Profile of the genes expressed in the human peripheral retina, macula, and retinal pigment epithelium determined through serial analysis of gene expression (SAGE) (original) (raw)
Abstract
We used the serial analysis of gene expression (SAGE) technique to catalogue and measure the relative levels of expression of the genes expressed in the human peripheral retina, macula, and retinal pigment epithelium (RPE) from one or both of two humans, aged 88 and 44 years. The cone photoreceptor contribution to all transcription in the retina was found to be similar in the macula versus the retinal periphery, whereas the rod contribution was greater in the periphery versus the macula. Genes encoding structural proteins for axons were found to be expressed at higher levels in the macula versus the retinal periphery, probably reflecting the large proportion of ganglion cells in the central retina. In comparison with the younger eye, the peripheral retina of the older eye had a substantially higher proportion of mRNAs from genes encoding proteins involved in iron metabolism or protection against oxidative damage and a substantially lower proportion of mRNAs from genes encoding proteins involved in rod phototransduction. These differences may reflect the difference in age between the two donors or merely interindividual variation. The RPE library had numerous previously unencountered tags, suggesting that this cell type has a large, idiosyncratic repertoire of expressed genes. Comparison of these libraries with 100 reported nonocular SAGE libraries revealed 89 retina-specific or enriched genes expressed at substantial levels, of which 14 are known to cause a retinal disease and 53 are RPE-specific genes. We expect that these libraries will serve as a resource for understanding the relative expression levels of genes in the retina and the RPE and for identifying additional disease genes.
The human retina is a highly specialized tissue that converts photons into neural signals that are communicated to the brain. This process involves different types of retinal cells, mainly neuronal cells (e.g., photoreceptors, bipolar cells, and others) and the nonneuronal retinal pigment epithelium (RPE). The set of genes expressed by the human neural retina has been elucidated partially, mainly through reported expressed sequence tag (EST) libraries [nonnormalized (1–3) or enriched (4,5)] that contain over 16,000 EST sequences derived from less than 5,000 genes (6). Our knowledge of the set of genes expressed by the RPE is more rudimentary, because only 624 ESTs have been sequenced (refs. 4 and 7, and GenBank Library 6359). Little is known about the variation in gene expression between different regions of the retina (e.g., the retinal periphery versus the macula); only a few genes with a preferential expression in the fovea have been identified (1). There is no counterpart to these different retinal regions in mouse retinas. Knowledge of the set of genes expressed in these regions is valuable, because some common diseases of the retina can affect one region preferentially (e.g., age-related macular degeneration).
The serial analysis of gene expression (SAGE) procedure, first described by Velculescu et al. (8), allows the compilation of thousands of transcripts from a tissue sample. With this technique, a short piece of each transcript (a SAGE tag that is 9–14 bases in length) is linked in series with 20–50 other such tags in a cloned DNA fragment. Sequencing of such a clone provides the sequence of 20–50 tags, and a set of a few thousand such clones represents a library of SAGE tags. Most individual SAGE tags can be assigned to specific genes by inspecting available NCBI sequences (SAGEmap). The relative abundance of each tag reliably reflects the level of gene expression based on a comparison with Northern blot analysis (9). The SAGE technique has been used to compare the profile of mRNA expression in different types of cancer cells (10, 11) and to identify genes that are up- or down-regulated in response to exposure to drugs or other stimuli (12, 13).
Here we describe SAGE libraries constructed from the human peripheral retina, macula, and RPE. We compared the expression profile of the retinal periphery versus the macula of the same eye and the expression profile of the peripheral retina from two individuals (ages 44 and 88 years). Finally, we evaluated the potential value of the library as a resource for identifying candidate genes for hereditary retinal diseases.
Methods
Procurement of Tissue.
The patients who donated eyes for this study had periocular malignancies that required exenteration of an otherwise normal eye as part of clinically indicated surgical therapy. Before surgery, the patients were contacted and gave their consent to donate parts of their to-be-enucleated eye for this research. The harvesting of the intraocular tissue fragments for this study did not interfere with the pathological evaluation of the extraocular malignancies. Each exenteration specimen was placed in saline after it was removed from the patient and promptly transported to the pathology laboratory. Each eye was cut open with a virgin razor blade, and retinal fragments were excised. The RPE was harvested by gentle scraping with a Pasteur pipette, and fragments of RPE were collected by aspiration in sterile 0.9% saline. Tissue fragments were collected and frozen within 45 min of the surgical removal of each specimen. The exenteration specimens, including the remaining ocular tissues, were subsequently placed in fixative and processed as required for diagnostic pathology.
The peripheral retinal fragments were derived from regions outside the central vascular arcades. The macular fragment was ≈6 mm in diameter and centered at the foveola. The RPE fragments were from the posterior pole and the periphery.
