Implications of CD94 deficiency and monoallelic NKG2A expression for natural killer cell development and repertoire formation (original) (raw)
Abstract
Natural killer (NK) cells are believed to achieve self-tolerance through the expression of self-MHC-specific inhibitory receptors, such as members of the Ly49 and CD94/NKG2 families. Individual Ly49 genes are stochastically expressed by NK subsets and are expressed in a monoallelic fashion, but little is known about the mechanisms underlying CD94/NKG2A expression. We show here that, like Ly49 genes, mouse Nkg2a is stochastically and monoallelically expressed. Thus, a single general mechanism controls expression of all known MHC-specific receptors by mouse NK cells. In addition, we find that DBA/2J mice are naturally CD94-deficient and do not express cell-surface CD94/NKG2A receptors, even on neonatal NK cells. Thus, self-tolerance of neonatal NK cells cannot be attributed to CD94/NKG2A expression. Taken together, the results lead to a reconsideration of current models of NK cell development and self-tolerance.
Cytolysis of infected, stressed, or transformed target cells by natural killer (NK) cells is regulated by several families of cell-surface receptors expressed by NK cells (1–3). In the mouse, a particular focus has been two families of cell-surface receptors that recognize MHC class I molecules—the Ly49 family and the CD94/NKG2 family. The Ly49 receptors are homodimers that bind directly to their class I ligands. CD94/NKG2 receptors, on the other hand, are heterodimers of CD94 and one of several NKG2 family members, i.e., NKG2A, NKG2C, or NKG2E. CD94/NKG2 receptors monitor classical class I expression “indirectly” by means of the recognition of classical class I signal peptides (4–8). These signal peptides are processed to a nonamer peptide called Qdm that is presented to CD94/NKG2+ NK cells by a nonclassical MHC class I molecule called Qa-1 (or HLA-E in humans). Ly49 and CD94/NKG2 receptors equip NK cells with the capacity to detect cells with abnormal MHC class I expression, such as often results from viral infection or transformation (9).
Ly49 receptors are expressed in a variegated pattern on NK cells, such that each NK cell stochastically expresses only a few of its receptor genes, and each receptor is expressed by only ≈10–50% of NK cells (3). Remarkably, NK cells expressing a given Ly49 gene usually (but not always) express only one of the two possible Ly49 alleles (10–12). Such “monoallelic” expression is believed to arise as a consequence of the stochastic mechanism that acts during NK development to choose a limited number of Ly49 alleles for expression. In contrast to Ly49 gene expression, little is known about the molecular mechanisms that control CD94/NKG2 expression. Nor is it understood how CD94/NKG2 expression would be coordinated with Ly49 expression to ensure that each NK cell expresses at least one self-specific inhibitory receptor, as is widely assumed to be required to prevent NK autoreactivity (13).
Interestingly, although neonatal NK cells express virtually no self-specific Ly49 receptors (14–16), the vast majority of these cells express CD94/NKG2 receptors (16–19). This finding led to the proposal that CD94/NKG2A expression is critical for assuring the self-tolerance of Ly49-negative cells in the neonatal period (16, 18).
Here we examine NKG2A expression with a specific mAb and provide evidence that the Nkg2a gene is monoallelically expressed. In addition, we describe a CD94-deficient mouse strain and its complementation by a CD94 transgene. Our studies lead to a reconsideration of current models of NK cell development and tolerance.
Materials and Methods
Mice.
DBA/2 substrains were purchased from Taconic Farms, Harlan, the National Cancer Institute, and Charles River Breeding Laboratories. All other strains of mice were obtained from The Jackson Laboratory. A backcross panel used to map the gene responsible for the lack of expression of CD94 in DBA/2J mice was generated by mating B6D2F1 mice to DBA/2J mice.
Abs and Flow Cytometry.
The 20d5 (anti-NKG2A/C/E) and 18d3 (anti-CD94) mAbs have been described (20). Purified 20d5 and 18d3 were conjugated to FITC (Pierce) or NHS-LC biotin (Pierce). Anti-DX5-phycoerythrin (PE), -NK1.1-PE, -NK1.1-biotin, -CD3-CyChrome, -Ly49D-FITC, -I-Ab-biotin, -I-Ad-biotin, and -CD3-PE were purchased from PharMingen. Isotype controls, anti-Kk-FITC, and tricolor conjugates of anti-CD4, -CD8, and streptavidin were purchased from Caltag (South San Francisco, CA). Streptavidin-Red613 was purchased from GIBCO/BRL.
The 16a11 anti-NKG2AB6 mAb was generated as described (20) except that 129/SvJ mice were immunized instead of rats. mAb 16a11 is a mouse IgG2bκ, purified over protein A agarose (Roche Molecular Biochemicals), and conjugated to NHS-LC biotin (Pierce). When costaining cells with 16a11 and 20d5, crossblocking was minimized by first incubating cells with 16a11 and then with 20d5.
Transient Transfections of COS-7 Cells.
