfGAP1 inserts hydrophobic residues between loosely packed lipids.
To determine if hydrophobic interactions indeed contribute to the recognition of curved lipid membranes by the ALPS motif, we compared the binding of [137–257]ArfGAP1 to neutral or anionic liposomes of decreasing radius. Figure 4A shows that [137–257]ArfGAP1 bound in a radius-dependent manner to pure PC liposomes and that the introduction of the anionic lipid phosphatidylserine (PS) had no effect on its avidity for liposomes. Moreover, the recruitment of [137–257]ArfGAP1 increased when palmitoyl-oleoyl lipids were replaced by di-oleoyl lipids. We concluded from these experiments that the interaction of the ALPS motif with the lipid bilayer is essentially hydrophobic and is driven by the increase in lipid spacing that occurs when membrane curvature or the ratio between conical and cylindrical lipids increases.
Figure 4.
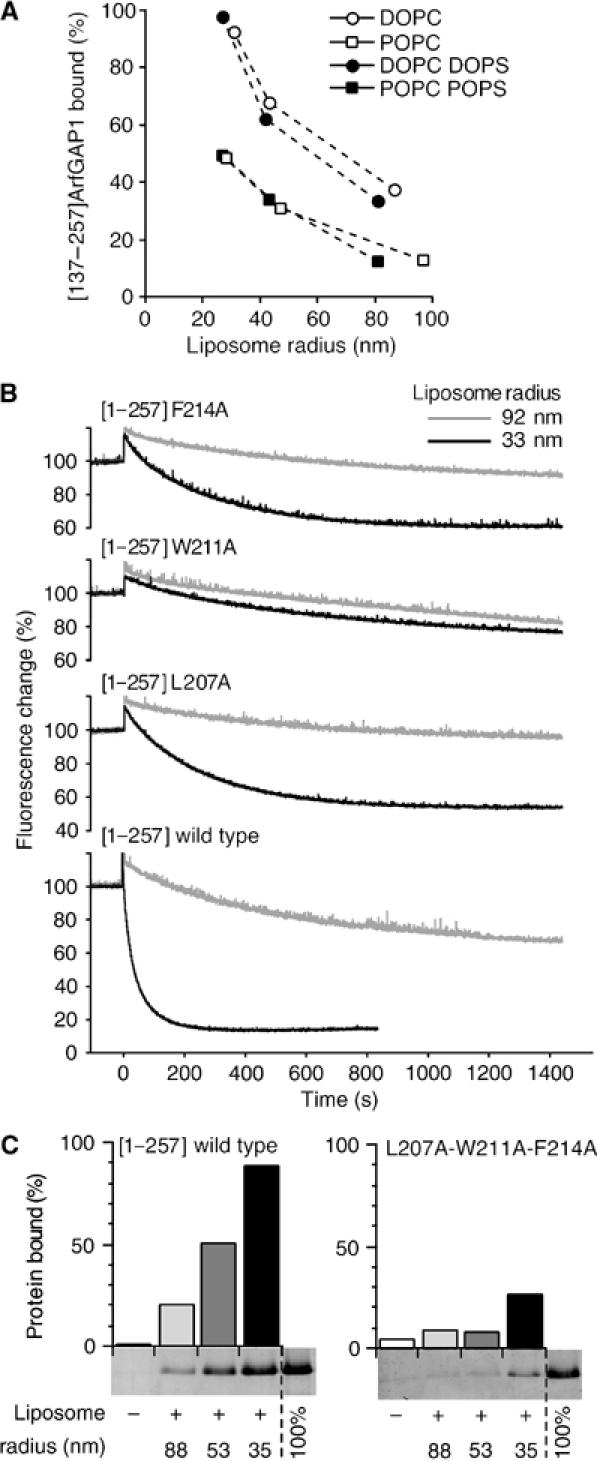
The interaction between the central region of ArfGAP1 and curved lipid membranes is exclusively hydrophobic. (A) Avidity of [137–257]ArfGAP1 for neutral or anionic liposomes of defined size as assessed by flotation experiments. The liposomes contained 100% DOPC (white circles), 100% POPC (white squares), 70% DOPC and 30% DOPS (black circles) or 70% POPC and 30% POPS (black squares). (B) GAP assay using Arf1-GTP bound to Golgi-mix liposomes extruded through 0.2 μm (grey traces) or 0.03 μm (black traces) polycarbonate filters. The hydrodynamic radius of the liposomes was 92 and 33 nm, respectively. GTP hydrolysis was initiated by the addition of 50 nM [1–257]ArfGAP1 carrying the indicated point mutations and was followed by tryptophan fluorescence. (C) Effect of the triple L207A–W211A–F214A mutation on the avidity of [1–257]ArfGAP1 for Golgi-mix liposomes of defined size. Protein concentration: 0.75 μM; lipid concentration: 0.75 mM.