SAGE Library Construction and Analysis.
We used a modification of the microSAGE protocol (14) to generate the four human SAGE libraries. The sequences were analyzed by using the SAGE2000 software (courtesy of Victor Velculescu and Ken Kinzler, Johns Hopkins University School of Medicine). Tags with ambiguous bases, duplicate ditags, and abnormally short tags (<14 bases) were eliminated automatically. The identity of the mRNAs corresponding to the SAGE tags was determined through inspection and comparison with the SAGEmap (www.ncbi.nlm.nih.gov/SAGE/SAGEtag.cgi) and UniGene (www.ncbi.nlm.nih.gov/UniGene/) databases.
To identify tags in genomic sequences, we developed a computer program that we have named TAGSEARCH. This program searches the genomic sequence at the 3′ end of a specified gene for each 14-bp tag, including an _Nla_III recognition sequence (CATG), represented in SAGE libraries. After the program identifies a tag, it searches for a nearby polyadenylation signal sequence (AATAAA or AATTAA). This software, written by John Keene of D. H. Keene Associates**,** runs on a Microsoft WINDOWS platform and will be available upon request ateyegene.meei.harvard.edu.
The χ2 statistic was calculated for all comparisons of tag frequency, taking into account the frequency of each tag and the total number of tags per library. To identify tissue-specific tags, the most common tags were compared with tags from 100 nonocular libraries (www.ncbi.nlm.nih.gov/SAGE/). The assignment of a protein's function was, for the most part, based on the database LocusLink (www.ncbi.nlm.nih.gov/LocusLink/).
Results and Discussion
SAGE Libraries.
We harvested mRNA from one eye from each of two humans without retinal disease, an 88-year-old female (patient 1) and a 44-year-old male (patient 2). SAGE libraries were prepared from the peripheral retina (one from each eye), the macula (from eye 2), and the RPE (from eye 1). SAGE tags from 53,666 to 105,312 mRNA molecules were sequenced from each library for a total of 320,998 tags from the four SAGE libraries (Table 1; also deposited in NCBI database). Approximately 15% of the retinal tags and 52% of the RPE tags were observed only once. Some of these singleton tags may be due to sequencing errors with the remainder corresponding to mRNAs expressed at very low levels. The small proportion of spurious tags caused by sequencing errors is likely to be approximately the same in the different libraries we compiled. Thus, it seems likely that the large number of singleton tags in the RPE reflects an abundance of rare transcripts in that cell type. We have confined the analyses presented in this paper to tags encountered at least twice.
Table 1.
SAGE tags in the human retinal and RPE libraries
| Peripheral retina | Macula, Eye 2* | Retina totals | RPE, Eye 1* | |||
|---|---|---|---|---|---|---|
| Eye 1* | Eye 2* | Combined | ||||
| Total no. of tags | 59,661 | 105,312 | 164,973 | 102,359 | 267,332 | 53,666 |
| Tags for analysis†, (%) | 50,193 (84) | 90,444 (86) | 140,637 (85) | 87,555 (86) | 228,192 (85) | 25,712 (48) |
| Different transcripts | 13,344 | 19,199 | 23,112 | 18,660 | 26,355 | 10,404 |
| UniGenes‡, (%) | 10,942 (82) | 15,373 (80) | 18,119 (78) | 15,023 (81) | 20,251 (77) | 6,401 (62) |
After excluding singletons and combining tags derived from the same transcript, the total number of different transcripts was 26,355 in the combined retinal libraries and 10,404 in the RPE library. Approximately 77% of the retinal tags could be assigned to UniGene entries, but only 62% of the RPE tags could be assigned. The high proportion of unassigned RPE tags (38%), together with the high proportion of singleton RPE tags mentioned in the previous paragraph, suggest that a large proportion of genes expressed by the RPE are tissue-specific. Many RPE-specific tags are from unknown genes. This may be due in part to the small number of ESTs that have been sequenced from RPE libraries (4, 7).
New Tag Assignments.
Most of the genes that were known previously to be expressed in the retina or the RPE had corresponding tags in our SAGE libraries. However, we came across a number of genes known to be expressed in the retina but for which there initially appeared to be no representative tag in our libraries. We considered the possibility that these genes may produce mRNAs with 3′ ends different than those reported previously. By using the TAGSEARCH software, we tentatively assigned some of the tags in our libraries to known genes for which no tags had otherwise been found. Examples are the genes encoding red and green cone opsins (OPN1LW and_OPN1MW_), which appear to have the same, newly identified tag AGGTCTGCCT. The program also found a putative rhodopsin tag, CTCACCCGCC, expressed at a low level and possibly derived from a rare alternative transcript not yet catalogued in EST sequence databases.