COS-7 cells were transfected with supercoiled DNA (Qiagen, Chatsworth, CA), using Lipofectamine reagent (GIBCO/BRL), and analyzed after 2 days. Transfected cDNAs were cloned into the pME18S expression vector (GenBank accession no. AB009864), as described (4, 20–22).
Reverse Transcription (RT)-PCR to Distinguish NKG2ABALB and NKG2AB6.
Radioactive RT-PCR was performed as described (20), except that primers NKG2A 5′ex3 (5′-TTAATTGTCGCTGTGGTTGTAATTACTAC-3′) and NKG2 3′BALB (5′-TCAGATGGG(A/G)AATTTACACTTACAAAGATATGG-3′) were used, and the PCR product was digested with _Mbo_I, which cuts the B6 PCR product in two places (nucleotides 308 and 540) and the BALB product only once (at nucleotide 308).
Sequencing.
Primers NKG2 5′437 (5′-GAAAATCTTGGAATGACAGTTTGG-3′) and NKG2 3′BALB were used to amplify a portion (nucleotides 461–702) of NKG2 cDNAs from lymphokine-activated killer cell RNA by PCR with Taq polymerase (Promega). PCR products were cloned into the T easy vector (Promega) and sequenced by using standard T7 and SP6 primers. A few clones contained a small number of nucleotide substitutions [compared with the canonical B6 or BALB NKG2A sequence; 5 substitutions seen in 19 cDNAs (4,579 bp) sequenced]. These were likely introduced by Taq, because consistent substitutions did not occur at reproducible positions, except for two clones, in which changes at nucleotide 686 were observed: in clone DP-9 there was a G to A change in an otherwise BALB-like sequence, and in clone DP-10 there was an A to G change in an otherwise B6-like sequence. These reciprocal changes are likely the result of chimeric B6/BALB cDNAs generated during PCR by annealing of a truncated NKG2A product derived from one strain to the NKG2A product of the other strain.
Northern Blotting.
RNA blots of lymphokine-activated killer cell cultures were prepared as described (21) as were cDNA probes (4, 21).
CD94 Transgenic Mice.
The CD94 cDNA (21) was subcloned into the class I promoter/Igμ enhancer expression cassette (23) and injected into fertilized (B6 × CBA/J)F2 eggs. Transgenic founder mice were identified by Southern blotting with a CD94 cDNA probe and by staining peripheral blood lymphocytes with 18d3. The mice were first backcrossed 3 times to C57BL/6, and then 2 times to DBA/2J. After the first cross to DBA/2J, mice that carried the B6 NK complex (NKC) (i.e., _Nkrp1c_B6+, as assessed with the PK136 mAb) were selected for the second cross to DBA/2J, thereby ensuring that subsequent generations did not carry the CBA/J NKC.
Results and Discussion
A Specific Anti-NKG2A mAb.
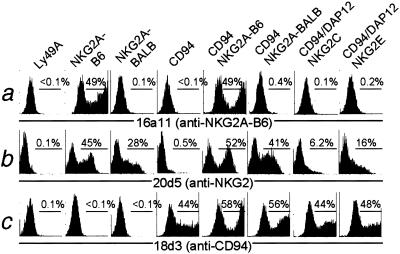
A hybridoma, 16a11, was obtained by immunization of 129Sv/J mice with CD94/NKG2AB6-expressing Chinese hamster ovary (CHO) cells. mAb 16a11 stained COS7 cells transfected with NKG2AB6 alone and also recognized COS7 cells transfected with both CD94 and NKG2AB6 (Fig. 1a). In contrast, 16a11 did not react appreciably with COS7 cells transfected with Ly49A, NKG2ABALB, CD94 alone, or with CD94 and NKG2ABALB together (Fig. 1a). Moreover, 16a11 did not react appreciably with COS cells expressing CD94/NKG2CB6/DAP12 or CD94/NKG2EB6/DAP12. Staining with 18d3 (anti-CD94) and 20d5 (anti-NKG2) mAbs confirmed expression of the transfected molecules (Fig. 1 b and c). Although the expression of NKG2C is weak on transiently cotransfected COS cells (Fig. 1 and ref. 20), stably transfected CHO cells expressing high levels of CD94/NKG2C (20) were also not stained by 16a11 mAb (data not shown). We infer that 16a11 is an NKG2AB6-specific mAb.
Figure 1.
A mAb, 16a11, distinguishes NKG2AB6 from other NKG2 molecules. COS7 cells were transfected with expression vectors encoding the indicated cDNAs, and 48 h later, aliquots of the transfectants were stained with (a) biotinylated 16a11 mAb and streptavidin-PE, (b) FITC-conjugated 20d5 mAb, which recognizes all tested NKG2 molecules, or (c) biotinylated 18d3 (anti-CD94), followed by streptavidin-PE. Transfection of CD94/NKG2C/DAP12 was relatively inefficient in this experiment, but the absence of staining by 16a11 was confirmed in two other transient transfection experiments and in an experiment that used stable Chinese hamster ovary cell transfectants expressing high levels of CD94/NKG2C (data not shown).
