Gcn4p, a Master Regulator of Gene Expression, Is Controlled at Multiple Levels by Diverse Signals of Starvation and Stress (original) (raw)
All cells undergo rapid transcriptional reprogramming in response to environmental changes by mobilizing transcriptional activators and repressors. Transcriptional activator proteins function by binding to specific DNA sequences and recruiting the transcriptional machinery to the promoters of genes under their control. Various mechanisms have been elucidated for stimulating activator function in response to environmental signals. For example, when cells of the yeast Saccharomyces cerevisiae are shifted to medium with galactose as a carbon source, the activator Gal4p is released from the inhibitory effects of the repressor Gal80p, which binds to the Gal4p activation domain (43). Other yeast transcriptional activators are regulated at the level of nuclear localization. This group includes Pho4p, Gln3p, and Yap1p, whose presence in the nucleus is coupled to the levels of inorganic phosphate, the quality of the nitrogen source, and oxidative stress, respectively (3, 45). The activator Gcn4p is regulated by a unique translational control mechanism that increases the cellular concentration of Gcn4p in amino acid-starved cells, where increased transcription of amino acid biosynthetic genes under its control is essential to maintaining cell growth (36).
It has been known for many years that Gcn4p stimulates the transcription of more than 30 amino acid biosynthetic genes, representing 12 different pathways, in response to starvation for any of several amino acids. This regulatory response is known as general amino acid control (GAAC) (33, 44). Two recent studies in which cDNA microarrays were used to conduct a genome-wide analysis of gene expression showed that Gcn4p induces a much larger set of genes, encompassing 1/10 or more of the yeast genome (42, 68). In addition, there is increasing evidence that Gcn4p is induced under conditions of starvation or stress besides amino acid deprivation. Hence, GAAC is much broader with regard to the range of stimuli that elicit the response and the ensemble of genes that are transcriptionally induced. We begin by reviewing the molecular mechanisms that regulate the expression and activity of Gcn4p and the stimuli to which they respond, and then we describe the breadth of the Gcn4p transcriptome as revealed by cDNA microarray studies.
TRANSLATIONAL INDUCTION OF GCN4 BY PK Gcn2p
The principal means of inducing Gcn4p expression in amino acid-starved cells operates at the level of GCN4 mRNA translation. This response is complete within 1 to 2 h of the onset of starvation and increases Gcn4p abundance by a factor of 2 to 10, depending on the strain background. An additional 2-fold increase in Gcn4p levels occurs after several hours of starvation through an increase in the GCN4 mRNA level (1, 34). The induction of GCN4 translation requires protein kinase (PK) Gcn2p, whose only known substrate is the α subunit of translation initiation factor 2 (eIF2) (Fig. 1). eIF2 is responsible for delivering charged methionyl initiator tRNA (Met-tRNAiMet) to the small (40S) ribosomal subunit in the first step of translation initiation. It binds to the ribosome as a ternary complex (TC) containing Met-tRNAiMet and GTP and is subsequently released as an inactive eIF2-GDP binary complex. Recycling of inactive eIF2-GDP to active eIF2-GTP requires the guanine nucleotide exchange factor eIF2B. Phosphorylation of eIF2α on serine-51 by Gcn2p converts eIF2-GDP from a substrate to an inhibitor of eIF2B, impeding the formation of the TC. The degree of reduction in TC levels evoked by Gcn2p activation in amino acid-starved cells does not substantially inhibit general protein synthesis but does activate GCN4 translation (35).
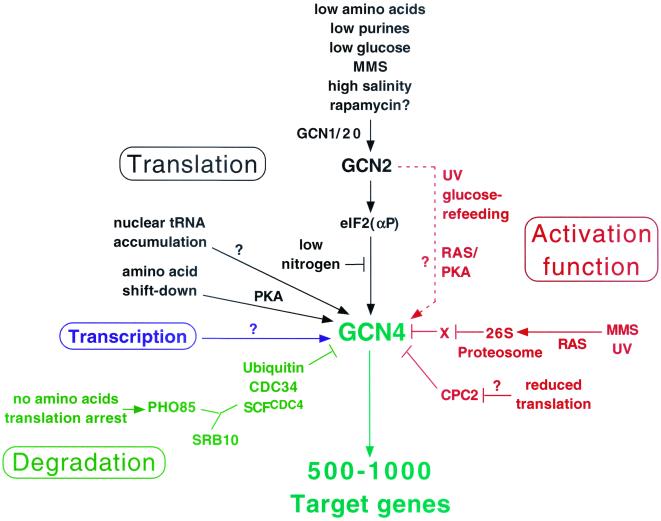
FIG. 1.
Summary of diverse regulatory mechanisms controlling Gcn4p expression or activity and the signals to which they respond. Activation is depicted with arrows; inhibition is depicted with bars. The environmental or physiological conditions regulating these responses are color coded according to the aspect of Gcn4p being regulated, including its expression at the level of translation, transcription, or degradation or its function as an activator. The role of Gcn2p in stimulating Gcn4p function in response to UV or glucose refeeding needs to be verified and thus is depicted with a broken line. Question marks indicate a lack of knowledge concerning the regulatory factors involved. X, hypothetical repressor of Gcn4p that is negatively regulated by the proteosome. See the text for details.
Translational induction of GCN4 is mediated by four short upstream open reading frames (uORFs) in the GCN4 mRNA leader, of which the first (uORF1) and fourth (uORF4) are sufficient for nearly wild-type translational control. According to the current model, essentially all ribosomes scanning from the 5" end of the mRNA translate uORF1, and ∼50% of these resume scanning as 40S subunits. Under nonstarvation conditions, virtually all of these reinitiating 40S ribosomes rebind the TC and reinitiate at uORF4, after which they dissociate from the mRNA. Under starvation conditions, when phosphorylation of eIF2α lowers the TC level, ∼50% of the ribosomes scanning from uORF1 will reach uORF4 before rebinding the TC, and lacking Met-tRNAiMet bypass uORF4. Subsequently, these ribosomes rebind the TC before reaching GCN4 and reinitiate translation there instead. Thus, the moderate reduction in TC levels elicited by Gcn2p activation allows a fraction of reinitiating ribosomes to bypass the inhibitory uORF4 and reinitiate at GCN4 instead (35).
The kinase activity of Gcn2p is stimulated in amino acid-starved cells by binding of uncharged tRNA that accumulates under these conditions to a regulatory domain related to histidyl-tRNA synthetase (HisRS) and located C terminal to the PK domain in Gcn2p (96). Starvation for any single amino acid elicits the activation of Gcn2p (96) and, consistently, Gcn2p binds several types of uncharged tRNA with similar affinities but shows a reduced affinity for the charged form of a given tRNA. It was proposed that tRNA binding releases the HisRS-like domain, along with the extreme C-terminal portion of Gcn2p, from inhibitory interactions with the PK domain (19). More recent findings suggest that tRNA modulates association of the PK domain with the HisRS and C-terminal regions, converting inhibitory to stimulatory interactions (74). The activation of Gcn2p by uncharged tRNA also requires interactions between the N terminus of Gcn2p and the Gcn1p-Gcn20p protein complex (22, 51, 52, 84), and there is increasing evidence that this association occurs near the decoding (A) site on translating ribosomes. We have suggested that Gcn1p-Gcn20p facilitates the binding of uncharged tRNA to the ribosomal A site or its transfer from the A site to the HisRS-like domain in Gcn2p for kinase activation (Fig. 2). Detection in the A site of uncharged tRNA base paired to the cognate codon in mRNA would facilitate the activation of Gcn2p by a single species of uncharged tRNA in cells starved for only one amino acid (60, 84). This process would be analogous to the mechanism for the activation of RelA protein, which mediates the general control of amino acid and ribosome biosyntheses during amino acid starvation in Escherichia coli (stringent response) (8).