Even after this search for new tag assignments, there remained a few genes we expected to be represented but for which no matching tag was encountered (e.g., CRX, USH2A,RHOK in the retina, and MERTK in the RPE). These tags may be absent because the corresponding genes are expressed at a very low level (for example, no ESTs have been reported from_RHOK_ in the retina and MERTK in the RPE), because they produce unstable mRNA molecules that were degraded in the 45 min between enucleation and the harvesting of tissues, or because of an erroneous prediction of the expected tag through inaccurate knowledge of the 3′ end of the mRNA sequence.
Validity of the Libraries.
We assessed the validity of the SAGE libraries by evaluating tags from genes previously known to be expressed specifically in the retina or RPE. For example, the genes for rhodopsin (RHO), the rod transducin subunits (GNB3, GNAT1, and_GNGT1_), the rod cGMP phosphodiesterase subunits (PDE6A, PDE6B, and PDE6G), the subunits of the rod cGMP-gated cation channel (CNGA1 and_CNGB1_), other proteins involved in rod phototransduction (RCV1, SAG, GUCA1A, and_SLC24A1_), and other rod photoreceptor proteins (HRG4, ABCR, RDS, and ROM1) are known to be expressed in the retina. As expected, tags corresponding to all these genes were present in the retinal SAGE libraries derived from the peripheral retina and the macula. Similarly, the genes RPE65, 11-_cis_-retinol dehydrogenase (RDH5), transthyretin (TTR), bestrophin (VMD2), cathepsin D (CTSD), and cystatin C (CST3) are all known to be expressed by the RPE; all were detected in the RPE library.
We also evaluated the purity of the RPE library by examining whether tags corresponding to genes known to be expressed in the retina and not the RPE were present only in the retinal libraries and not in the RPE library. Genes encoding photoreceptor proteins (e.g.,RHO, GNAT1, GNB1, GNGT1,PDE6A, PDE6B, PDE6G, ROM1,RDS, RCV1, SAG, CNGA1,CNGB1, GUCA1A, and SLC24A1) were ideal for this evaluation, because they are expressed by photoreceptor cells immediately adjacent to the RPE but not by the RPE itself. In total, these genes accounted for 1.7% of the tags in the retinal libraries overall, although they only accounted for 0.05% of the tags in the RPE library. The small percentage of photoreceptor-specific tags in the RPE library probably is the result of minor contamination of the RPE with retina during dissection.
Evaluating the amount of RPE that contaminated the retinal fragments used to construct SAGE libraries was more difficult because the few RPE-specific genes of which we are aware (e.g.,RPE65, RDH5, and VMD2) are not expressed at a high level in the RPE and therefore could not serve as sensitive markers of contamination. Our evaluation of the tags corresponding to these genes suggested some contamination (these tags represented 0.05% of the tags in the RPE versus 0.005% of the tags in the combined retinal libraries). However, it is possible that one or more of these genes is actually expressed by the neural retina at low levels. Other evidence that the amount of RPE contamination in the retinal libraries is low came from our observation of seven tags (tag sequences ACAAAACCAA, ATAACACATA, GGGTTGGGGA, GCTCCAGCTG, CTCCCGCTGG, ATACTGACAC, and TGGGTGCTGG) that were abundant in the RPE library (each was at a frequency >100 tags per 105) but were absent or very rare (less than 2 tags per 105) in the peripheral retinal library of the same eye.
Gene Expression in the Retina.