Expression of NKG2A in B6 and BALB/c Mice.
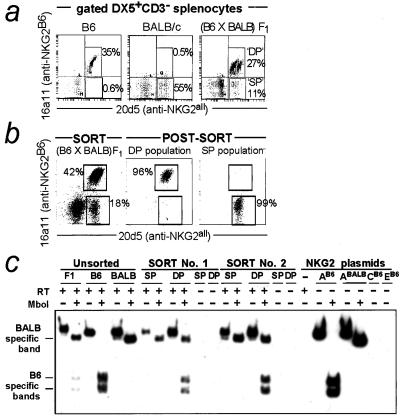
A population of DX5+CD3− spleen cells from B6 mice stained with 16a11 mAb (Fig. 2a). As expected, virtually all 16a11+ (NKG2A+) NK cells in B6 mice costained with the 20d5 mAb (anti-NKG2A/C/E), resulting in a diagonal staining pattern (Fig. 2a) that is characteristically observed with two mAbs that recognize the same antigen. Surprisingly, almost all 20d5+ cells were 16a11+ (Fig. 2a), implying that there are very few NKG2C/E+A− NK cells. A few cells were present in the 16a11−20d5+ quadrant in multiple experiments, but because we lack a specific anti-NKG2C/E mAb, it remains unclear whether these are bona fide NKG2C/E+NKG2A− cells. On the basis of the very low levels of NKG2C/E mRNA in NK cells (20), it is tempting to conclude that there are very few NKG2C/E+ cells among freshly isolated NK cells in B6 mice. However, it remains possible that there is a substantial number of NKG2C/E+ cells that also coexpress NKG2A or that express NKG2C/E at low cell-surface levels.
Figure 2.
Monoallelic expression of NKG2A. (a) Nylon wool-passed spleen cells were stained with 16a11-biotin (anti-NKG2AB6), followed by DX5-PE, CD3-CyChrome, streptavidin-613, and 20d5-FITC (anti-NKG2B6 and BALB). DP, 16a11/20d5+; SP, 20d5+, as indicated. (b) DP and SP lymphokine-activated killer cells from (BALB/c × C57BL/6)F1 mice were sorted to >95% purity. One representative sort of three is shown. (c) NKG2A was amplified by radioactive RT-PCR. Reverse transcriptase (RT) was omitted in certain reactions as a control for DNA contamination of the RNA samples, as indicated. The BALB and B6 alleles of NKG2A share one _Mbo_I site, and in addition, NKG2AB6 contains a second site not found in NKG2ABALB. The results obtained from two independent sorts (of three) are shown. A very rapidly migrating fragment (corresponding to the cDNA fragment 5′ of the first _Mbo_I site) is visible on the autoradiograph for _Mbo_I-digested samples but is not shown.
The allele specificity of 16a11 implied by the transfection data (Fig. 1) was confirmed by the failure of the 16a11 mAb to stain NK cells from BALB/c mice (Fig. 2a). Approximately one-half of BALB/c NK cells stained well with the 20d5 mAb, indicating that these cells express at least one NKG2-like molecule, likely NKG2ABALB (22).
Monoallelic and Biallelic Expression of NKG2A.
Strikingly, when NK cells from (B6 × BALB/c)F1 (abbreviated as CB6F1) mice were costained with 20d5 and 16a11 mAb, two distinct populations of 20d5+ NK cells were evident (Fig. 2a): a single-positive (SP) population that was 20d5+16a11− and a double-positive (DP) population that was 20d5+16a11+. We hypothesized that SP cells express the BALB but not the B6 allele of NKG2A, whereas the DP population includes cells that express only the B6 NKG2A allele and/or cells that express both the B6 and BALB alleles of NKG2A. To test this possibility, RNA was isolated from sorted IL-2-cultured SP and DP NK cells (Fig. 2b). RT-PCR was conducted in the presence of [α32P]dCTP, using primers capable of amplifying NKG2A from B6 or BALB mice. The resulting PCR products were digested with _Mbo_I, a restriction enzyme that differentially digests NKG2AB6 (2 cuts) and NKG2ABALB (1 cut). _Mbo_I-digested PCR products derived from sorted DP cells included both B6 and BALB/c NKG2A species (Fig. 2c). However, the NKG2A PCR product derived from SP cells was solely of BALB/c origin. Thus, the RNA analysis suggested that the SP cells express NKG2A monoallelically.