Conserved hydrophobic residues in the ALPS motif contribute to the recognition of curved membranes
Among the most conserved residues of the ALPS motif are several hydrophobic residues (Figure 1A). We postulated that these residues could sense defects in lipid packing induced by membrane curvature. Three of these residues, L207, W211 and F214, were mutated individually or in combination into alanine. The mutations were introduced in the [1–257]ArfGAP1 construct as this form could be easily expressed in Escherichia coli and could be used both in the flotation assay and in the GAP assay.
The L207A, W211A and F214A mutations had drastic effects on the sensitivity of ArfGAP1 to membrane curvature. Figure 4B shows the time course of Arf1 inactivation initiated by the addition of the various [1–257]ArfGAP1 mutants when Arf1-GTP was prebound to small liposomes (black traces) or large liposomes (grey traces). For the wild-type form of [1–257]ArfGAP1, decreasing the liposome radius from 92 to 33 nm increased the rate of Arf inactivation 25-fold. Under the same conditions, a five-fold increase was observed for the L207A and F214A mutants. The W211A mutation had an even more severe effect as it almost abolished the sensitivity of [1–257]ArfGAP1 to liposome curvature (Figure 4B). The results of the flotation assay agreed qualitatively with what was observed in GAP assay. The W211A mutation reduced more strongly the avidity of [1–257]ArfGAP1 for liposomes than the L207A and F214A mutations (data not shown). However, the three mutants still exhibited significant binding to liposomes, probably because the flotation assay was performed at higher lipid concentration than the GAP assay. To confirm the importance of the L207–W211–F214 triplet in the adsorption of ArfGAP1 onto curved lipid membranes, we constructed a triple mutant in which these three hydrophobic residues were replaced simultaneously by alanine. Figure 4C shows that the triple mutant remained essentially soluble in the flotation assay even with small liposomes. This further highlights the essential contribution of L207, W211 and F214 in the recognition of curved membrane.
Limited proteolysis experiments
The hydrophobic residues identified above might directly contact the lipid membrane or alternatively contribute to the folding of the ALPS motif. However, we suspected that this region was actually not well structured in solution. First, we noticed that wild-type [1–257]ArfGAP1 showed longer retention on a gel filtration column as compared to the triple L207A–W211A–F214A mutant, suggesting that the three hydrophobic residues were exposed to the gel matrix (Figure 3C). Second, several constructs containing the central region of ArfGAP1 were partially degraded during protein expression and purification. This degradation was more readily observed for the small GST fusions (see Figure 3B), which were soluble in bacteria and therefore more exposed to proteolysis than the His-tagged constructs, which accumulated in inclusion bodies. The apparent molecular weight of some of the degraded forms suggested that the ALPS motif was a major site for proteolysis.
To probe the folding of the ALPS motif, we conducted limited proteolysis experiments. In agreement with previous findings (Goldberg, 1999), we observed that full-length ArfGAP1 is highly sensitive to proteolysis and that no structural domain, except the N-terminal Zn-finger GAP domain, could be inferred on the basis of resistance to proteolysis (data not shown). However, interesting observations were made using chymotrypsin, which cleaves after aromatic residues. In solution or in the presence of large liposomes, chymotrypsin cleaved [1–257]ArfGAP1 into two major fragments (Figure 5A). The largest fragment (band 1) appeared rapidly and then slowly disappeared (Figure 5A and B). As determined by Western blot and mass spectrometry, this fragment includes the N-terminal polyhistidine tag and ends at residue 208. Tyrosine 208 is right in the middle of the ALPS motif. The smallest fragment (band 2), which appeared more slowly, includes the N-terminal His tag and ends at residue F144, just after the N-terminal Zn-finger domain. Strikingly, the addition of small liposomes had no effect on cleavage at position 144 but abolished cleavage at position 208. Moreover, an additional fragment (band 3) accumulated in the presence of small liposomes with the same kinetics as band 2. This new fragment was identified as the complement of the 1–144 fragment (aa 145–257). Obviously, the 145–257 fragment could not accumulate in solution, as cleavage at position 208 precedes cleavage at position 144. In conclusion, the high susceptibility of the ALPS motif to proteolysis suggests that this region is not a structurally defined domain in solution. Yet this region is capable of interacting directly and specifically with highly curved membranes, which leads to its protection from proteolysis.
Figure 5.