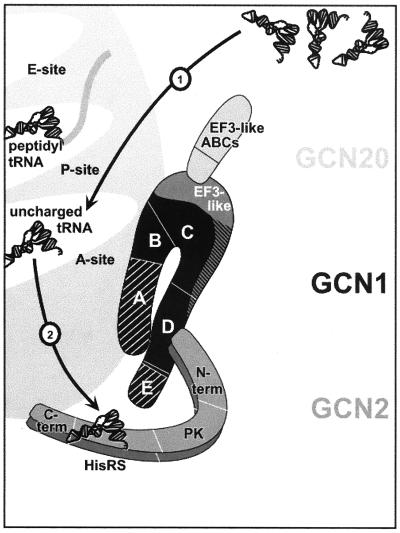
FIG. 2.
Hypothetical model for the stimulatory role of the GCN1-GCN20 complex in the activation of GCN2 by uncharged tRNA in the ribosomal A site. GCN1 is shown as a black ribbon containing near its center the EF3-like region. GCN20 is shown as a light grey rod, with the EF3-related ATP binding cassettes (ABCs) located toward the C terminus (term) and the N-terminal segment bound to the EF3-like region of GCN1. A GCN2 dimer is shown as a pair of medium grey ribbons with tRNA bound to the HisRS-like domains. Both GCN1-GCN20 and GCN2 have ribosome binding activities, which for GCN2 are conferred by its C-terminal domain. By analogy with the activation of E. coli RelA protein by uncharged tRNA, we propose that uncharged tRNA bound to the ribosomal A site and base paired with the cognate codon in mRNA is the activating ligand for GCN2. Based on their similarity to EF3, GCN1 and GCN20 may facilitate the binding of uncharged tRNA to the A site (arrow 1) or the transfer of tRNA from the A site to the HisRS-like domain in GCN2 (arrow 2). The physical association between GCN1-GCN20 and the N-terminal portion of GCN2 is consistent with the second mechanism. (Taken from reference 84.)
Gcn2p has an ortholog in mammalian cells (5, 88), and a recent study indicated that its kinase activity is activated specifically by amino acid limitation, whereas a different eIF2α kinase, known as PERK, is activated in response to unfolded proteins in the endoplasmic reticulum (81). The activation of either kinase leads to a general inhibition of translation initiation (although the effect was not pronounced for Gcn2p), consistent with the suppression of TC levels, but it also specifically stimulates the translation of ATF4 mRNA, encoding a transcriptional activator of the stress-inducible transcription factor CHOP/GADD153 (29). Thus, it seems that eIF2α phosphorylation is a conserved mechanism used from yeasts to mammals to reduce the rate of general protein synthesis while simultaneously increasing the expression of transcriptional activators whose functions are required under conditions of starvation or stress. Orthologs of Gcn1p and Gcn20p are present in mammals (60, 94), suggesting that the mechanism for Gcn2p activation by uncharged tRNA on the ribosome also may be highly conserved between yeasts and mammals.
Gcn4p synthesis is induced under conditions besides amino acid deprivation, including starvation for purines (79), glucose limitation (98), growth on the nonfermentable carbon source ethanol (98), high salinity in the growth medium (25), treatment with the alkylating agent methyl methanesulfonate (MMS) (68), and treatment with rapamycin (inhibitor of kinases Tor1p and Tor2p) (93). Except for rapamycin treatment (which was not extensively analyzed), the induction of GCN4 translation under all of these conditions is dependent on Gcn2p kinase function (Fig. 1). Surprisingly, it also requires the tRNA binding activity of the HisRS-like regulatory domain in Gcn2p (25, 68, 79, 98). One explanation for the latter requirement is that the synthesis of amino acids would be impaired under each of these starvation or stress conditions, increasing the concentration of uncharged tRNA and activating Gcn2p via the conventional mechanism that operates in amino acid-deprived cells. A related possibility is that each of these conditions interferes with amino acid uptake, preventing supplementation of an amino acid auxotrophy in the yeast strains used for the studies. For purine limitation and MMS treatment, both explanations are unlikely because experiments have been conducted with prototrophs and medium supplemented with all amino acids (68, 79, 98). The latter explanation has not been ruled out for high salinity, however, since a trp1 auxotroph was used and it was not investigated whether tryptophan uptake was inhibited by high salt concentrations (25). For glucose limitation, it was shown that cytoplasmic amino acid pools were depressed and that the addition of all amino acids to the medium diminished but did not eliminate the derepression of GCN4 translation. In addition, there was a reduced requirement for ribosome binding by Gcn2p and for Gcn20p function in the derepression of GCN4 translation. Thus, it appears that Gcn2p is activated in glucose-deprived cells growing on minimal medium partly through the conventional mechanism involving decreased amounts of amino acid pools and increased amounts of deacylated tRNA and partly through an unknown mechanism that does not involve ribosomal sensing of uncharged tRNAs dependent on Gcn20p (98). An interesting way to explain why deacylated tRNA is required as an activating ligand for Gcn2p in cells starved for purines or glucose or in cells treated with MMS is to propose that Gcn2p is covalently modified in a way that increases its affinity for uncharged tRNA. This process would effectively lower the concentration of uncharged tRNA required for high-level binding to Gcn2p and allow kinase activation by the basal levels of tRNA present in amino acid-replete cells.
Gcn2p-INDEPENDENT INDUCTION OF GCN4 TRANSLATION
GCN4 translation can be induced independently of Gcn2p under certain conditions, including a shift from amino acid-rich to minimal medium (92) (Fig. 1). This transient induction in a nutritional shift-down is impaired in a mutant containing low, constitutive PK A (PKA) activity, suggesting that PKA activation is required for the response (21). Consistently, mutants with hyperactive PKA function (bcy1 and RAS2val) show constitutively derepressed GCN4 translation that is at least partly independent of Gcn2p (21, 59). It is not understood how high-level PKA activity induces GCN4 translation independently of eIF2α phosphorylation, except that it depends on the uORFs in GCN4 mRNA. One possibility is that eIF2B activity is impaired by hyperactive PKA, leading to decreased TC levels, possibly through the phosphorylation of an eIF2B subunit (97).
GCN4 translation is also induced independently of Gcn2p in mutants with defects in tRNA processing or nuclear export (Fig. 1). This response is elicited by overexpressing mutant tRNAs that cannot be processed or exported efficiently from the nucleus or by overexpressing gene products that interfere with the 5" processing of tRNAs by RNase P (75, 95). Because defective or unprocessed tRNAs are trapped in the nucleus, it was proposed that a signaling pathway can detect aberrant tRNA accumulation in the nucleus and evoke the inhibition of TC formation or function in the cytoplasm, derepressing GCN4 translation in the process (75).
A recent study showed that the derepression of GCN4 translation and transcriptional activation of Gcn4p target genes in amino acid-deprived cells was prevented under conditions of general nitrogen limitation, even though Gcn2p was activated and eIF2α was phosphorylated on Ser-51 at high levels (27) (Fig. 1). Presumably, phosphorylated eIF2 (P-eIF2) does not inhibit eIF2B in nitrogen-starved cells; alternatively, the reduction in TC levels produced by eIF2α phosphorylation does not permit ribosomes to bypass GCN4 uORF2 to uORF4 after translating uORF1. In the latter instance, the rate of scanning might be reduced to the point where all of the reinitiating 40S ribosomes have time to rebind TC before reaching uORF4 even though TC levels are reduced, ensuring reinitiation at uORF4 instead of at GCN4. There were also indications that Gcn4p stability was reduced in nitrogen-starved cells. The repressing effect of nitrogen limitation on Gcn4p expression was independent of several known sensors of nitrogen availability, including Mep2p, Gpa2p, Ure2p, and Gln3p (27).