Control experiments using cloned cDNA templates and cDNA templates from B6 or BALB/c NK cells demonstrated that the RT-PCR assay distinguished NKG2AB6 and NKG2ABALB and did not amplify NKG2C or NKG2E under the conditions used (Fig. 2c). All PCR products were cut completely by _Mbo_I at the nonpolymorphic site, confirming that _Mbo_I enzyme was added and was efficacious. To ensure that the observed cDNA species were properly identified, we cloned and sequenced partial NKG2 cDNAs obtained from SP and DP cells. Of nine SP-derived clones, eight matched the NKG2ABALB sequence exactly, and one differed at only one nucleotide, probably because of a Taq polymerase error. These sequences all differed at several (4–7) nucleotides from the homologous portion of other NKG2 sequences, including NKG2AB6. The data strongly support the conclusion that the SP population expresses NKG2ABALB but not NKG2AB6. The formal possibility that SP cells express a novel Nkg2 gene that is identical to _Nkg2a_BALB in the sequenced region is unlikely, because Southern blotting of _Eco_RI-digested or _Bam_HI-digested BALB/c genomic DNA with an NKG2A-specific probe revealed only a single band (data not shown).
As expected, sequencing of NKG2 cDNAs derived from DP NK cells yielded a mixture of B6 and BALB-like NKG2A cDNAs. Because all DP cells stain with the NKG2AB6-specific 16a11 mAb, the data suggest that the DP population is either homogeneously biallelic or contains both biallelic and monoallelic (NKG2AB6 only) cells. Although we cannot definitively adjudicate between these two possibilities, we favor the latter possibility. We noted that the DP population appeared heterogeneous, consisting of two overlapping subpopulations that were arranged in a “double-diagonal” configuration (Fig. 2 a and b). In a prior study of mono/biallelic Ly49G2 expression (12), two similar double-diagonal subpopulations were proven to correspond to a biallelic (right-most) and a monoallelic (left-most) population. In the present case, the dimmer staining of the left-most population may occur because of the lower expressed Nkg2a gene dosage, and/or because the 16a11 mAb may partially block binding of the 20d5 mAb to NKG2AB6. Because the two populations do not cleanly separate in the flow cytometric analysis, it is difficult to test this hypothesis directly by examining the mRNA content of each population.
Only a few unrearranged genes have previously been shown to exhibit monoallelic expression (e.g., the Ly49, olfactory receptor, Il-2, and Il-4 genes). Although Ly49 genes were known to be monoallelically expressed (10), monoallelic expression of Nkg2a was not necessarily anticipated. NKG2A and Ly49 receptors are both C-type lectins, but they are no more related to each other than they are to other C-type lectins (e.g., CD69; see ref. 4). It is therefore interesting that Ly49 and CD94/NKG2 receptors seem to be regulated by a fundamentally conserved gene expression mechanism. We speculate that monoallelic expression of MHC-specific receptors may turn out to be a general phenomenon that will be extended to human NKG2A and perhaps even to the human killer cell Ig-like receptors.
Why is NKG2A expressed monoallelically? NKG2A exhibits little polymorphism and different NKG2A alleles likely have the same ligand specificity (i.e., for Qa-1/Qdm). It is thus unlikely that monoallelic expression evolved to prevent coexpression in the same cell of two allelic forms of NKG2A. Instead, we propose that monoallelic NKG2A expression arises as a consequence of the particular stochastic gene expression mechanism that acts during NK development to distribute class I-specific receptors to NK subsets (3). In this model, the two NKG2A alleles are regulated independently such that expression of one allele does not prevent nor assure the expression of the alternate allele. Thus, most NKG2A+ NK cells will express NKG2A monoallelically, although, as observed here, some cells expressing both NKG2A alleles will also arise.
Expression of NKG2A on NK and T Cell Subsets.
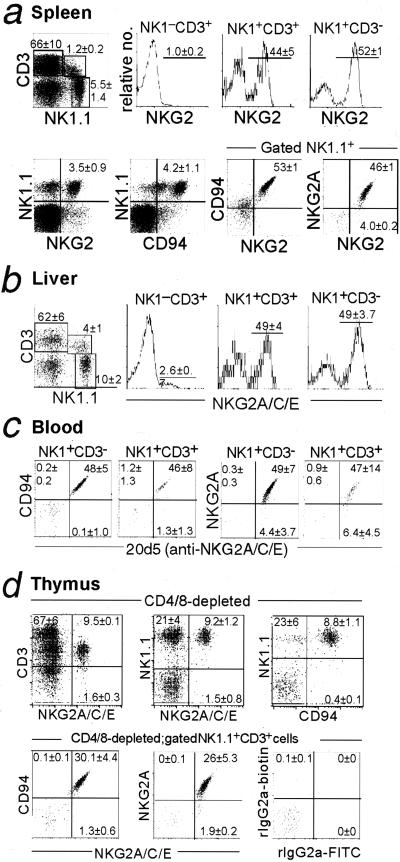
Expression of CD94/NKG2 by splenic NK and T cells was investigated. As previously described for Ly49 receptors, CD94/NKG2 receptors were largely limited to subsets of NK1.1+CD3− and NK1.1+CD3+ cells in the spleen, liver, and blood (Fig. 3 a_–_c). Very few NK1.1−CD3+ T cells expressed CD94/NKG2 receptors, and in the spleen, most of these cells were CD8+CD44+ memory phenotype T cells (data not shown), a population of cells that also expresses Ly49 receptors (24). Our results with mAbs confirm studies using Qa-1 tetramers (4, 20, 25) or an anti-NKG2A/C/E mAb (16) and allow for a specific assessment of CD94 and NKG2A expression. We found that virtually all NKG2+ cells were NKG2A+ and CD94hi, and that many, if not all, of the NKG2− NK cells stained dimly with CD94 mAb (Figs. 2 and 3a). This pattern is reminiscent of the expression of CD94/NKG2 receptors in humans (26).