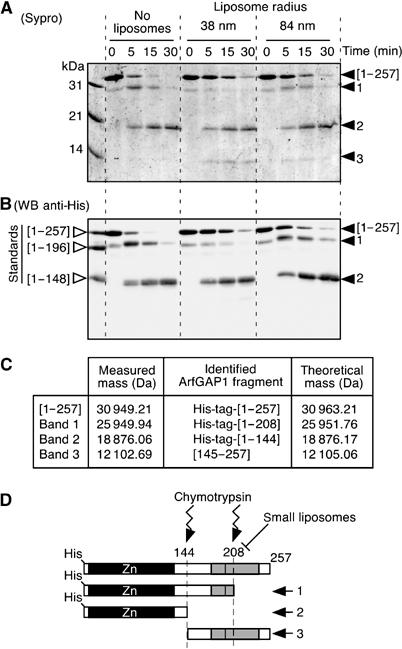
The central region of ArfGAP1 is highly susceptible to proteolysis in solution but is protected in the presence of small liposomes. ArfGAP1[1–257] was incubated with no liposomes, small DOPC:DOPS liposomes or large DOPC:DOPS liposomes. At time zero, chymotrypsin was added. The reaction was stopped at the indicated times and analysed by SDS–PAGE using sypro-orange staining (A) and by Western blot using an anti-polyhistidine tag antibody (B). The first line in the Western blot is a mixture of the indicated ArfGAP constructs (see Figure 3A), which were used as standards. Note that ArfGAP1[1–257] is slightly contaminated at time zero by a band that migrates between the first fragment that is generated by chymotrypsin (band 1) and the [1–196] standard. Similar results were observed with Golgi-mix liposomes. (C) Mass spectrometry analysis of the various fragments generated by chymotrypsin. (D) Schematic view of the proteolysis reaction.
The ALPS motif becomes structured at the surface of highly curved membranes
Several hydrophobic residues of the ALPS motif are regularly spaced every three or four residues (199-FLNNAMSSLYSGWSSFTTGASKFAS-223), a feature suggestive of an amphipathic α-helical structure (Figure 6B). However, the sequence contains several glycine, serine and threonine residues, which have a low propensity to form α-helices. We reasoned that the ALPS motif could be unstructured in solution but could fold as an amphipathic α-helix on curved membranes through the insertion of hydrophobic residues between loosely packed lipids.
Figure 6.
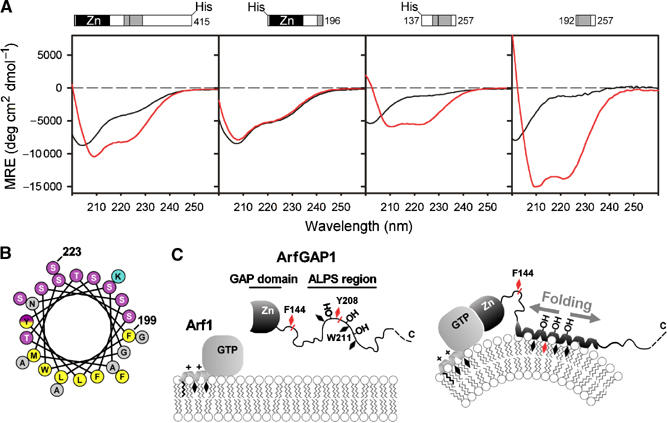
The ALPS motif adopts a helical structure at the surface of highly curved membrane. (A) Far-UV CD spectra of full-length ArfGAP1 and of truncated constructs in the absence (black traces) or presence (red traces) of small DOPC/DOPS liposomes (hydrodynamic radius 24–30 nm). From left to right: ArfGAP1 (8 μM) with and without 4.6 mM liposomes (lipid/protein ratio=575); [1–196]ArfGAP1 (13 μM) with and without 3.225 mM liposomes (L/P=250); [137–257]ArfGAP1 (30 μM) with and without 7.5 mM liposomes (L/P=250); [192–257]ArfGAP (14 μM) with and without 3.125 mM liposomes (L/P=223). (B) Helical-wheel representation of the 199–223 sequence of rat ArfGAP1. The same colour code as Figure 1 was used. The yellow–purple gradation used for tyrosine illustrates the dual character of this residue (hydrophobic and hydroxylated). (C) Schematic view of the coupled folding binding process by which the ALPS motif recognizes curved membranes. Black diamonds represent hydrophobic residues. Chymotrypsin cleavage sites are shown in red. Note that ArfGAP1 interacts also with transmembrane proteins and with coatomer, which have been omitted in this scheme for clarity.