REGULATION OF Gcn4p STABILITY
The amount of Gcn4p in the cell is determined by its rate of degradation as well as its rate of synthesis, and Gcn4p is a very short-lived protein with a half-life of 5 min or less in nutrient-replete cells. Rapid degradation of Gcn4p by the 26S proteosome is dependent on the ubiquitin-conjugating enzyme Cdc34p (49) and the SCFCDC4 ubiquitin ligase complex (61). It also requires the phosphorylation of specific residues in the Gcn4p activation domain, notably, Thr-105 and Thr-165, and the cyclin-dependent PKs Pho85p (61) and Srb10p (11) (Fig. 1). These kinases have additive effects on Gcn4p degradation in vivo, and the inactivation of both proteins is required to stabilize Gcn4p to the extent seen for the inactivation of SCFCDC4. Similarly, the removal of multiple phosphorylation sites in Gcn4p is required for pronounced stabilization of the protein (11). Thus, it appears that the phosphorylation of Gcn4p by either PK enhances its ubiquitination and degradation by the proteosome. Consistently, deletion of PHO85 derepressed the transcription of the Gcn4p target gene HIS4 by about twofold (61). Inactivation of SRB10 increased Gcn4p levels by about twofold (11), but only a few Gcn4p target genes were derepressed in an srb10 mutant (38). The latter result might be explained by proposing that Srb10p, as a component of the polymerase II holoenzyme, is needed for transcriptional activation by Gcn4p in addition to rapid degradation of Gcn4p, such that inactivation of SRB10 has offsetting positive and negative effects on Gcn4p-dependent transcription.
Interestingly, the rate of Gcn4p degradation is substantially reduced under severe amino acid starvation conditions or when protein synthesis is impaired, and this regulation has been attributed to the inhibition of Gcn4p phosphorylation by Pho85p (49, 61) (Fig. 1). Stabilization of Gcn4p under starvation conditions makes sense because it would enhance the transcriptional activation of amino acid biosynthetic genes by Gcn4p. In studies conducted thus far, Gcn4p was stabilized under conditions of severe amino acid starvation imposed by culturing an amino acid auxotroph on minimal medium. In contrast, the translational induction of GCN4 expression has been studied primarily with leaky auxotrophs (bradytrophs) or by treatment of wild-type cells with inhibitors of a biosynthetic enzyme, in such a way that protein synthesis and cell growth still proceed at a reduced rate. It is possible that Gcn4p stabilization occurs under only more severe starvation conditions, where protein synthesis is fully arrested.
The Srb10p-dependent degradation of Gcn4p that remains in a pho85 mutant occurs constitutively. Considering that Gcn4p can interact with the polymerase II mediator complex (20, 28, 53, 67) and that Srb10p is associated with the mediator (66), it is tempting to propose that the recruitment of the mediator by Gcn4p to the promoter results in phosphorylation and, ultimately, degradation of Gcn4p by the proteosome. This process would provide a constitutive mechanism for limiting the number of transcripts that a promoter-bound Gcn4p molecule could produce before being destroyed (11). In this way, sustained induction of Gcn4p target genes would require continuous high-level translation of GCN4 mRNA.
REGULATION OF Gcn4p FUNCTION
There are several indications that Gcn4p function, in addition to expression or stability, is also subject to regulation. Deletion of CPC2, encoding a Gβ-like WD repeat protein, leads to increased Gcn4p-dependent transcriptional activation under nonstarvation conditions in wild-type cells and in gcn2 mutant cells under starvation and nonstarvation conditions alike. As the _cpc2_Δ mutation did not increase the Gcn4p level, it was proposed that Cpc2p negatively regulates the nuclear localization or activation function of Gcn4p. Cpc2p still exerted its negative effect in nonstarved cells that were moderately overexpressing Gcn4p (by a factor of about three), suggesting that the absence of Cpc2p-dependent repression in starved cells cannot be attributed to the titration of Cpc2p by the excess Gcn4p produced under these conditions. The fact that Cpc2p is functional as a negative regulator in amino acid-starved _gcn2_Δ cells might indicate that the activation of Gcn2p by uncharged tRNA is required to down-regulate Cpc2p-repressing activity in starved wild-type cells (37) (Fig. 1). If so, it must be stipulated that Cpc2p inhibitory function can be overcome in _gcn2_Δ cells by the extremely high levels of Gcn4p produced by mutations in eIF2B that fully derepress GCN4 translation and the expression of Gcn4p target genes (36). Interestingly, the cpc-2 gene in Neurospora was first identified by a point mutation that impairs GAAC in this organism (50), and the same phenotype was observed when the mutant Neurospora gene was expressed in yeast (37). Apparently, this mutant form of Cpc2p cannot be inactivated properly in starved cells.
Inactivation of CPC2 also permits heme-deficient growth of hap1 mutant strains, suggesting that Cpc2p may regulate multiple systems in yeast (10). Interestingly, Cpc2p is an ortholog of mammalian RACK-1, a receptor of activated PK C (PKC) involved in localizing PKC to the cell particulate fraction (80). Mammalian RACK-1 can complement the _cpc2_Δ defect in general amino acid control (37), indicating conservation of a regulatory function of Cpc2p/RACK-1 between fungi and mammals. Surprisingly, Cpc2p was identified as a stoichiometric component of yeast 40S ribosomes that remains associated with 40S subunits even in the presence of high salt concentrations (10, 55). A nonsense mutation in CPC2 impairs 40S-60S subunit joining in the initiation phase of translation, suggesting an important role for Cpc2p in ribosome function (10). RACK-1 was also found associated with human 40S subunits (55). In view of its association with translating ribosomes and the involvement of RACK-1 in PKC regulation, Cpc2p may function in a signal transduction pathway that is linked to the rate of translation (10). Perhaps the Gcn4p-antagonizing function of Cpc2p is activated on ribosomes translating at the maximum rate in cells growing on rich medium and then deactivated by Gcn2p in starved cells (Fig. 1).
It is known that refeeding glucose to glucose-starved cells triggers a transient increase in cAMP synthesis that is dependent on the RAS proteins and the G protein subunit Gpa2p (13). Interestingly, this regimen induced Gcn4p-dependent transcription that was dependent on Ras2p and Gpa2p and, unexpectedly, also required Gcn2p. Surprisingly, no increase in Gcn4p expression was observed, and eIF2α phosphorylation did not increase on refeeding glucose to starved cells. Hence, it was proposed that Gcn2p has a novel role in stimulating Gcn4p activity rather than inducing its translation, independently of its function as an eIF2α kinase (59) (Fig. 1). Although no increase in the level of P-eIF2α occurred when glucose was supplied, P-eIF2α was already present at an elevated level in the glucose-starved cells. Hence, it is possible that Gcn4p could not be synthesized in glucose-starved cells despite an elevated level of eIF2α phosphorylation because protein synthesis was blocked under these extreme conditions. The readdition of glucose would allow the translation of GCN4 mRNA to proceed without a further increase in the level of eIF2α phosphorylation. Gcn4p levels are very low in cells grown on the rich medium used by these workers (59), and a small, transient increase in Gcn4p levels sufficient to account for the induction of Gcn4p target genes by glucose refeeding may have been difficult to detect. High salinity is another situation where increased eIF2α phosphorylation failed to stimulate GCN4 translation until several hours had elapsed, and this result was attributed to severe inhibition of all translation during the lag period in salt-stressed cells (25).