Figure 3.
Expression of CD94 and NKG2 on NK cells and NK T cells. Lymphocytes obtained from the indicated organs of B6 mice (n = 3) were preblocked with anti-FcR mAb 2.4G2 and stained with PK136-PE, CD3-CyChrome, 20d5-FITC (anti-NKG2) mAb, 18d3-biotin (anti-CD94) mAb, and/or 16a11-biotin (anti-NKG2AB6) mAb (developed with streptavidin-613). (a) Nylon wool-passed spleen cells. (b) Livers were dissociated and nylon wool-passed, and lymphocytes were recovered at the interface of a 44% and 60% Percoll step gradient. (c) Blood lymphocytes were obtained at the interface of media and Lympholyte M. (d) Thymocytes were partially depleted of CD4+ and/or CD8+ thymocytes by mAb and complement treatment, before staining with the indicated mAbs.
CD94/NKG2 Receptor Expression in the Thymus.
Ly49 receptor expression has been reported on a small subset of thymic NK1.1+ T cells (27). We found that a small percentage of CD4−CD8− thymocytes also express CD94/NKG2 receptors, and that the majority of these CD94/NKG2+ T cells are NK1.1+ and CD3low (Fig. 3d). Most of the CD94/NKG2+ T cells are αβ T cells, although some are γδ T cells (data not shown). Costaining with 16a11 and 20d5 indicated that virtually all CD94/NKG2+ NK1.1+ T cells express the inhibitory NKG2A isoform (Fig. 3d). These results establish that in addition to their expression of Ly49 inhibitory receptors, NK1.1+CD3+ thymocytes also express inhibitory CD94/NKG2A receptors, which may play an important role in NK T cell development and function.
Overlap of Ly49 and NKG2A Expression.
To a first approximation, it was found that the overlap of Ly49 receptors and NKG2 receptors was largely stochastic (Table 1); there were significant numbers of NK cells expressing each possible combination of NKG2A and Ly49 receptors, and the percentage of NKG2A+Ly49+ cells observed was always within 2-fold of the percentage expected if NKG2A and Ly49 receptors were expressed independently. By using our specific mAbs, we confirmed a study with Qa-1 tetramers (28) that showed the NKG2A-negative population is modestly enriched for cells expressing Ly49 receptors, especially Ly49D (Table 1 and data not shown). An inverse correlation of CD94/NKG2A expression and killer cell Ig-like receptor expression was similarly noted among human NK cells (29). We extended these findings by demonstrating that the inverse correlation of CD94/NKG2A and Ly49 expression cannot be explained by receptor signaling caused by engagement by MHC, because a similar enrichment existed in class I-deficient β2-microglobulin−/− mice (Table 1). The results suggest that an intrinsic bias in the gene expression machinery reduces the likelihood of NKG2A and Ly49 coexpression.
Table 1.
Overlap of Ly49 and NKG2A expression
| Subset | Percentage of splenic CD3−NK1.1+NK cells | |||
|---|---|---|---|---|
| C57BL/6 | C57BL/6 β2m−/− | |||
| Observed* | Predicted† | Observed* | Predicted† | |
| %Ly49A+NKG2A+ | 6.4 ± 1.3 | 7.8 ± 1.5 | 6.7 ± 0.4 | 9.2 ± 0.4 |
| %Ly49D+NKG2A+ | 15.0 ± 0.1 | 24.6 ± 0.3 | 10.9 ± 1.5 | 21.1 ± 1.5 |
| %Ly49F+NKG2A+ | 3.3 ± 0.7 | 4.1 ± 0.7 | 5.4 ± 0.7 | 6.4 ± 0.5 |
| %Ly49G2+NKG2A+ | 20.2 ± 2.5 | 24.8 ± 3.1 | 20.6 ± 2.0 | 24.9 ± 1.9 |
Overall, we conclude that expression of NKG2A alleles in a given NK cell is regulated largely by, although not entirely independently of, the inhibitory Ly49 receptors expressed by that cell. The significance of independent expression is two-fold. First, stochastic expression of Nkg2a and Ly49 genes confers a general advantage to the NK receptor repertoire by maximizing its combinatorial diversity, and thus its ability to discriminate target cells expressing distinct combinations of MHC molecules (3). Second, that a general stochastic mechanism controls the expression of NKG2A and Ly49 receptors implies that by chance some NK cells will be generated that express no self-specific inhibitory receptors. Indeed, in B6 mice, we find that ≈25% of NK cells fail to express CD94/NKG2A and/or Ly49C/I/A, the only known H-2b-reactive receptors in B6 mice. Current models of NK tolerance cannot adequately explain the existence of these cells. It is possible that these cells express hitherto unidentified inhibitory receptors. Alternatively, it is possible that NK cells need not express “at least one” self-MHC-specific receptor, but can instead achieve self-tolerance through adopting a state of “hyporesponsiveness” or by other means (3). Thus, the observation that expression of both NKG2A and Ly49 receptors is under general stochastic control may necessitate revision of our current models of NK development and tolerance.