To assess the effect of membrane curvature on the secondary structure of the ALPS motif, we performed far-UV circular dichroism (CD) spectroscopy. The experiments were carried out with full-length ArfGAP1 as well as with three truncated constructs: one including the Zn-finger GAP domain ([1–196]ArfGAP1) and two including the ALPS motif ([137–257]ArfGAP1 and [192–257]ArfGAP1). The [192–257]ArfGAP1 construct was a lyophilized peptide purified by HPLC and solubilized in CD buffer before measurement. Figure 6A shows that full-length ArfGAP1 and the two constructs encompassing the ALPS motif undergo a dramatic change in secondary structure upon binding to small liposomes. In contrast and as expected from previous experiments (Figure 3D and E), [1–196]ArfGAP1 showed no change in secondary structure. In solution, [137–257]ArfGAP1 and [192–257]ArfGAP1 do not show a well-defined secondary structure as indicated by a CD minimum at 202 nm (black traces). When incubated with a saturating amount of small liposomes, the two constructs shifted to a typical α-helical pattern with two CD minima at 210 and 222 nm (red traces). The mean residue ellipticity reached more negative values for the smallest construct, suggesting a higher percentage of α-helix. By fitting the experimental spectra, we estimated the helical content of liposome-bound [137–257]ArfGAP1 and of liposome-bound [192–257]ArfGAP1 at 20 and 48%, respectively. Considering the length of the two constructs, this indicates that 25–33 residues have adopted an α-helical structure, a number that fits well with the size of the 199–223 amphipathic sequence. The CD spectra of full-length ArfGAP1 somehow recapitulated the features of the truncated forms. In solution, ArfGAP1 showed a high content of disordered secondary structure. However, ordered secondary structures, also observed in [1–196]ArfGAP1 and which probably corresponded to the Zn-finger GAP domain, were detected. In the presence of small liposomes, the CD spectrum of full-length ArfGAP1 was clearly dominated by α-helical structures. We concluded from the CD experiments that the adsorption of ArfGAP1 to curved membranes is accompanied by a dramatic structural change and that this change is due to the folding of the ALPS motif into an α-helical structure.
Discussion
We have shown that ArfGAP1 responds to membrane curvature through the membrane adsorption and folding of a motif, which acts as an ArfGAP1 Lipid Packing Sensor (ALPS). This motif is conserved from yeast to mammals (Figure 1A) and is the hallmark of the ArfGAP1/Gcs1 subfamily. Intriguingly, ArfGAP3 and Glo3, which display redundant functions with ArfGAP1 and Gcs1, do not have this motif (Poon et al, 1999; Lewis et al, 2004; Watson et al, 2004). The ALPS motif includes regularly spaced hydrophobic residues, which probably insert directly between lipids provided that the membrane is sufficiently bent. This insertion is accompanied by the folding of the ALPS motif into an amphipathic helical structure. By detecting lipid-packing defects, the ALPS motif would enable tuning the rate of GTP hydrolysis in Arf1 according to the membrane deformation induced by the COPI coat (Figure 6C). The ALPS motif shows overlap with the region of ArfGAP1 (aa 203–334) that has been shown to be required for its cellular localization (Yu and Roth, 2002). Thus, the targeting of ArfGAP1 to the Golgi may involve several determinants, of which the ALPS motif may serve mostly to control the orientation of the GAP domain towards membrane-bound Arf1GTP.
The ALPS motif is centred on a tryptophan residue (W211) (Figure 1A). This residue is strictly conserved and its replacement by an alanine abolished the sensitivity of ArfGAP1 to membrane curvature (Figure 4B). Close to W211 is Y208, which becomes protected from proteolysis as a result of membrane binding (Figure 5), and two well-conserved hydrophobic residues, L207 and F214, which are also important for the recognition of curved membranes (Figure 4B and C). We propose that the side chain of L207, W211 and F214 (and perhaps that of neighbouring hydrophobic residues) inserts between lipids provided that the membrane is sufficiently bent. This insertion explains the strict hydrophobic character of the ArfGAP1/membrane interaction and its sensitivity to both lipid geometry and membrane curvature (Figure 4A) (Antonny et al, 1997b; Bigay et al, 2003).
CD spectroscopy demonstrates that the binding of the ALPS motif to curved lipid membranes is accompanied by its folding into an α-helical structure (Figure 6A). According to secondary-structure prediction algorithms, the 195–230 sequence of ArfGAP1 could form α-helices, but a drop in the probability is found in the middle of the sequence due to the presence of two glycines and several serines and threonines. Consequently, the propensity of this region to adopt an α-helical structure may be exquisitely sensitive to the environment. In the absence of a favourable hydrophobic surface, this region is mostly unstructured as indicated by its high susceptibility to proteolysis (Figure 5) and by its CD spectrum (Figure 6A). However, at the surface of highly curved lipid membranes where defects in lipid packing facilitate the penetration of hydrophobic residues, the ALPS region clearly adopts an α-helical structure (Figure 6A) and becomes protected from proteolysis (Figure 5).