UV irradiation induces Gcn4p-dependent transcription, and this response is dampened (but not eliminated) in a _ras2_Δ mutant, which has reduced PKA activity. Based on these findings and the fact that the activation of PKA stimulates the transcription of Gcn4p target genes in _gcn2_Δ cells, it was proposed that RAS/PKA mediates the Gcn2p-independent stimulation of Gcn4p activity by UV. This process was likened to the known involvement of the Ras pathway in UV activation of the transcription activator AP-1 in mammalian cells (21). However, more recently it was reported that Gcn2p is essential for the UV induction of Gcn4p-dependent transcription in yeast (59). In fact, this conclusion is consistent with the previous finding that the constitutive derepression of GCN4 translation by elimination of the uORFs nearly eliminated the stimulatory effect of UV on HIS gene transcription (21). Despite the requirement for Gcn2p in transcriptional induction by UV, no increase in Gcn4p levels was observed following UV irradiation, again prompting the suggestion that Gcn2p has a role in stimulating Gcn4p function independently of eIF2α phosphorylation and GCN4 translational control (59) (Fig. 1). To prove this hypothesis, it is crucial to show that the phosphorylation site on eIF2α and the uORFs in GCN4 mRNA are not required for the induction of Gcn4p-dependent transcription in response to UV irradiation (or the refeeding of glucose to glucose-starved cells). Given that the UV induction of Gcn4p target genes was dampened but not eliminated in a _ras2_Δ mutant (21), the RAS/PKA pathway could make a constitutive contribution to transcriptional activation by Gcn4p on rich medium whereas Gcn2p, acting alone or in combination with other factors, could mediate the stimulatory effect of UV on Gcn4p synthesis or function.
Mutations inactivating each of three different subunits of the 19S regulatory cap of the 26S proteosome, Rpn11p, Cim3p, or Cim5p, abolished Gcn4p-dependent transcriptional activation in response to UV or MMS treatment of wild-type cells and eliminated the derepression that occurs in RAS2val mutant cells, all growing in rich medium. In contrast, these mutations had no effect on transcriptional activation by Gcn4p in cells starved for histidine. The effects of rpn11 mutations on transcriptional activation in MMS-treated cells could not be explained by decreased Gcn4p levels (90), indicating that Gcn4p activation function was induced by UV or MMS in a manner regulated by the proteosome. Recently, it was shown that other mutations affecting the proteolytic activity of the catalytic 20S core particle of the proteosome also impaired activation by Gcn4p in MMS-treated cells (J. Reese, personal communication). It was proposed that UV irradiation or MMS treatment would stimulate a signal transduction pathway that involves RAS/PKA and that triggers proteosomal degradation of an unknown negative regulator of Gcn4p activity (factor X in Fig. 1). This putative repressor would be effective only in rich medium, perhaps being titrated out in starved cells when Gcn4p is produced at high levels through increased translation. The recent conclusions that Gcn2p is also required for the UV induction of Gcn4p-dependent transcription and that it functions without increasing Gcn4p levels (59) raise the interesting possibility that UV-stimulated phosphorylation of the putative repressor by Gcn2p triggers the degradation of the repressor by the proteosome, with attendant stimulation of Gcn4p function.
TRANSCRIPTIONAL PROFILING SHOWS THAT Gcn4p IS A MASTER REGULATOR OF GENE EXPRESSION DURING AMINO ACID STARVATION
Because Gcn4p induces genes encoding amino acid biosynthetic enzymes in multiple pathways and the inactivation of GCN4 impairs cell growth on media containing amino acid imbalances or inhibitors of amino biosynthesis (32), it was generally assumed that stimulating amino acid biosynthesis to maintain high levels of acylated tRNAs for protein synthesis was the only function of Gcn4p. It was then discovered that certain adenine biosynthetic genes also were induced by Gcn4p in amino acid-starved cells and that at least ADE8 was induced in purine-deprived cells in a Gcn4p-dependent fashion (44, 65, 79). These findings and the realization that GCN4 translation was induced by purine or glucose deprivation led us to conduct a genome-wide analysis with cDNA microarrays to determine whether Gcn4p has additional targets besides amino acid biosynthetic genes.
Expression profiling with cDNA microarrays (56, 85) was used to identify all genes that are induced by a factor of two or more in cells treated with 3-aminotriazole (3-AT), a competitive inhibitor of the histidine biosynthetic enzyme His3p. Histidine limitation imposed by 3-AT leads to high-level induction of GCN4 expression (31). We found that 1,400 genes were induced by 3-AT, representing ∼23% of the genome surveyed. To evaluate the contribution of Gcn4p to this massive response, the expression profile of the wild-type strain was compared to the profiles obtained with a _gcn4_Δ mutant and with a strain expressing Gcn4p constitutively at high levels (GCN4c). Not all of the 1,400 genes induced by 3-AT in wild-type cells yielded significant data in the experiments involving the _gcn4_Δ and GCN4c strains. Of the 897 genes that did, ∼60% were dependent on Gcn4p for maximal induction by 3-AT. These 539 genes were regarded as a minimum estimate of the number of Gcn4p target genes in the yeast genome (Fig. 3). (If 60% of the entire set of 1,400 genes induced by 3-AT in wild-type cells are Gcn4p dependent, then there may be 840 Gcn4p target genes, or ∼14% of the genome.) Interestingly, a small subset of these genes was very weakly induced by 3-AT in wild-type cells but was repressed by 3-AT in the _gcn4_Δ mutant; hence, these genes depend on Gcn4p simply to avoid being repressed under starvation conditions. Such genes might contain promoter elements that mediate reduced transcription in response to histidine starvation and would require activation by Gcn4p to counteract this repressive effect (68).
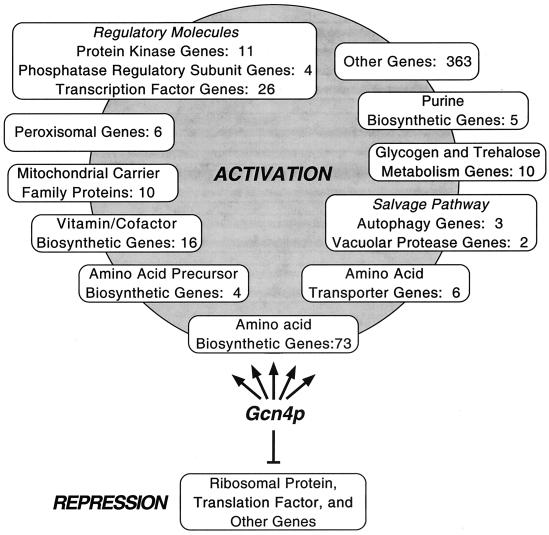
FIG. 3.
Schematic representation of functional categories of Gcn4p target genes. When Gcn4p is induced under conditions of histidine starvation, it elicits the transcriptional activation of at least 539 genes, designated Gcn4p targets (shown above Gcn4p in the activation group). These genes were induced ≥2-fold by 3-AT treatment in wild-type cells and showed a ≥2.0-fold higher level of expression in wild-type cells than in _gcn4_Δ cells (in the presence of 3-AT) or in GCN4c cells than in GCN4 cells (in the absence of 3-AT). The numbers of Gcn4p targets in different functional categories are indicated. Many genes were repressed by a factor of ≥2.0 in response to 3-AT treatment and were dependent on Gcn4p for maximal repression under these conditions (depicted below Gcn4p in the repression group). Prominent among the repressed genes are those encoding RPs or translation initiation factors.
Surprisingly, more than 500 genes were induced by a factor of two or more by 3-AT in the _gcn4_Δ mutant. Many genes in this group were classified as Gcn4p targets because they were induced to a greater degree by 3-AT in the wild-type strain than in the _gcn4_Δ strain, and this subgroup includes canonical Gcn4p target genes encoding amino acid biosynthetic enzymes, e.g., HIS5, ARG4, and MET28. About 25% of the Gcn4p target genes are subject to this dual regulation via Gcn4p-dependent and -independent mechanisms that respond to amino acid limitation (68). To explain the Gcn4p-independent mechanism, it could be proposed that some component of the transcriptional machinery is altered under starvation conditions to permit increased transcription from certain promoters to augment their activation by Gcn4p. For example, an altered form of SAGA, the transcriptional coactivator complex containing histone acetyltransferase Gcn5p, was detected in histidine-starved cells and appears to facilitate promoter binding of TBP more effectively than does conventional SAGA (4). Another sizable subgroup of genes induced by 3-AT in the _gcn4_Δ mutant was induced to nearly the same degrees in GCN4 and _gcn4_Δ strains (68). Such genes might be activated exclusively by the Gcn4p-independent pathway(s) discussed above. It remains to be seen whether the latter genes were induced by histidine starvation or by some other deleterious effect of 3-AT on cell physiology (68).