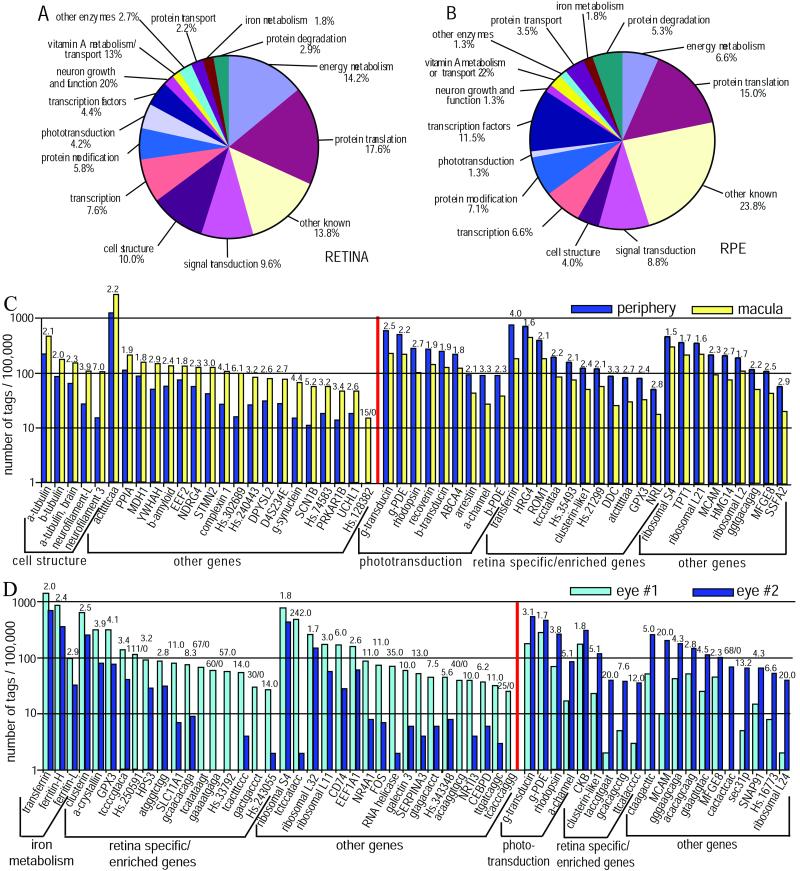
Of the 1,000 most frequent tags in the combined peripheral retina and macular libraries, 216 were derived from mitochondrial DNA or were ambiguous tags that could not be assigned to a single gene. The remaining 784 tags correspond to 690 distinct, known genes and 47 unknown tags in the UniGene database. A provisional function could be assigned to the proteins expressed by 65.2% of the known genes, which together accounted for 77.5% of the tags from the 690 known genes. The largest functional categories are proteins involved in protein translation, accounting for 17.6% of the genes (23.1% of the tags), and proteins involved in energy metabolism, accounting for 14.2% of the genes (19.4% of the tags). Among the genes involved in energy metabolism are those encoding 10 of the 28 enzyme subunits in the glycolytic pathway. Approximately 4.2% of the genes (8.5% of the tags) encode proteins that participate in phototransduction (Fig.1A).
Figure 1.
(A) A pie chart depicting the percentage of distinct genes encoding proteins in various functional categories out of the total number of genes encoding proteins with known function. Data from the macula library and the two peripheral retinal libraries were combined. (B) A pie chart similar to that shown in_A_ but based on data from the RPE library. (C) A comparison of the expression levels of the genes that are expressed at higher levels in the macula than in the retinal periphery (left of the vertical red line) or higher in the periphery than in the macula (right of the vertical red line). (D) A comparison of the expression levels of the genes that are expressed at higher levels in the peripheral retina of eye 1 than in the peripheral retina of eye 2 (left of the vertical red line) or higher in the peripheral retina of eye 2 than in the peripheral retina of eye 1 (right of the vertical red line). In both C and_D_, the number above each pair of bars is the ratio of tags from that gene in the two SAGE libraries unless there are zero tags in one library, in which case the actual tag frequencies (tags per 105) are shown. Tags that do not correspond to any known genes are represented by their 10-base sequence.
We separately tabulated the 50 most highly expressed tags in the combined retinal libraries, including tags from genes with unknown function (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). Six tags in this set have as-yet-unknown gene assignments including the most common tag sequence representing 1.59% of all tags in the retinal libraries. In this list are many tags from genes expected to be expressed at high levels based on previously reported EST libraries (2, 3) such as those encoding proteins involved in phototransduction and other proteins including_HRG4_, ABCR, and ROM1. Genes encoding two proteins involved in iron metabolism, transferrin and ferritin, are represented among the 10 most highly expressed tags; these genes will be discussed below. Many genes encoding proteins involved in protein translation and enzymes involved in energy metabolism are also in the top-50 list.
We further compared the expression levels of all 28 genes encoding the subunits of the 10 enzymes of the glycolytic pathway (Table 6, which is published as supporting information on the PNAS web site) in the retinal libraries with the levels of those genes in five nonocular SAGE libraries from the following normal human tissues: cerebellum, thalamus, kidney, prostate, and lung. In comparison with these other tissues, the retina devotes the highest proportion of mRNA synthesis to these genes. The total number of these tags in the retina (21,610 per 106) is 2.7–4.2 times greater than in the cerebellum (7,376), thalamus (8,086), kidney (5,347), prostate (5,180), or lung (6,666). The high expression level of genes encoding proteins involved in energy metabolism is consistent with the high metabolic rate of the retina (in particular of the photoreceptor cells) and is consistent with data from a retinal EST library (3).
Gene Expression in the RPE.