Several proteins interact with lipid membranes through the adsorption of amphipathic α-helices, without necessarily requiring the bending of the lipid membrane. So what prevents the ALPS motif from interacting with flat membranes? We suggest that the ability of the amphipathic helix of the ALPS motif to sense membrane curvature lies on its remarkable polar face, which is composed almost exclusively of serine and threonine residues (Figure 6B). The polar face of membrane-adsorbing amphipathic helices generally contains charged residues, which contribute to membrane adsorption by making electrostatic interactions with lipid polar heads. Thus, the N-terminal amphipathic helix of epsin directly contacts the polar head of PIP2 through its basic polar face (Ford et al, 2002; Stahelin et al, 2003). Other amphipathic helices contain positively and negatively charged residues distributed in a well-defined manner in the polar face to make electrostatic interactions with the polar head of zwitterionic lipids such PC and phosphatidylethanolamine (PE) (Segrest et al, 1992; Cornell and Northwood, 2000; Ulmer et al, 2005). For Arf-GTP, a myristoyl group acts in concert with an N-terminal amphipathic helix to help anchor the protein at the membrane surface (Antonny et al, 1997a; Pasqualato et al, 2002). These examples illustrate that several interactions must work in concert to overcome the cost of spreading apart lipid molecules that should accompany the deep insertion of an amphipathic helix. Consequently, epsin (and perhaps Arf1-GTP) can induce curvature of an initially flat membrane (Ford et al, 2002; Farsad and De Camilli, 2003). In contrast, the ALPS motif can only sense membrane curvature because, except for the presence of hydrophobic residues, it has no additional feature that could help its membrane interaction. Thus, the abundance of threonines and serines may simply arise from the need of having a hydrophilic face without having charged residues that could contribute to membrane binding.
The ALPS motif does not recognize membrane curvature per se, that is, a curved geometry, but loose lipid packing, which is a consequence of membrane curvature. As such, the ALPS motif is very different from the BAR domain, a recently discovered structural domain that can sense and induce membrane curvature (Peter et al, 2004). The BAR domain consists of an elongated six-strand coiled-coil with an overall banana shape, the concave face of which is basic and therefore adapted to interact with acidic and curved lipid membranes (Peter et al, 2004). In vitro, both the BAR domain (Peter et al, 2004) and the ALPS motif (this study) interact preferentially with small liposomes. In vivo however, they may be adapted to different membrane deformations. Thus, high lipid packing should exclude the ALPS motif from the neck of a budding vesicle, whereas the complex surface geometry of such a membrane region, with a dual curvature (negative and positive), could be well adapted to the BAR domain.
From deletion mapping experiments, the boundaries of the ALPS motif are not clear-cut. If the GST-192–231 construct displays significant avidity for small liposomes, larger constructs encompassing this region bound significantly better (Figure 3D). The lack of clear boundaries is not surprising for an intrinsically unstructured region, but it will be important to determine whether the structural change that accompanies the adsorption of ArfGAP1 to curved lipid membranes is limited to the amphipathic sequence around W211 or whether it propagates beyond. ArfGAP1 interacts not only with Arf1-GTP, but also with coatomer and transmembrane proteins (Aoe et al, 1999; Goldberg, 1999; Lanoix et al, 2001; Majoul et al, 2001; Szafer et al, 2001; Rein et al, 2002; Yang et al, 2002), and a large structural change imposed by membrane curvature may have profound effect on this interaction network in the confined environment of a coated membrane bud. Also of note is the fact that many proteins that are involved in the formation of transport vesicles contain large regions that are predicted to be intrinsically unfolded. This feature is very important for the rapid and reversible formation of multiple protein/protein interactions (Dafforn and Smith, 2004). In addition, these regions may also be affected by membrane-induced folding events.
Materials and methods
Liposome preparation
Lipids in chloroform were purchased from Avanti Polar Lipids except egg PC (Sigma) and NBD-PE (Molecular Probes). Liposomes used in flotation and GAP activity experiments were produced by extrusion. A dried film was prepared by evaporation of a mixture of the indicated lipids in chloroform and resuspended in 50 mM Hepes pH 7.2 and 120 mM K-acetate. After five steps of thawing and freezing in liquid nitrogen, the liposome suspension was extruded sequentially through (pore size) 0.4, 0.2, 0.1, 0.05 and 0.03 μm polycarbonate filters using a hand extruder (Avanti) at a final lipid concentration of 1–2.5 mM. The liposome radius was estimated by DLS in a Dyna Pro instrument. Liposomes were stored at room temperature and used within 2 days after preparation. The composition of Golgi-mix liposomes was (mol%) egg PC (50), egg PE (19), brain PS (5), liver phosphatidylinositol (10), cholesterol (16) and NBD-PE (0.2). (C18:1–C18:1) Golgi-mix liposomes had the same polar head composition but were prepared with dioleoyl-PC, dioleoyl-PE and dioleoyl-PS. The small liposomes used for CD spectroscopy were obtained by sonication of DOPC:DOPS (70:30) suspension (15 mM) in 10 mM Tris pH 7.5 and 150 mM KCl with a titanium tip sonicator. Titanium and lipid debris were removed by centrifugation at 100 000 g for 20 min to obtain a homogeneous population of liposomes with an average radius of 24–30 nm (as determined by DLS on several samples).