Remarkably, about 1,000 genes were repressed by a factor of two or more in 3-AT-treated cells, and two-thirds of these were dependent on Gcn4p for strong repression. Many of these genes displayed residual repression in the _gcn4_Δ mutant, indicating Gcn4p-dependent and -independent repression mechanisms. The 90 genes encoding ribosomal proteins (RPs) and many genes encoding translation factors formed the largest group with a common function among the repressed genes (68). This finding can be rationalized as a mechanism for coordinating decreased ribosome production and protein synthesis with the induction of amino acid biosynthesis in amino acid-starved cells, analogous to the stringent response of E. coli (8). As most RP genes lack a recognizable Gcn4p binding site, Gcn4p probably contributes indirectly to their repression. Interestingly, very few genes were repressed under nonstarvation conditions in the GCN4c mutant, suggesting that a combination of a high level of Gcn4p and amino acid starvation is needed for widespread Gcn4p-dependent repression in histidine-starved cells (68). To explain this dual requirement, we propose that promoters of RP genes are down-regulated in starved cells by a mechanism involving antagonism of the activator Rap1p (64) and signal transduction by PKA (47) and the Tor proteins (72). Induction of Gcn4p in amino acid-starved cells would intensify the response by sequestering one or more transcription factors required at RP promoters (squelching) (73). In this view, squelching alone by overexpressing Gcn4p is insufficient for strong repression in nonstarved cells.
Only 235 of the 539 Gcn4p target genes contain a predicted Gcn4p binding site between positions −20 and −600 relative to the start codon. The remaining Gcn4p targets may contain degenerate sites that were not recognized by our computer search for consensus binding sites. For example, Gcn4p can bind to cAMP response elements (CREs) in vitro (87), and it activates transcription from at least one CRE in vivo (70). Alternatively, many target genes may be regulated indirectly through induction of other transcriptional activators by Gcn4p (see below). The latter explanation is consistent with the fact that a larger number of canonical Gcn4p target genes encoding amino acid biosynthetic enzymes contain a Gcn4p binding site (∼84%) compared to target genes at large (∼44%). Moreover, genes induced by Gcn4p are much more likely to contain a binding site in the 5"-proximal 300 nucleotides than are those that are repressed in a Gcn4p-dependent fashion, consistent with the idea that Gcn4p-mediated repression is indirect (68). Recent studies combining the chromatin immunoprecipitation technique with DNA microarray analysis have made it possible to identify all promoters bound by a given transcriptional activator in vivo (39, 54, 77). Application of this method for Gcn4p should allow a determination of which genes are regulated directly through Gcn4p binding to promoters.
Jia et al. used microarray analysis to determine the expression profile for a subset of the yeast genome (1,529 genes) in cells treated with sulfometuron methyl (SM), an inhibitor of branched-chain amino acid biosynthesis (42). Treatment of yeast cells with SM, like treatment with 3-AT, induces GCN4 translation (32). In accordance with the findings obtained with 3-AT, SM induced multiple amino acid and vitamin biosynthetic genes. However, SM repressed genes encoding enzymes of methionine and purine biosyntheses and sulfur assimilation pathways. Additionally, SM treatment altered the expression of fewer genes in a _gcn4_Δ mutant (42) than did 3-AT treatment (68). These differences in the Gcn4p-dependent and -independent responses to 3-AT and SM remain to be explained.
Microarray studies on the transcriptional responses to the activators Gal4p, Yap1p, Pdr1p/Pdr3p, Mac1p, and Ace1p (12, 17, 18, 26, 57, 82) yielded only a few target genes. The activator Ndt80p has been implicated in the regulation of ∼200 genes, and Zap1p has 111 target genes (57), still well below the 539 Gcn4p target genes. Rap1p binds to 294 different loci in vivo and is regarded as a general regulatory factor (54). While it remains to be seen how many genes Gcn4p binds to directly, based on the large number of genes it can induce, Gcn4p may be one of the most pervasive sequence-specific transcriptional activators functioning in the yeast genome.
SCOPE OF THE Gcn4p TRANSCRIPTOME
Amino acid and purine biosynthetic enzymes.
In accordance with previous results, our cDNA microarray analysis showed that Gcn4p activates 78 genes encoding amino acid and purine biosynthetic enzymes (68) (Fig. 3). Gcn4p target genes were identified in every amino acid biosynthetic pathway except for cysteine, and even for the Cys pathway, genes involved in the biosynthesis of its precursors serine and homocysteine are under Gcn4p control. Genes involved in glutamate and glutamine biosyntheses showed very modest Gcn4p-dependent induction by 3-AT, a result which was surprising considering that Gln and Glu are the principal amino group donors in amino acid biosynthesis (58). However, genes encoding certain enzymes that produce citrate or convert citrate to α-ketoglutarate were induced by Gcn4p, and α-ketoglutarate is a precursor of Glu and Gln biosyntheses (68).
Interestingly, GLN3, encoding a key activator of glutamine synthetase and glutamate synthase (products of GLN1 and GLT1, respectively), was induced by Gcn4p; however, many of the Gln3p target genes (including GLN1) were not (68). Gln3p is excluded from the nucleus in a manner dependent on the TOR PKs in cells containing an adequate nitrogen supply (3). As starvation for histidine alone would not limit general nitrogen availability, Gln3p should be excluded from the nucleus in histidine-starved cells. Moreover, URE2, a negative regulator of Gln3p, also was induced by Gcn4p (68); hence, the Gln3p induced by Gcn4p was probably unable to activate transcription. Apparently, Gcn4p cannot induce GLN1 transcription on its own, although there are conflicting data on this point (63). The pathway-specific transcriptional activators LYS14, LEU3, MET4, and MET28 also are induced by Gcn4p (68, 99), such that induction of genes in these pathways by Gcn4p could be indirect. However, there is evidence that Gcn4p also activates at least some genes in the Lys, Leu, and Met biosynthetic pathways in a direct manner (32, 76).
Not every gene in a particular amino acid biosynthetic pathway is regulated by Gcn4p (68); however, enzymes that are not induced transcriptionally can still be activated by Gcn4p indirectly through allosteric control. For example, Aro7p catalyzes the first step in the biosynthesis of Phe and Tyr (44) and is allosterically stimulated by tryptophan (6). Although Gcn4p does not induce ARO7 transcription (6), Aro7p activity should be elevated as a consequence of increased Trp biosynthesis resulting from the transcriptional activation of TRP genes by Gcn4p.
A number of the adenine biosynthetic genes are Gcn4p targets (65, 68, 79) (Fig. 3). The induction of adenine biosynthetic enzymes by Gcn4p could be viewed as a strategy to support increased histidine biosynthesis, as the purine ring of ATP is partially consumed in the histidine pathway. On the other hand, the pathway to AMP consumes PRPP, glycine, aspartate, glutamine, and tetrahydrofolate derivatives, and its induction could be counterproductive to amino acid biosynthesis. Mutations in GCN4 impair cell growth under adenine starvation conditions (79), suggesting that Gcn4p-dependent activation of ADE genes (ADE8 in particular) is important for cell survival under adenine starvation conditions and thus may have little to do with boosting amino acid biosynthesis.
The ADE genes have one or more consensus Gcn4p binding sites (5"TGACTC3") in their promoters, consistent with a direct role for Gcn4p in their activation. However, Bas1p is an activator of multiple ADE genes (16), and it also binds to TGACTC elements (15, 91). Interestingly, BAS1 is induced by Gcn4p, so that Gcn4p might indirectly activate ADE genes by inducing BAS1 (68). As Bas1p additionally activates numerous amino acid biosynthetic genes (HIS4 [2, 91], HIS7 [89], and SHM2 and MTD1 [16]), Gcn4p-dependent activation of these latter genes could involve a contribution from induced levels of Bas1p.