CD94 Deficiency in DBA/2J Mice.
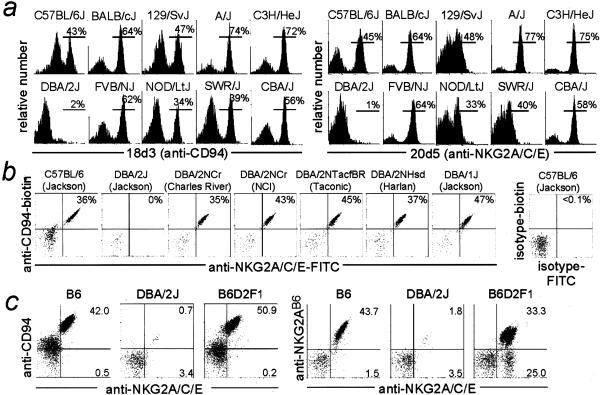
The 18d3 and 20d5 mAbs detected CD94 and NKG2 molecules on a variable percentage of NK cells in most inbred strains (Fig. 4a). Remarkably, DBA/2J mice appeared to lack a discrete population of CD94/NKG2+ NK cells. To confirm and extend this finding, we examined CD94/NKG2 cell surface expression in various DBA/2 substrains from different vendors (Fig. 4b). Strikingly, DBA/2J (The Jackson Laboratory) was unique among the five DBA/2 substrains in apparently failing to express CD94/NKG2 receptors.
Figure 4.
Expression of CD94/NKG2 receptors by inbred mouse strains other than DBA/2J. (a) Spleen cells were nylon wool-passed and stained with DX5-biotin, CD3-PE, streptavidin-tricolor, and purified 18d3 (anti-CD94) or 20d5-FITC(anti-NKG2) mAbs. The 18d3 staining was developed with an FITC-goat anti-rat IgG secondary. Gated DX5+CD3− NK cells from one representative mouse of two analyzed for each strain are shown. (b) Gated NK cells from the indicated strains, obtained from vendors indicated in parentheses, are shown. (c) Nylon wool-passed spleen cells from individual female 8–9-week-old mice were stained first with biotinylated 18d3 or 16a11, followed by FITC-20d5, streptavidin tricolor, and DX5-PE.
(B6 × DBA/2J)F1 mice (abbreviated as B6D2F1) expressed CD94/NKG2 receptors (Fig. 4c), and interestingly, contained 20d5+16a11−NK cells, which likely express an NKG2 molecule from the DBA/2J chromosome. These data suggested that the DBA/2J defect affects expression of CD94 rather than NKG2A, which was confirmed by Northern blotting (Fig. 5a). CD94 mRNA was absent in DBA/2J NK cells but easily detected in B6D2F1 NK cells (which express normal levels of CD94/NKG2). NKG2A transcripts were easily detected in NK cells from both strains. We hypothesized that the defective expression of CD94/NKG2 receptors in DBA/2J mice is largely or exclusively the result of a defect in Cd94 gene expression.
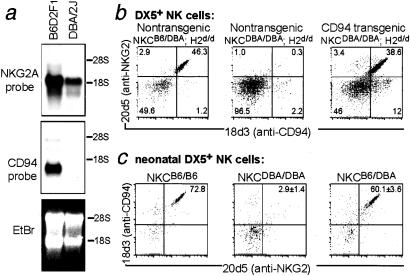
Figure 5.
DBA/2J mice are deficient in CD94 expression. (a) Twenty micrograms of IL-2-cultured B6D2F1 or DBA/2J NK cell RNA was run on two identical formaldehyde gels and blotted to nylon membrane. One blot was probed with an NKG2A-specific cDNA probe, derived from the 5′ half of mouse NKG2A. The second blot was probed with a cDNA probe derived from the 3′ half of mouse CD94. The integrity and quantity of the RNA samples was assessed by ethidium bromide staining of a third identical gel. It appeared that there was slightly more RNA in the B6D2F1 sample than in the DBA/2 sample. A lack of CD94 mRNA in DBA/2J was confirmed in an additional Northern blotting experiment (data not shown). (b) CD94 transgenic mice were generated on a mixed genetic background and backcrossed twice to DBA/2J mice to generate mice of the indicated genotypes, all H-2d/d littermates. NKC denotes the portion of mouse chromosome six that contains the Ly49 and Nkg2 gene clusters and the _Nkrp1c_B6 (NK1.1) gene. DX5+ NK cells were stained with 20d5 (anti-NKG2A/C/E)-FITC and 18d3 (anti-CD94)-biotin/streptavidin Tricolor. (c) Splenic cells were isolated from 1–3-day-old [B6D2F1 × DBA/2J] backcross mice or from a B6 control mouse. Gated DX5+ NK cells were analyzed for CD94/NKG2 expression as in b. The presence of the B6 NKC was ascertained in a separate stain with the PK136 mAb (anti-NKRP1CB6) and the NKC genotype is indicated for each dotplot. The percentage of CD94/NKG2+ among DX5+ NK cells is indicated (mean ± SD) for NKCDBA/DBA (n = 6) and NKCB6/DBA (n = 5) mice.