Protein expression and purification
Full-length myristoylated Arf1 and Gcs1p were purified as described (Franco et al, 1995; Poon et al, 2001). Full-length rat ArfGAP1 with a polyhistidine tag was purified from SF9 cells (generous gift from J Premont) by MonoQ chromatography as described (Premont and Vitale, 2001).
The [1–257], [1–148], [1–196] and [137–257] fragments of rat ArfGAP1 were PCR-amplified from the pKM260 construct (Cukierman et al, 1995) using primers that incorporated _Nde_I and _Bam_HI restriction sites and ligated into the pET16b expression vector (Novagen). This vector adds a polyhistidine tag and a cleavage site for factor X. Point mutations (L207A, W211A and F214A) were introduced in the ArfGAP1[1–257]-pET16b vector using the site-directed mutagenesis kit from Stratagene. The pET16b ArfGAP1 constructs were expressed in E. coli at 37°C and purified from inclusion bodies by nickel chromatography (Qiagen) under denaturing conditions as described (Huber et al, 2001). Renaturation was performed by gentle dialysis against a mixture of two solutions; the first contained 25 mM Tris pH 7.5, 150 mM NaCl, 4 M guanidine, 2 mM DTT and 1 mM PMSF and was gradually replaced (overnight) by the second solution containing 25 mM Tris pH 7.5, 50 mM NaCl, 50 μM ZnCl2, 2 mM DTT and 1 mM PMSF. Aggregated material was discarded by centrifugation. For the [1–148], [1–196] and [1–257] constructs, the supernatant was further purified by MonoQ chromatography (Amersham) using a linear 0–1 M NaCl gradient. For the [137–257] construct, the supernatant was further purified by gel filtration on a sephacryl HR 200 column (Amersham) in 50 mM Tris pH 7.5, 100 mM NaCl and 1 mM MgCl2.
For the GST fusions, the [192–231] or [192–257] sequence of ArfGAP1 was PCR-amplified as _Bam_HI/_Eco_RI inserts and cloned into the pGEX-2T expression vector (Roche), which includes a thrombin cleavage site. After expression, bacteria were lysed in 50 mM Tris pH 7.4 and 150 mM NaCl, supplemented with 1 mM PMSF, 1 mM pepstatin, 10 mM bestatin, 10 mM phosphoramidon and a cocktail of anti-proteases (Roche). The supernatant was incubated for 1 h with a glutathione Sepharose 4B gel (Amersham) and the beads were submitted to several washes. GST fusions were eluted in the same buffer supplemented with 10 mM glutathione. Alternatively, the beads with GST-[192–257]ArfGAP1 were incubated with thrombin to allow the elution of the ArfGAP1 peptide. The thrombin eluate was further purified by HPLC on a chromolith C18 column (Merck) with an acetonitrile gradient. By mass spectrometry, we identified one HPLC peak as the [192–257]ArfGAP1 peptide and a second peak as the [192–214]ArfGAP1 peptide. The two peptides also contained the GS sequence from the thrombin cleavage site of the pGEX-2T expression vector. The [192–257]ArfGAP1 peptide was lyophilized and stored at −20°C before use.
Analytical gel exclusion chromatography
Aliquots of the various purified ArfGAP1 constructs (0.7–2 nmol in a total volume of 100 μl) were applied to a superose 12 column and eluted at a flow rate of 0.5 ml/min in 25 mM Hepes pH 7.5, 100 mM KCl, 1 mM MgCl2 and 1 mM DTT. The column was calibrated using the following standards: bovine serum albumin (66.0 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (29.0 kDa) and cytochrome c (12.4 kDa).
Tryptophan fluorescence GAP assay
Tryptophan fluorescence was measured at 340 nm (bandwidth 20 nm) upon excitation at 297.5 nm (bandwidth 3 nm) in a Shimadzu R5301 fluorimeter equipped with stirring and injection facilities. All experiments were performed at 37°C. The cylindrical quartz cuvette initially contained extruded liposomes (0.2 mM) in HKM buffer (50 mM Hepes pH 7.2, 120 mM KAc, 1 mM MgCl2, 1 mM DTT). Arf1-GDP (0.5 μM) was added and activated by the sequential addition of 40 μM GTP and 2 mM EDTA (giving 1 μM [Mg2+]free). After 10 min, 2 mM MgCl2 (giving 1 mM [Mg2+]free) was added. GTP hydrolysis was initiated by the addition of Gcs1p, ArfGAP1 or the indicated constructs. For simplicity, the fluorescence level of Arf1-GDP was arbitrary set at zero and the fluorescence level after the GTP loading step was set at 100%. Therefore, the fluorescence scale shown in the figures directly reflects the fraction of Arf1 that undergoes GTP hydrolysis.