Vitamin biosynthetic enzymes, mitochondrial carrier proteins, and peroxisome biogenesis proteins.
A number of genes newly identified as Gcn4p targets in the microarray studies can be regarded as peripherally involved in amino acid biosynthesis. These include 16 genes for biosynthesis of the vitamins biotin, NAD, tetrahydrofolate, riboflavin, pyridoxal phosphate, and coenzyme A (42, 68) (Fig. 3). As vitamins function as cofactors for many enzymes, vitamin biosynthesis might be induced by Gcn4p to support increased amino acid production. Portions of certain amino acid biosynthetic pathways, including Arg, Lys, Ile, Val, and Leu, take place in mitochondria; hence, precursors and intermediates in these pathways must be shuttled between the two cellular compartments (44). Interestingly, Gcn4p induces the transcription of 10 members of the mitochondrial carrier family (68) (Fig. 3), proteins involved in the transport of small molecules between the cytosol and mitochondria (71). Mutation of one of these genes, ARG11/ORT1, confers leaky arginine auxotrophy (14), consistent with its importance in arginine biosynthesis.
It was surprising that five PEX genes and the transcriptional activator, PIP2, required for peroxisome formation (46, 83), were identified as Gcn4p targets (68) (Fig. 3), as this organelle is the site of β oxidation in the catabolism of fatty acids. However, recent evidence indicates that the lysine biosynthetic enzymes Lys1p and Lys4p are localized in peroxisomes and that lysine prototrophy depends on Pex8p and Pex15p (24). Thus, the induction of peroxisomes may indirectly promote lysine biosynthesis.
Amino acid transporters and autophagy proteins.
The members of two other groups of newly identified Gcn4p target genes provide nonbiosynthetic means of increasing amino acid levels. Yeast cells import amino acids through general and specific amino acid permeases, and Gcn4p induces several genes encoding these transporters (68) (Fig. 3). Gcn4p-mediated activation of GAP1 and AGP1, encoding general amino acid permeases, would stimulate transport of most amino acids under starvation conditions. Genes encoding permeases involved in the uptake of biotin, purines, and pyrimidines also are induced by Gcn4p (68).
In response to starvation for various essential nutrients, cytosolic proteins are targeted to the vacuole for bulk degradation in membrane-bound autophagosomes in a process known as autophagy (48, 69). Three APG genes required for autophagy were identified as Gcn4p targets (Fig. 3), suggesting that Gcn4p enhances this response under amino acid starvation conditions. The induction of genes encoding a vacuolar aminopeptidase (LAP4) and an alanine-arginine aminopeptidase (AAP1) by Gcn4p might accelerate the degradation of proteins transported to the vacuole by the autophagy pathway (68). However, as histidine-starved _gcn4_Δ cells still produced autophagic vesicles, it seems that Gcn4p is not critically required for the response. Moreover, various apg mutants were not impaired for growth under histidine starvation conditions, at least when cell division was still occurring (68). Perhaps the salvage of amino acids from proteins by autophagy lessens the impact of more severe starvation, in which cell division is arrested.
Transcription factors, PKs, and protein phosphatases.
Gcn4p induces numerous regulatory proteins and thus may evoke a cascade of regulatory events leading to gene activation or repression. Twenty-six of the 159 known or predicted DNA binding transcription factors in the MIPS functional catalog (62) are Gcn4p targets (Fig. 3). These include the regulators of amino acid and purine biosyntheses and peroxisome biogenesis already discussed above. Additionally, transcriptional activators involved in the utilization of poor nitrogen sources, catabolism of maltose, heat shock response, copper homeostasis, meiosis, and Ty transcription are induced by Gcn4p. For most of the latter, known target genes were not induced in histidine-starved cells; this activity might occur only under the physiological conditions that normally stimulate these activators and their target genes, e.g., in heat-shocked amino acid-starved cells (68).
Eleven genes in the MIPS PK group and four protein phosphatase regulatory subunits were found to be Gcn4p targets (68) (Fig. 3). The PK Npr1p is activated in nutrient-poor medium and increases the stability of general amino acid permease Gap1p (86); hence, the observed induction of NPR1 by Gcn4p could augment the effect of increased GAP1 transcription evoked by Gcn4p in histidine-starved cells. Two of the three genes encoding PKA catalytic subunits, TPK1/SRA3 and TPK2, were identified as Gcn4p targets (68), and several genes dependent on TPK1 or TPK2 for high-level expression (78) were identified as Gcn4p targets. Thus, Gcn4p might indirectly stimulate the expression of a subset of PKA-dependent genes.
Glycogen metabolic enzymes.
Ten genes encoding enzymes involved in biosynthesis or breakdown of the storage carbohydrate glycogen were identified as Gcn4p targets (Fig. 3). Since _gcn4_Δ cells were found to accumulate higher levels of glycogen than wild-type cells under histidine starvation conditions, it appears that Gcn4p induction under these circumstances leads to a net breakdown of glycogen (68). The glucose thus released might stimulate amino acid precursor biosynthesis via glycolysis and early steps of the tricarboxylic acid cycle. Interestingly, glucose deprivation in cells replete with amino acids leads to Gcn2p-dependent glycogen accumulation (98) (see below).
Biological significance of the extensive Gcn4p transcriptome.
There are 375 Gcn4p target genes that do not fall into the functional categories described above and have no obvious role in the production of amino acids or purines (Fig. 3). Many of these genes are among the most highly induced Gcn4p targets, suggesting that their induction serves an important biological function (68). Given that Gcn4p is induced by conditions other than amino acid deprivation (Fig. 1), perhaps many Gcn4p target genes enhance cell survival in response to other starvation or stress conditions. This notion seems quite likely for purine deprivation, as adenine biosynthetic enzymes are Gcn4p targets (Fig. 3) and gcn4 cells are more sensitive than wild-type cells to an inhibitor of purine biosynthesis (79). The activation of Gcn2p during prolonged incubation in medium containing a low glucose concentration serves to maintain vacuolar amino acid pools and glycogen levels and to minimize the lag time required before the resumption of growth when cells are transferred to medium containing abundant glucose (98). Perhaps the enhanced glycogen accumulation under these conditions results from Gcn4p-dependent induction of glycogen biosynthetic enzymes (68). It is puzzling, however, that the amino acid biosynthetic genes are not induced following the diauxic shift when cells have consumed all of the glucose in the medium and are respiring on ethanol (18). Perhaps the induction of Gcn4p during growth on medium containing a low glucose concentration merely prevents repression of its target genes. Another positive effect of activating Gcn2p under these conditions may be to slow down protein synthesis by eIF2α phosphorylation.
Many amino acid biosynthetic genes are induced by MMS (41). Given that MMS induces GCN4 translation (68) and that it also stimulates Gcn4p activation function at several HIS genes (90) (Fig. 1), the induction of amino acid biosynthetic genes by MMS is probably mediated by Gcn4p. Interestingly, gamma irradiation, which causes DNA damage, did not induce Gcn4p target genes (40). Moreover, the induction of GCN4 translation by MMS did not require the DNA damage checkpoint proteins Dun1p, Mec1p, and Rad53p, and _gcn4_Δ cells were not hypersensitive to MMS (68). Thus, protein methylation rather than DNA damage may be the stimulus that triggers increased GCN4 translation in MMS-treated cells. In any case, the induction of Gcn4p target genes is not critical for survival in the presence of MMS (68). On the other hand, UV irradiation also stimulates the activation function of Gcn4p and possibly GCN4 translation (Fig. 1), and gcn4 mutants show a modest increase in UV sensitivity (21). Certain Gcn4p target genes are involved in the repair of DNA damage (68), so that Gcn4p may enhance the efficiency of DNA repair in UV- and MMS-treated cells.