The RPE exhibits an expression profile very different from the neural retina. We scanned the 1,000 most highly expressed tags (each represented three or more times) to obtain a list of the 798 most highly expressed tags for which an unambiguous gene assignment could be made. These 798 tags correspond to 445 distinct, known genes and 333 unknown tags in the UniGene database. Of the 445 known genes, no function could be assigned to 49% (accounting for 50.2% of the tags from the 445 known genes). Of the remaining 227 genes with a function assigned to the corresponding gene product, ≈15% (15.4% of tags from the 227 known genes) encode proteins involved in translation, and 11.5% (11.2% of tags) encode transcription factors (Fig.1B). Approximately 5.3% (9.5% of tags) encode proteins involved in protein degradation. One of the major functions of the RPE is the phagocytosis and digestion of the photoreceptor outer segments. This task requires metabolic pathways to catabolize the phagocytosed proteins and other components of the outer segment disks and plasma membrane. Thus, the relatively high percentage of tags from genes encoding proteins involved in protein degradation might be expected. Indeed, although the RPE devotes more than 9.5% of its transcripts to produce proteins involved in protein degradation, the neural retina devotes less than 3%.
Of the 50 most common tags in the RPE library, 29 do not correspond to any known gene. Of these 29 orphan tags, 22 appear to be expressed exclusively by the RPE, because they could not be found in any of the 100 available SAGE libraries derived from other cell types. Some of these orphan tags are highly expressed: 5 of the 7 most abundant tags are orphans, and 3 of the 4 most abundant tags are RPE-specific orphans (Table 5, which is published as supporting information on the PNAS web site). Of the 21 most frequent tags for which a gene assignment could be made, some were from genes previously known to be expressed in the RPE, including transthyretin [a protein involved in vitamin A transport (7, 15)], cathepsin D [a lysosomal enzyme important in the proteolysis of opsin and in the digestion of phagocytosed photoreceptor outer segments (16)], and β-amyloid binding protein [a protein implicated in Alzheimer's disease (17)].
The Retinal Periphery Versus the Macula.
The posterior pole of the human retina has a specialized region, called the macula, that has the highest density of photoreceptor cells and provides the best visual acuity. The anatomic boundary of the macula is not standardized. Here it is considered to be a 6-mm-diameter disk centered at the foveola; this area processes visual information from the central 20° of visual field diameter. The macula occupies less than 1.4% of the surface of the retina, yet it contains ≈8.4% of the cone photoreceptors, 3.4% of the rod photoreceptors, and ≈60% of the ganglion cells of the retina (18). Furthermore, the ratio of rods to cones is lower in the macula (≈8:1) than in the peripheral retina [≈20:1 (19)].
We compared the set of 87,555 nonsingleton tags in the macula library with the set of 90,444 nonsingleton tags in the peripheral retinal library derived from the same eye (donor 2). For this comparison, the number of tags in the libraries first was adjusted mathematically (i.e., normalized) to account for the fact that we had sequenced slightly fewer tags from the macula than from the peripheral retina. We considered that the abundance of a tag to be statistically significantly different between the two libraries if a χ2 test yielded a P value < 3.2 × 10−5. This value would roughly correspond to a P value threshold of 0.01 after a Bonferroni correction for the 15,625 comparisons that were performed.
Twenty-four genes were statistically significantly more frequent in the macula than in the periphery (Fig. 1C and Table 7, which is published as supporting information on the PNAS web site). None of these genes are expressed exclusively by the retina. Five of the genes that showed a preferential expression in the macula encode cell-structural proteins found in neuronal axons [neurofilament 3 (NEF3), light neurofilament (NEFL), brain-specific α-tubulin (TUBA3), α-tubulin (_K_-α-1), and β-tubulin (FKBP1A)]. Their relative abundance in the macula is likely to reflect the high density of ganglion cells in the retina. Ganglion cells are the only cells in the retina with axons more than a few millimeters in length; their axons extend many centimeters and end in the central nervous system. Also, the axons of photoreceptor cells in the macula (which form Henle's nerve fiber layer) are much longer than elsewhere in the retina (20). mRNAs from genes encoding cell-trafficking and signal-transduction proteins also account for a higher percentage of transcription in the macula versus the retinal periphery.
Although the cone-specific genes encoding cone α-transducin (GNAT2), the α′ and γ subunits of cone cGMP-phosphodiesterase (PDE6C and PDE6H), and the cone opsins (OPN1SW and_OPN1LW_/OPN1MW) as a group were more abundant in the macula than the periphery, the difference was not statistically significant (P = 0.06). The lack of a substantially higher number of cone-specific tags in the macula versus the periphery simply may indicate that the fraction of all cells that are cones may not be very different in the two regions. In other words, not only cones but also other retinal cell types such as ganglion cells are at their highest densities in the macula, and the tags from these other cell types dilute the contribution from cones.