Flotation experiments
Proteins (0.5–1 μM) and liposomes (0.5–1 mM) were incubated in HKM buffer at room temperature for 5 min in a total volume of 150 μl. The suspension was adjusted to 30% sucrose by adding and mixing 100 μl of a 75% w/v sucrose solution in HKM buffer. The resulting high-sucrose suspension was overlaid with 200 μl HKM containing 25% w/v sucrose and 50 μl HKM containing no sucrose. The sample was centrifuged at 55 000 r.p.m. (240 000 g) in a Beckman swing rotor (TLS 55) for 1 h. The bottom (250 μl), middle (150 μl) and top (50 μl) fractions were manually collected from the bottom using a Hamilton syringe and analysed by SDS–PAGE using the fluorescent dye sypro-orange (Molecular Probes). The gels were visualized and quantified using a FUJI LAS-3000 fluorescence imaging system. We checked the recovery of the liposomes during centrifugation and after fraction collection by following NBD-PE fluorescence. A fluorescence image of the tubes before and after centrifugation was taken in the fluorescence imaging system using a small mirror at 45°, which directed the side image of the tubes to the vertically positioned CCD camera. After fraction collection, the amount of NBD in the various collected fractions was quantified using the same imaging system.
Limited proteolysis
[ArfGAP1] (0.8 μM) was incubated in HKM buffer with or without liposomes (0.8 mM) at 25°C in a total volume of 150 μl. At time zero, 0.08 μg/ml chymotrypsin was added. At the times indicated, 30 μl aliquots were withdrawn, supplemented with 0.5 mM PMSF and stored on ice. The proteolysis reaction was analysed by SDS–PAGE using sypro-orange staining and by Western blot using an anti-polyhistidine tag antibody.
Mass spectrometry
Mass measurements were made on a Perseptive Biosystems Voyager-DE PRO MALDI-TOF mass spectrometer equipped with a 1.3 m ion flight tube, delayed-extraction technology and a reflector analyser (Framingham, MA). Prior to mass spectrometry, protein samples were desalted and concentrated using microscale reversed-phase column (ZipTipC4, Millipore, Bedford, USA) by washing with 0.1% trifluoroacetic acid (TFA) and eluting with 80% acetonitrile and 0.1% TFA. The samples were mixed (1:10) with sinapinic acid matrix (3,5-dimethoxy-4-hydroxycinamic acid, 10 mg/ml; Sigma, St Louis, USA), and 2 μl (20 pmol) of the mixture was applied to the sample plate. All spectra were acquired in a positive linear mode with a 25 kV acceleration voltage. For each sample, mass spectra were obtained by accumulating 200 shots and by averaging five measurements, and a close external calibration was performed using a protein mixture with mass range from 5734.59 Da to 44 613 Da. Processing of mass spectra was performed with the Data Explorer 4.0 software.
Circular dichroism measurements
CD spectroscopy was performed on a Chirascan spectropolarimeter (Applied Photophysics). Before measurements, the proteins were dialysed against 10 mM Tris pH 7.5, 150 mM KCl and 1 mM DTT. The experiments were performed at room temperature in a HELLMA quartz cell with an optical path length of 0.02 cm. Each spectrum was obtained by averaging several scans recorded from 200 to 260 nm with a bandwidth of 2 nm, a step of 1 nm and a scan speed of 20 nm/min. Control spectra of liposomes in buffer were subtracted from the protein spectra. The spectra were analysed in the 200–240 nm range with the CDPro software (Sreerama and Woody, 2000), which contains three fitting algorithms and a set of reference spectra (SMP50). The percentages of secondary structure given in Results are those estimated with the CONTIN/LL algorithm, which gave the best fits.
Acknowledgments
We thank Sabine Scarzello for mass spectrometry analysis and the plateforme RIO (Centre de Biochimie Structurale, Montpellier) for sharing the CD instrument. We thank Richard Premont for reagents and Dan Cassel, Marc Chabre, Michel Franco, Harvey McMahon and Sonia Paris for discussions. This work was supported by the CNRS (ACI ‘jeune chercheur' and ACI ‘dynamique et réactivité des assemblages biologiques') and the EMBO Young Investigator Programme. G Drin is supported by a CNRS postdoctoral fellowship.