GCN4 is required for survival in the presence of high NaCl concentrations, and this requirement can be attributed at least partly to the fact that Gcn4p induces the transcription of HAL1, whose product plays an important role in maintaining Na+-K+ ion homeostasis. It appears that Gcn4p competes with the repressor Sko1p for binding to a CRE in the HAL1 promoter to effect HAL1 induction by high salt concentrations (70). Surprisingly, _gcn2_Δ cells are more resistant than wild-type cells to salt stress, and this phenotype results partly from the inability of _gcn2_Δ cells to derepress Gcn4p to high levels. Hence, it is possible that a modest induction of GCN4 enhances cell survival in moderate salt concentrations by inducing HAL1 transcription, whereas the hyperexpression of GCN4 and the extensive eIF2α phosphorylation that results from the strong activation of Gcn2p at high salinities will impede growth (25).
Gcn4p is induced by rapamycin (93), and this activity may contribute to the widespread gene induction evoked by this drug, as 31% of the 228 genes induced twofold or more by rapamycin (30) are Gcn4p target genes (68). Rapamycin inhibits Tor1p and Tor2p, which negatively regulate Gln3p function; hence, a subset of rapamycin-induced genes includes the targets of Gln3p that are involved in the utilization of poor nitrogen sources and that are subject to nitrogen catabolite repression (NCR) (7, 30). Interestingly, Gcn4p is required for full induction by rapamycin of the Gln3p target genes DAL5 and DAL1, of which DAL5 is a known Gcn4p target (68). Rapamycin induces GCN4 expression on rich medium to only ∼1/10 the level achieved by 3-AT on minimal medium (93). This relatively small amount of Gcn4p may not be sufficient to activate most Gcn4p target genes, explaining why only a small fraction of the latter are induced by rapamycin, but it may contribute substantially to the activation of NCR-regulated promoters that contain both Gcn4p and Gln3p binding sites. The contribution of Gcn4p to the activation of NCR-regulated promoters may be nullified when nitrogen starvation becomes severe enough to override the translational induction of GCN4 (27) (Fig. 1). The induction of Gcn4p by rapamycin may also contribute to the drastic repression of RP genes elicited by this drug (72), although the modest level of Gcn4p induced by rapamycin makes this notion doubtful (93).
Two recent studies identified about 500 to 600 genes that were regulated in a stereotypical manner in response to diverse stress conditions; this response is called the environmental stress response (ESR) (23) or the common environmental response (CER) (9). Fifty percent of the genes induced in the ESR also were induced in the CER, and 32% of the genes common to the ESR and the CER were identified as Gcn4p target genes (68). It is possible that Gcn4p contributes to the induction of these coregulated genes under stress conditions besides amino acid starvation that induce the expression or activity of Gcn4p, akin to the contribution of Gcn4p to gene induction by rapamycin described above. Many of the genes repressed in histidine-starved cells overlap the genes repressed in the ESR or the CSR, so that Gcn4p may also contribute to gene repression under diverse stress conditions. It will be interesting to determine the full range of stress conditions that activate the expression or function of Gcn4p and to determine where transcriptional activation (or repression) by Gcn4p makes an important contribution to cell survival. Careful consideration of the diverse groups of genes belonging to the Gcn4p transcriptome combined with an understanding of the stress conditions that induce Gcn4p and its target genes should lead eventually to a full appreciation of the scope of general amino acid control.
Acknowledgments
We thank Tom Dever for helpful comments and John Reese for personal communiation of unpublished results.
REFERENCES
- 1.Albrecht, G., H. U. Mosch, B. Hoffmann, U. Reusser, and G. H. Braus. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. **273:**12696-12702. [DOI] [PubMed]
- 2.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237**:**874-880. [DOI] [PubMed] [Google Scholar]
- 3.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402**:**689-692. [DOI] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20**:**634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlanga, J. J., J. Santoyo, and C. De Haro. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 265**:**754-762. [DOI] [PubMed] [Google Scholar]
- 6.Braus, G. H. 1991. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55**:**349-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13**:**3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel, M., and K. E. Rudd. 1987. The stringent response, p. 1410-1438. In F. C. Neidhardt, J. L. Ingraham, B. Magasanik, K. B. Low, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 9.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12**:**323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chantrel, Y., M. Gaisne, C. Lions, and J. Verdiere. 1998. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 148**:**559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15**:**1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282**:**699-705. [DOI] [PubMed] [Google Scholar]
- 13.Colombo, S., P. Ma, L. Cauwenberg, J. Winderickx, M. Crauwels, A. Teunissen, D. Nauwelaers, J. H. de Winde, M. F. Gorwa, D. Colavizza, and J. M. Thevelein. 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17**:**3326-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabeel, M., O. Soetens, M. De Rijcke, R. Pratiwi, and R. Pankiewicz. 1996. The ARG11 gene of Saccharomyces cerevisiae encodes a mitochondrial integral membrane protein required for arginine biosynthesis. J. Biol. Chem. 271**:**25011-25018. [DOI] [PubMed] [Google Scholar]
- 15.Daignan-Fornier, B., and G. R. Fink. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89**:**6746-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis, V., and B. Daignan-Fornier. 1998. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 259**:**246-255. [DOI] [PubMed] [Google Scholar]
- 17.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470**:**156-160. [DOI] [PubMed] [Google Scholar]
- 18.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278**:**680-686. [DOI] [PubMed] [Google Scholar]
- 19.Dong, J., H. Qiu, M. Garcia-Barrio, J. Anderson, and A. G. Hinnebusch. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6**:**269-279. [DOI] [PubMed] [Google Scholar]
- 20.Drysdale, C. M., B. M. Jackson, R. McVeigh, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, P. A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18**:**1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelberg, D., C. Klein, H. Martinetto, K. Struhl, and M. Karin. 1994. The UV response involving the ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell 77**:**381-390. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Barrio, M., J. Dong, S. Ufano, and A. G. Hinnebusch. 2000. Association of GCN1/GCN20 regulatory complex with the conserved N-terminal domain of eIF2α kinase GCN2 is required for GCN2 activation in vivo. EMBO J. 19**:**1887-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11**:**4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geraghty, M. T., D. Bassett, J. C. Morrell, G. J. Gatto, Jr., J. Bai, B. V. Geisbrecht, P. Hieter, and S. J. Gould. 1999. Detecting patterns of protein distribution and gene expression in silico. Proc. Natl. Acad. Sci. USA 96**:**2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Goossens, A., T. E. Dever, A. Pascual-Ahuir, and R. Serrano. 2001. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 76**:**30753-30760. [DOI] [PubMed]
- 26.Gross, C., M. Kelleher, V. R. Iyer, P. O. Brown, and D. R. Winge. 2000. Identification of the copper regulon in saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275**:**32310-32316. [DOI] [PubMed] [Google Scholar]
- 27.Grundmann, O., H. U. Mosch, and G. H. Braus. 2001. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 276**:**25661-25671. [DOI] [PubMed] [Google Scholar]
- 28.Han, S. J., Y. C. Lee, B. S. Gim, G. H. Ryu, S. J. Park, W. S. Lane, and Y. J. Kim. 1999. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol. 19**:**979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6**:**1099-1108. [DOI] [PubMed] [Google Scholar]
- 30.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96**:**14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinnebusch, A. G. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81**:**6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Hinnebusch, A. G. 1986. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. Crit. Rev. Biochem. 21**:**277-317. [DOI] [PubMed] [Google Scholar]
- 34.Hinnebusch, A. G. 1985. A hierarchy of _trans_-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5**:**2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Hinnebusch, A. G. 1996. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2, p. 199-244. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Hoffmann, B., H. U. Mosch, E. Sattlegger, I. B. Barthelmess, A. Hinnebusch, and G. H. Braus. 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 31**:**807-822. [DOI] [PubMed] [Google Scholar]
- 38.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Goulb, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95**:**717-728. [DOI] [PubMed] [Google Scholar]