Twenty-nine genes were significantly more abundant in the peripheral retinal library than the macula library (Fig. 1C and Table 8, which is published as supporting information on the PNAS web site). Approximately a third of these tags corresponds to genes encoding proteins in the rod phototransduction cascade including rhodopsin (RHO), the β and γ subunits of transducin (GNB1 and GNGT1), the β and γ subunits of rod cGMP-phosphodiesterase (PDE6B and PDE6G), the α subunit of the cGMP-gated cation channel (CNGA1), and recoverin (RCV1). The relative abundance of rod-specific tags in the periphery is likely a reflection of the higher proportion of rod photoreceptors there compared with the macula. However, tags from some genes that are not specific to rod photoreceptors also were more abundant in the periphery [e.g., transferrin (TF) and dopa decarboxylase (DDC), among others].
Comparison of the Peripheral Retina in Two Individuals.
We compared the mRNA expression profile in the peripheral retinal library derived from the 88-year-old donor 1 with the peripheral retinal library derived from the 44-year-old donor 2. As with the comparison of the retinal periphery versus the macula, the number of tags in the two libraries first was normalized. The same χ2 test and the same threshold for statistical significance were used.
The 36 genes and 20 orphan tags with unknown gene assignment that were expressed at significantly different levels in the two donors are presented in Fig. 1D and Tables 9 and 10, which are published as supporting information on the PNAS web site. This set of genes does not appear to be distributed randomly among the two donors: 24 genes and 12 orphan tags are expressed at higher levels in donor 1, whereas 12 genes and 8 orphan tags were expressed at higher levels in donor 2.
Some genes have exceptionally different frequencies between the two donors. For example, donor 1 has six tags from genes with unknown function that are each found at a frequency of 25–111 per 105 yet are absent from donor 2; donor 2 has one such high-frequency tag that is absent from donor 1. Other remarkably discrepant genes are those encoding RNA helicase (tag frequency of 70 per 105 in donor 1 but only 2 per 105 in donor 2) and the unassigned tag TCTCCATAAC (484 per 105 in donor 1 but only 2 per 105 in donor 2). It seems unlikely that the striking differences in the expression of these genes in the two donors is simply due to polymorphisms in the 3′ untranslated regions of the genes causing alternative polyadenylation signal sequences to be used. If that were the case, one would expect some symmetry in the occurrence of such tags in the two donors. That is, for every highly discrepant tag that is overrepresented in one donor, there should be, on average, another discrepant tag that is overrepresented in the other donor. This is not the case: the majority of the strikingly discrepant genes are overrepresented in donor 1. The discrepancies in the expression of these genes may be caused by the difference in age between the two donors.
Most of the genes that are expressed at statistically significantly different levels in the two donors are not as strikingly discrepant as those discussed in the previous paragraph but instead are only 2–20 times more abundant in one donor versus the other. Three of the genes that are expressed at a higher level in the older donor 1 versus donor 2 encode proteins involved in iron metabolism (ferritin light polypeptide, ferritin heavy polypeptide, and transferrin). The heavy and light polypeptides of ferritin are expressed at high levels in many tissues including the retina. Transferrin is expressed also in many tissues but achieve the highest known expression level in the retina (>5 times higher than any of the 100 SAGE libraries in SAGEmap). Iron's role in the retina is important, as evidenced by the observation that null mutations in the ceruloplasmin gene, encoding a protein important for iron homeostasis, produce a phenotype that includes retinal degeneration (21). The expression of iron metabolism genes can increase synchronously in response to the increasing iron concentrations (22). The higher expression of these three iron metabolism genes in the older donor 1 could be related to the increase in iron stores with age. Iron in the body generally increases ≈2-fold in the 44–88-year age interval (23). However, adult males generally have much greater iron stores than adult females, and one might expect the iron stores in donor 1, an 88-year-old female, to be comparable to those in donor 2, a 44-year-old male. Thus, we are uncertain as to the actual basis for the greatly increased expression of these iron metabolism genes in the retina of the older donor.
A few other genes that show a preferential expression in the retina from the older donor are of particular interest. Glutathione peroxidase 3 (GPX3) catalyzes the reduction of hydrogen peroxide, organic hydroperoxide, and lipid peroxides by reduced glutathione and functions in the protection of cells against oxidative damage (Online Mendelian Inheritance in Man database no. 138321). The high level of expression of this gene in the retina compared with nonocular SAGE libraries and the higher expression levels in the older eye makes this gene an attractive candidate for age-related retinal diseases. The gene is localized to 5q32–33 and was suggested previously as a candidate for age-related macular degeneration (24). Another gene showing a higher expression level in the older donor encodes clusterin (also known as apolipoprotein J), which is speculated to be involved in apoptosis, lipid transport, and neurodegeneration (Online Mendelian Inheritance in Man database no. 185430). A related gene that encodes retinal clusterin (clusterin-like protein, or CLUL1) is expressed at a significantly higher level in the younger donor.