References
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M (1997a) N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36: 4675–4684 [DOI] [PubMed] [Google Scholar]
- Antonny B, Huber I, Paris S, Chabre M, Cassel D (1997b) Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J Biol Chem 272: 30848–30851 [DOI] [PubMed] [Google Scholar]
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol 3: 531–537 [DOI] [PubMed] [Google Scholar]
- Antonny B, Schekman R (2001) ER export: public transportation by the COPII coach. Curr Opin Cell Biol 13: 438–443 [DOI] [PubMed] [Google Scholar]
- Aoe T, Huber I, Vasudevan C, Watkins SC, Romero G, Cassel D, Hsu VW (1999) The KDEL receptor regulates a GTPase-activating protein for ADP-ribosylation factor 1 by interacting with its non-catalytic domain. J Biol Chem 274: 20545–20549 [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R (1994) COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77: 895–907 [DOI] [PubMed] [Google Scholar]
- Bigay J, Gounon P, Robineau S, Antonny B (2003) Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature 426: 563–566 [DOI] [PubMed] [Google Scholar]
- Cornell RB, Northwood IC (2000) Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem Sci 25: 441–447 [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D (1995) The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270: 1999–2002 [DOI] [PubMed] [Google Scholar]
- Dafforn TR, Smith CJ (2004) Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep 5: 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13: 83–117 [DOI] [PubMed] [Google Scholar]
- Donaldson JG (2000) Filling in the GAPs in the ADP-ribosylation factor story. Proc Natl Acad Sci USA 97: 3792–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K, De Camilli P (2003) Mechanisms of membrane deformation. Curr Opin Cell Biol 15: 372–381 [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Franco M, Chardin P, Chabre M, Paris S (1995) Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J Biol Chem 270: 1337–1341 [DOI] [PubMed] [Google Scholar]
- Futai E, Hamamoto S, Orci L, Schekman R (2004) GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J 23: 4146–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J (1999) Structural and functional analysis of the ARF1–ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96: 893–902 [DOI] [PubMed] [Google Scholar]
- Huber I, Rotman M, Pick E, Makler V, Rothem L, Cukierman E, Cassel D (2001) Expression, purification, and properties of ADP-ribosylation factor (ARF) GTPase activating protein-1. Methods Enzymol 329: 307–316 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T (2000) Three ways to make a vesicle. Nat Rev Mol Cell Biol 1: 187–198 [DOI] [PubMed] [Google Scholar]
- Lanoix J, Ouwendijk J, Stark A, Szafer E, Cassel D, Dejgaard K, Weiss M, Nilsson T (2001) Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J Cell Biol 155: 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Poon PP, Singer RA, Johnston GC, Spang A (2004) The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol Biol Cell 15: 4064–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Duden R, Phair RD, Lippincott-Schwartz J (2005) ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J Cell Biol 168: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Straub M, Hell SW, Duden R, Soling HD (2001) KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev Cell 1: 139–153 [DOI] [PubMed] [Google Scholar]
- Mandiyan V, Andreev J, Schlessinger J, Hubbard SR (1999) Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta. EMBO J 18: 6890–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Mills IG (2004) COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol 16: 379–391 [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back' communication. EMBO Rep 3: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Huber I, Singer RA, Johnston GC (2001) Expression, analysis, and properties of yeast ADP-ribosylation factor (ARF) GTPase activating proteins (GAPs) Gcs1 and Glo3. Methods Enzymol 329: 317–324 [DOI] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC (1999) Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer RA, Johnston GC (1996) Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA 93: 10074–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont RT, Vitale N (2001) Purification and characterization of GIT family of ADP-ribosylation factor (ARF) GTPase-activating proteins. Methods Enzymol 329: 335–343 [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Hirsch DS (2004) Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal 16: 401–413 [DOI] [PubMed] [Google Scholar]
- Rein U, Andag U, Duden R, Schmitt HD, Spang A (2002) ARF-GAP-mediated interaction between the ER–Golgi v-SNAREs and the COPI coat. J Cell Biol 157: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Schweikert M, Wieland FT, Nickel W (2003) Functional reconstitution of COPI coat assembly and disassembly using chemically defined components. Proc Natl Acad Sci USA 100: 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res 33: 141–166 [PubMed] [Google Scholar]
- Sreerama N, Woody RW (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287: 252–260 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W (2003) Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J Biol Chem 278: 28993–28999 [DOI] [PubMed] [Google Scholar]
- Szafer E, Rotman M, Cassel D (2001) Regulation of GTP hydrolysis on ADP-ribosylation factor-1 at the Golgi membrane. J Biol Chem 276: 47834–47839 [DOI] [PubMed] [Google Scholar]
- Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms JB, Rothman JE (1993) Hydrolysis of bound GTP by ARF protein triggers uncoating of Golgi-derived COP-coated vesicles. J Cell Biol 123: 1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer TS, Bax A, Cole NB, Nussbaum RL (2005) Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 280: 9595–9603 [DOI] [PubMed] [Google Scholar]
- Watson PJ, Frigerio G, Collins BM, Duden R, Owen DJ (2004) Gamma-COP appendage domain—structure and function. Traffic 5: 79–88 [DOI] [PubMed] [Google Scholar]
- Yang JS, Lee SY, Gao M, Bourgoin S, Randazzo PA, Premont RT, Hsu VW (2002) ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol 159: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Roth MG (2002) Casein kinase I regulates membrane binding by ARF GAP1. Mol Biol Cell 13: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Traub LM, Kornfeld S (1998) ADP-ribosylation factor 1 transiently activates high-affinity adaptor protein complex AP-1 binding sites on Golgi membranes. Mol Biol Cell 9: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]