- 39.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409**:**533-538. [DOI] [PubMed] [Google Scholar]
- 40.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20**:**8157-8167.11027285 [Google Scholar]
- 41.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96**:**1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia, M. H., R. A. Larossa, J. M. Lee, A. Rafalski, E. Derose, G. Gonye, and Z. Xue. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3**:**83-92.11015603 [Google Scholar]
- 43.Johnston, M., and M. Carlson. 1992. Regulation of carbon and phosphate utilization, p. 193-282. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression, vol. II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 44.Jones, E. W., and G. R. Fink. 1982. Regulation of amino acid and nucleotide biosynthesis in yeast, p. 181-300. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15**:**291-339. [DOI] [PubMed] [Google Scholar]
- 46.Karpichev, I. V., Y. Luo, R. C. Marians, and G. M. Small. 1997. A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of beta-oxidation enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 17**:**69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14**:**1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky, D. J., and S. D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290**:**1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13**:**6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kruger, D., J. Koch, and I. B. Barthelmess. 1990. cpc-2, a new locus involved in general control of amino acid synthetic enzymes in Neurospora crassa. Curr. Genet. 18**:**211-215. [DOI] [PubMed] [Google Scholar]
- 51.Kubota, H., K. Ota, Y. Sakaki, and T. Ito. 2001. Budding yeast GCN1 binds the GI domain to activate the eIF2α kinase GCN2. J. Biol. Chem. 276**:**17591-17596. [DOI] [PubMed] [Google Scholar]
- 52.Kubota, H., Y. Sakaki, and T. Ito. 2000. GI domain-mediated association of the eIF2α kinase GCN2 with its activator GCN1 is required for general amino acid control in budding yeast. J. Biol. Chem. 275**:**20243-20246. [DOI] [PubMed] [Google Scholar]
- 53.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19**:**2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28**:**327-334. [DOI] [PubMed] [Google Scholar]
- 55.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and, J. R. Yates, 3rd. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17**:**676-682. [DOI] [PubMed] [Google Scholar]
- 56.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14**:**1675-1680. [DOI] [PubMed] [Google Scholar]
- 57.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97**:**7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magasanik, B. 1992. Regulation of nitrogen utilization, p. 283-317. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Marbach, I., R. Licht, H. Frohnmeyer, and D. Engelberg. 2001. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J. Biol. Chem. 276**:**16944-16951. [DOI] [PubMed] [Google Scholar]
- 60.Marton, M. J., C. R. Vazquez de Aldana, H. Qiu, K. Chakraburtty, and A. G. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on enlongating ribosomes in activation of the eIF2α kinase GCN2. Mol. Cell. Biol. 17**:**4474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol. Biol. Cell 11**:**915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mewes, H. W., J. Hani, F. Pfeiffer, and D. Frishman. 1998. MIPS: a database for protein sequences and complete genomes. Nucleic Acids Res. 26**:**33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell, A. P., and B. Magasanik. 1984. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol. Cell. Biol. 4**:**2767-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moehle, C. M., and A. G. Hinnebusch. 1991. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 11**:**2723-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mosch, H. U., B. Scheier, R. Lahti, P. Mantsala, and G. H. Braus. 1991. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J. Biol. Chem. 266**:**20453-20456. [PubMed] [Google Scholar]
- 66.Myer, V. E., and R. A. Young. 1998. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273**:**27757-27760. [DOI] [PubMed] [Google Scholar]
- 67.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with SWI/SNF complex and SRB/mediator. Mol. Cell 4**:**657-664. [DOI] [PubMed] [Google Scholar]
- 68.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21**:**4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohsumi, Y. 1999. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos. Trans. R. Soc. London B. Biol. Sci. 354**:**1577-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascual-Ahuir, A., R. Serrano, and M. Proft. 2001. The Sko1p repressor and Gcn4p activator antagonistically modulate stress-regulated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 21**:**16-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paulsen, I. T., M. K. Sliwinski, B. Nelissen, A. Goffeau, and M. H. Saier, Jr. 1998. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 430**:**116-125. [DOI] [PubMed] [Google Scholar]
- 72.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10**:**987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ptashne, M., and A. F. Gann Alexander. 1990. Activators and targets. Nature 346**:**329-331. [DOI] [PubMed] [Google Scholar]
- 74.Qiu, H., J. Dong, C. Hu, C. S. Francklyn, and A. G. Hinnebusch. 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J. 20**:**1425-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiu, H., C. Hu, J. Anderson, G. R. Bjork, S. Sarkar, A. K. Hopper, and A. G. Hinnebusch. 2000. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 20**:**2505-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramos, F., E. Dubois, and A. Pierard. 1988. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Eur. J. Biochem. 171**:**171-176. [DOI] [PubMed] [Google Scholar]
- 77.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290**:**2306-2309. [DOI] [PubMed] [Google Scholar]
- 78.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97**:**5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13**:**5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ron, D., C. H. Chen, J. Caldwell, L. Jamieson, E. Orr, and D. Mochly-Rosen. 1994. Cloning of an intracellular receptor for protein kinase C: a homolog of the á subunit of G proteins. Proc. Natl. Acad. Sci. USA 91**:**839-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ron, D., and H. P. Harding. 2000. PERK and translational control by stress in the endoplasmic reticulum, p. 547-560. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 82.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16**:**939-945. [DOI] [PubMed] [Google Scholar]
- 83.Rottensteiner, H., A. J. Kal, M. Filipits, M. Binder, B. Hamilton, H. F. Tabak, and H. Ruis. 1996. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 15**:**2924-2934. [PMC free article] [PubMed] [Google Scholar]
- 84.Sattlegger, and Hinnebusch. 2000. Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 19**:**6622-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270**:**467-470. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17**:**6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sellers, J. W., A. C. Vincent, and K. Struhl. 1990. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol. Cell. Biol. 10**:**5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sood, R., A. C. Porter, D. Olsen, D. R. Cavener, and R. C. Wek. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics 154**:**787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Springer, C., M. Kunzler, T. Balmelli, and G. H. Braus. 1996. Amino acid and adenine cross-pathway regulation act through the same 5"-TGACTC-3" motif in the yeast HIS7 promoter. J. Biol. Chem. 271**:**29637-29643. [DOI] [PubMed] [Google Scholar]
- 90.Stitzel, M. L., R. Durso, and J. C. Reese. 2001. The proteasome regulates the UV-induced activation of the AP-1-like transcription factor Gcn4. Genes Dev. 15**:**128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246**:**931-935. [DOI] [PubMed] [Google Scholar]
- 92.Tzamarias, D., I. Roussou, and G. Thireos. 1989. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell 57**:**947-954. [DOI] [PubMed] [Google Scholar]
- 93.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183**:**2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vazquez de Aldana, C. R., M. J. Marton, and A. G. Hinnebusch. 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 14**:**3184-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vazquez de Aldana, C. R., R. C. Wek, P. San Segundo, A. G. Truesdell, and A. G. Hinnebusch. 1994. Multicopy tRNA genes functionally suppress mutations in yeast eIF-2α kinase GCN2: evidence for separate pathways coupling GCN4 expression to uncharged tRNA. Mol. Cell. Biol. 14**:**7920-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15**:**4497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Welsh, G. I., C. M. Miller, A. J. Loughlin, N. T. Price, and C. G. Proud. 1998. Regulation of eukaryotic initiation factor eIF2B: glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin. FEBS Lett. 421**:**125-130. [DOI] [PubMed] [Google Scholar]
- 98.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20**:**2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou, K., P. R. G. Brisco, A. E. Hinkkanen, and G. B. Kohlhaw. 1987. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 15**:**5261-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]