A number of genes specifically expressed by rod photoreceptors are expressed at significantly higher levels in the younger donor (e.g., rhodopsin and the α subunit of the rod cGMP-gated channel). The number of rod photoreceptors in the human retina decreases with age, with ≈32% loss from the second to the ninth decade (25), and there is a concomitant fall in average electroretinogram (ERG) amplitudes during this interval (26). It is reasonable to suppose that the reduction of tags from rod-specific genes in the older retina reflects this phenomenon.
SAGE as a Tool to Identify Candidate Genes.
Aiming to identify retinal and RPE-specific transcripts, we compared the expression level of the 1,000 most common retinal tags to 100 nonocular SAGE libraries (SAGEmap), and we separately compared the 100 most common RPE tags to the same nonocular libraries. The analysis revealed 89 retinal tags and 53 RPE tags that were either specific to those tissues or highly enriched in them (Table2 and Tables 11 and 12, which are published as supporting information on the PNAS web site). Fourteen of the 89 retinal tags correspond to genes already known to cause a retinal disease. All but 6 of the 53 RPE tags are unassigned tags, and 1 of these 6 corresponds to a gene (VMD2) known to cause a retinal disease. Most of the remaining high frequency retina- or RPE-enriched genes have not been evaluated for pathogenic mutations among patients with hereditary retinal diseases. Many are within chromosomal regions that are known to harbor unidentified retinal disease genes (Table 3 and Table 13, which is published as supporting information on the PNAS web site), and it is reasonable to consider them as candidates for these or other retinal diseases. We expect that the candidate genes presented here, in combination with candidate genes based on mouse SAGE retinal libraries (9), will be useful for identifying retinal disease genes.
Table 2.
Transcripts specific to or enriched in the retina and RPE
| Retina | RPE | Total | |
|---|---|---|---|
| Specific tags | 68 | 45 | 113 |
| Enriched tags | 21 | 8 | 29 |
| Total | 89 | 53 | 142 |
| Tags corresponding to known retinal or RPE genes | 37 | 2 | 39 |
| Tags corresponding to known retinal disease genes | 14 | 1 | 15 |
Table 3.
Candidate genes for retinal disease
| Gene (symbol) | Chromosomal localization |
|---|---|
| Hs. 43586 | 1p31–32 |
| Hs. 310689 | 1q25 |
| Frizzled-related protein (FRZB) | 2q24–31 |
| Palmitoylated 4 | 2q33.2 |
| Hs. 66803 | 3 |
| HT017 protein | 3p14.3 |
| Transferrin (TF) | 3q21 |
| Hermansky–Pudlak syndrome-3 (HPS3) | 3q24 |
| Hs. 300880 | 4p16 |
| Protein phosphatase, (rdgC homolog) (PPEF2) | 4q13–21 |
| Hs. 151710 | 5 |
| Glutathione peroxidase 3 (GPX3) | 5q23 |
| Hs. 246420 | 6p21 |
| Lysosomal apyrase-like 1 (LYSAL1) | 8 |
| Clusterin (CLU) | 8p21–12 |
| Hs. 154131 | 9p24 |
| MT-protocadherin (KIAA1775) | 10q22 |
| Cathepsin D (CTSD) | 11p15 |
| Hs. 239444 | 11q14 |
| Immunoglobulin superfamily (IGSF4) | 11q23.2 |
| Hs. 13768 | 12p13 |
| Sentrin/SUMO-specific protease (SENP1) | 12q12–13 |
| Hs. 33792 | 12q15 |
| Hs. 3821 | 13q13 |
| Hs. 21299 | 14q21 |
| Creatine kinase B (CKB) | 14q32 |
| Hs. 98927 (FLJ13993) | 15q22.3–23 |
| Cellular retinoic acid-binding protein 1 (CRABP1) | 15q24 |
| Hs. 261526 | 17 |
| Retinal clusterin 1 (CLUL1) | 18p11.3 |
| Hs. 122245 | 18p11.3 |
| Transthyretin (TTR) | 18q12.1 |
| Calcium-binding protein 5 (CABP5) | 19q13.3 |
| Hs. 98881 | 20q11 |
| Hs. 35493 | Multiple |
Supplementary Material
Supporting Tables
Acknowledgments
This work was supported by National Institutes of Health Grants EY08683 and EY09676, Massachusetts Lions Eye Research Fund, Inc., the Foundation for Retinal Research, the Ruth and Milton Steinbach Fund, Inc., and the Howard Hughes Medical Institute (to S.B.).
Abbreviations
RPE
retinal pigment epithelium
EST
expressed sequence tag
SAGE
serial analysis of gene expression
Footnotes
Data deposition: The tag sequences from the four SAGE libraries reported in this paper have been deposited in the Gene Expression Omnibus database (accession nos. GSM571–GSM574).