The gene(s) responsible for the lack of CD94 expression in DBA/2J mice was mapped by using 30 mice derived from the backcross of B6D2F1 mice to DBA/2J. The inheritance of the NKC from B6 mice in each backcross animal was determined by staining spleen cells for the NKC-linked B6-specific marker NK1.1. Of 30 mice, 17 were NK1.1+CD94/NKG2+, and 13 were NK1.1−CD94/NKG2−. Thus, CD94 expression cosegregated with NK1.1 expression in an all or none fashion in all 30 backcross animals, demonstrating that the defect in DBA/2J maps in or near the NK complex.
CD94 transgenic mice were generated on a mixed genetic background, and were then backcrossed twice to DBA/2J (see Materials and Methods). H-2d/d littermates that differed in NKC and transgene inheritance were compared. As expected, CD94/NKG2+ NK cells were not detected in nontransgenic NKCDBA/DBA mice (n = 3), whereas CD94/NKG2+ cells were abundant in nontransgenic NKCB6/DBA mice (n = 3; Fig. 5b). Strikingly, the CD94 transgene rescued CD94/NKG2 expression in NKCDBA/DBA mice (n = 6). These data demonstrate that the lack of CD94 expression itself, and not a defect in another NKC gene, is responsible for the NKG2-deficient phenotype of DBA/2J NK cells. The results establish that in vivo, CD94 is required to obtain detectable cell-surface levels of NKG2A expression. Overexpression of NKG2A, as in the transient transfection experiments in Fig. 1, can apparently override the requirement for CD94.
Importantly, neonatal NK cells from mice homozygous for the DBA/2J NK complex were CD94/NKG2-deficient, whereas most neonatal NK cells from mice with the B6 NKC expressed CD94/NKG2 receptors (Fig. 5c). The percentages of spleen cells in neonatal CD94+ and CD94− mice that were DX5+ were comparable (2.2 ± 0.3, n = 5 vs. 2.1 ± 0.4, n = 6, respectively). Taken together, the data suggest that functional CD94/NKG2 receptors are not required for NK cell development or tolerance in neonatal mice, in contrast to prior suggestions (16, 18). Because neonatal NK cells are also Ly49-deficient, we propose that CD94- and Ly49-independent mechanisms ensure the self-tolerance of neonatal NK cells. These mechanisms might include, for example, the expression of hitherto uncharacterized inhibitory receptors, a failure to express self-specific activating receptors, or the adoption of a “hyporesponsive” state (3, 30). The mechanisms that assure tolerance of neonatal NK cells might therefore resemble those that assure tolerance of adult NK cells that have failed to express CD94/NKG2 or Ly49 receptors (see above).
The identification of a mouse strain naturally deficient in CD94 expression is surprising. The implication is that although the CD94/NKG2 receptor system is conserved from mouse through humans (4), it is not conserved between inbred mouse strains. It is likely that laboratory mice are not exposed to the natural selective pressures that have maintained CD94 function in humans. We have not yet identified a molecular lesion within the DBA/2J Cd94 gene. Southern blotting suggests the genome of DBA/2J mice contains a CD94-like sequence (data not shown), consistent with the possibility that a point mutation, a rearrangement or a small deletion, rather than a large deletion, is responsible for the inactivation of the DBA/2J Cd94 gene. Our results may account for a report (25) that found diminished Qa-1 tetramer staining of NK cells from an unspecified DBA/2 substrain, although tetramer staining was not completely absent in that analysis. Our results may also account for complementary functional data suggesting that NK cells from DBA/2 mice are not inhibited by Qa-1 (31). The role of CD94/NKG2 receptors in immune defense has yet to be established. Thus, wild-type DBA/2J and CD94-transgenic DBA/2J mice should be useful in further genetically controlled experiments to elucidate the role of CD94/NKG2 receptors in the development and function of NK cells.
Acknowledgments
We thank Hector Nolla, Scot Liu, and Caelin White for expert technical assistance, and members of the Raulet laboratory for helpful discussions. This work was supported by National Institutes of Health Grant RO1-AI35021 (to D.H.R.). R.E.V. was supported by a Howard Hughes Predoctoral Fellowship.