References
- 1.Bernstein S L, Borst D E, Neuder M E, Wong P. Genomics. 1996;32:301–308. doi: 10.1006/geno.1996.0123. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Macke J P, Abella B S, Andreasson K, Worley P, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu-Matsumoto A, Adachi W, Mizuno K, Inazawa J, Nishida K, Kinoshita S, Matsubara K, Okubo K. Invest Ophthalmol Vis Sci. 1997;38:2576–2585. [PubMed] [Google Scholar]
- 4.den Hollander A I, van Driel M A, de Kok Y J, van de Pol D J, Hoyng C B, Brunner H G, Deutman A F, Cremers F P. Genomics. 1999;58:240–249. doi: 10.1006/geno.1999.5823. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S, Sharma A, Agarwal N, Swaroop A, Yang-Feng T L. Invest Ophthalmol Vis Sci. 2000;41:24–28. [PubMed] [Google Scholar]
- 6.Bortoluzzi S, d'Alessi F, Danieli G A. Invest Ophthalmol Vis Sci. 2000;41:3305–3308. [PubMed] [Google Scholar]
- 7.Paraoan L, Grierson I, Maden B E. Int J Biochem Cell Biol. 2000;32:417–426. doi: 10.1016/s1357-2725(99)00143-0. [DOI] [PubMed] [Google Scholar]
- 8.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 9.Blackshaw S, Fraioli R E, Furukawa T, Cepko C L. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson A T, Evron E, Umbricht C B, Pandita T K, Chan T A, Hermeking H, Marks J R, Lambers A R, Futreal P A, Stampfer M R, et al. Proc Natl Acad Sci USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. . (First Published May 16, 2000; 10.1073/pnas.100566997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 12.Angelastro J M, Klimaschewski L, Tang S, Vitolo O V, Weissman T A, Donlin L T, Shelanski M L, Greene L A. Proc Natl Acad Sci USA. 2000;97:10424–10429. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boon K, Caron H N, van Asperen R, Valentijn L, Hermus M C, van Sluis P, Roobeek I, Weis I, Voute P A, Schwab M, et al. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St. Croix B, Rago C, Velculescu V, Traverso G, Romans K E, Montgomery E, Lal A, Riggins G J, Lengauer C, Vogelstein B, et al. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 15.Cavallaro T, Martone R L, Dwork A J, Schon E A, Herbert J. Invest Ophthalmol Vis Sci. 1990;31:497–501. [PubMed] [Google Scholar]
- 16.Cavaney-Brooker D M, Rakoczy P E. Curr Eye Res. 1999;18:310–318. doi: 10.1076/ceyr.18.4.310.5357. [DOI] [PubMed] [Google Scholar]
- 17.Kajkowski E M, Lo C F, Ning X, Walker S, Sofia H J, Wang W, Edris W, Chanda P, Wagner E, Vile S, et al. J Biol Chem. 2001;276:18748–18756. doi: 10.1074/jbc.M011161200. [DOI] [PubMed] [Google Scholar]
- 18.Curcio C A, Allen K A. J Comp Neurol. 1990;300:5–25. doi: 10.1002/cne.903000103. [DOI] [PubMed] [Google Scholar]
- 19.Curcio C A, Sloan K R, Kalina R E, Hendrickson A E. J Comp Neurol. 1990;292:497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- 20.Fine B S, Yanoff M. Ocular Histology. Hagerstown, MD: Harper & Row; 1979. [Google Scholar]
- 21.Yamaguchi K, Takahashi S, Kawanami T, Kato T, Sasaki H. Ophthalmologica. 1998;212:11–14. doi: 10.1159/000027251. [DOI] [PubMed] [Google Scholar]
- 22.Beutler E, Bothwell T H. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Childs B C, Kinzler K W, Vogelstein B, editors. Ney York: McGraw–Hill; 2001. pp. 3127–3161. [Google Scholar]
- 23.Dallman P R, Siimes M A, Stekel A. Am J Clin Nutr. 1980;33:86–118. doi: 10.1093/ajcn/33.1.86. [DOI] [PubMed] [Google Scholar]
- 24.Weeks D E, Conley Y P, Mah T S, Paul T O, Morse L, Ngo-Chang J, Dailey J P, Ferrell R E, Gorin M B. Hum Mol Genet. 2000;9:1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 25.Gao H, Hollyfield J G. Invest Ophthalmol Vis Sci. 1992;33:1–17. [PubMed] [Google Scholar]
- 26.Birch D G, Anderson J L. Arch Ophthalmol. 1992;110:1571–1576. doi: 10.1001/archopht.1992.01080230071024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Tables