Abbreviations
NK
natural killer
NKC
NK complex
PE
phycoerythrin
RT
reverse transcription
SP
single positive (16a11−20d5+)
DP
double positive (16a11+20d5+)
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Long E. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 2.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 3.Raulet D H, Vance R E, McMahon C W. Annu Rev Immunol. 2001;19:291–330. doi: 10.1146/annurev.immunol.19.1.291. [DOI] [PubMed] [Google Scholar]
- 4.Vance R E, Kraft J R, Altman J D, Jensen P E, Raulet D H. J Exp Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braud V M, Allan D S J, O'Callaghan C A, Soderstrom K, D'Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, et al. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 6.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraft J R, Vance R E, Pohl J, Martin A M, Raulet D H, Jensen P E. J Exp Med. 2000;192:613–624. doi: 10.1084/jem.192.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ljunggren H G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 10.Held W, Roland J, Raulet D H. Nature (London) 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 11.Held W, Raulet D H. Eur J Immunol. 1997;27:2876–2884. doi: 10.1002/eji.1830271120. [DOI] [PubMed] [Google Scholar]
- 12.Tanamachi D M, Hanke T, Takizawa H, Jamieson A M, Raulet D H. J Exp Med. 2001;193:307–315. doi: 10.1084/jem.193.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janeway C A, Travers P, Walport M, Capra J D. Immunobiology: The Immune System in Health and Disease. London: Curr. Biol.; 1999. [Google Scholar]
- 14.Dorfman J R, Raulet D H. J Exp Med. 1998;187:609–618. doi: 10.1084/jem.187.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivakumar P V, Bennett M, Kumar V. Eur J Immunol. 1997;27:3100–3104. doi: 10.1002/eji.1830271204. [DOI] [PubMed] [Google Scholar]
- 16.Van Beneden K, Stevenaert F, De Creus A, Debacker V, De Boever J, Plum J, Leclercq G. J Immunol. 2001;166:4302–4311. doi: 10.4049/jimmunol.166.7.4302. [DOI] [PubMed] [Google Scholar]
- 17.Kubota A, Kubota S, Lohwasser S, Mager D L, Takei F. J Immunol. 1999;163:212–216. [PubMed] [Google Scholar]
- 18.Sivakumar P V, Gunturi A, Salcedo M, Schatzle J D, Lai W C, Kurepa Z, Pitcher L, Seaman M S, Lemonnier F A, Bennett M, et al. J Immunol. 1999;162:6976–6980. [PubMed] [Google Scholar]
- 19.Salcedo M, Colucci F, Dyson P J, Cotterill L A, Lemonnier F A, Kourilsky P, Di Santo J P, Ljunggren H G, Abastado J P. Eur J Immunol. 2000;30:1094–1101. doi: 10.1002/(SICI)1521-4141(200004)30:4<1094::AID-IMMU1094>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Vance R E, Jamieson A M, Raulet D H. J Exp Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance R E, Tanamachi D M, Hanke T, Raulet D H. Eur J Immunol. 1997;27:3236–3241. doi: 10.1002/eji.1830271222. [DOI] [PubMed] [Google Scholar]
- 22.Lohwasser S, Hande P, Mager D L, Takei F. Eur J Immunol. 1999;29:755–761. doi: 10.1002/(SICI)1521-4141(199903)29:03<755::AID-IMMU755>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Pircher H, Mak T M, Lang R, Ballhausen W, Ruedi E, Hengartner H, Zinkernagel R M, Burki K. EMBO J. 1989;8:719–727. doi: 10.1002/j.1460-2075.1989.tb03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coles M C, McMahon C W, Takizawa H, Raulet D H. Eur J Immunol. 2000;30:236–244. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 25.Salcedo M, Bousso P, Ljunggren H G, Kourilsky P, Abastado J P. Eur J Immunol. 1998;28:4356–4361. doi: 10.1002/(SICI)1521-4141(199812)28:12<4356::AID-IMMU4356>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Sivori S, Vitale M, Bottino C, Marcenaro E, Sanseverino L, Parolini S, Moretta L, Moretta A. Eur J Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 27.MacDonald H R, Lees R K, Held W. J Exp Med. 1998;187:2109–2114. doi: 10.1084/jem.187.12.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith H R C, Chuang H H, Wang L L, Salcedo M, Heusel J W, Yokoyama W M. J Exp Med. 2000;191:1341–1354. doi: 10.1084/jem.191.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valiante N, Uhberg M, Shilling H, Lienert-Weidenbach K, Arnett K, D'Andrea A, Phillips J, Lanier L, Parham P. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 30.Moretta L, Bottino C, Vitale M, Pende D, Cantoni C, Mingari M, Biassoni R, Moretta A. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 31.Jia S H, Kurepa Z, Bai A L, Forman J. J Immunol. 2000;165:6142–6147. doi: 10.4049/jimmunol.165.11.6142. [DOI] [PubMed] [Google Scholar]