A Haplotype Implicated in Schizophrenia Susceptibility Is Associated with Reduced COMT Expression in Human Brain (original) (raw)
Abstract
The gene encoding catechol-O-methyltransferase (COMT) is a strong candidate for schizophrenia susceptibility, owing to the role of COMT in dopamine metabolism, and the location of the gene within the deleted region in velocardiofacial syndrome, a disorder associated with high rates of schizophrenia. Recently, a highly significant association was reported between schizophrenia and a COMT haplotype in a large case-control sample (Shifman et al. 2002). In addition to a functional valine→methionine (Val/Met) polymorphism, this haplotype included two noncoding single-nucleotide polymorphisms (SNPs) at either end of the COMT gene. Given the role of COMT in dopamine catabolism and that deletion of 22q11 (containing COMT) is associated with schizophrenia, we postulated that the susceptibility COMT haplotype is associated with low COMT expression. To test this hypothesis, we have applied quantitative measures of allele-specific expression using mRNA from human brain. We demonstrate that COMT is subject to allelic differences in expression in human brain and that the COMT haplotype implicated in schizophrenia (Shifman et al. 2002) is associated with lower expression of COMT mRNA. We also show that the 3′ flanking region SNP that gave greatest evidence for association with schizophrenia in that study is transcribed in human brain and exhibits significant differences in allelic expression, with lower relative expression of the associated allele. Our results indicate that COMT variants other than the Val/Met change are of functional importance in human brain and that the haplotype implicated in schizophrenia susceptibility is likely to exert its effect, directly or indirectly, by down-regulating COMT expression.
Introduction
Schizophrenia (MIM 181500) is a common yet severe psychiatric condition, characterized by profound disturbances of cognition, emotion, and social functioning. Despite evidence for a substantial heritable component (Owen et al. 2002), the genetic loci that contribute to the disorder remain to be elucidated, and the underlying neurobiological mechanisms are largely unknown.
Recently, a highly significant association was reported between variants within the gene encoding catechol-O-methyltransferase (COMT [MIM 116790]) and schizophrenia in a large Israeli sample (Shifman et al. 2002). The COMT gene is a strong candidate for schizophrenia susceptibility, owing to the role of the encoded enzyme in the metabolism of released dopamine (Axelrod and Tomchick 1958), and its physical localization to chromosome 22q11 (Grossman et al. 1992). Hyperdopaminergic function has long been postulated in schizophrenia (van Rossum 1966; Carlsson 1978), although direct evidence for a primary dopaminergic abnormality has proved difficult to substantiate. Chromosome 22q11 has been implicated in linkage studies of schizophrenia (Pulver 2000) and is the site of microdeletions that are associated with velocardiofacial syndrome (VCFS [MIM 192430]), a condition in which high rates of schizophrenia have been reported (Murphy et al. 1999).
The COMT protein occurs as two distinct forms: a soluble form found in the cell cytoplasm (S-COMT) and a longer, membrane-bound form (MB-COMT). In most assayed tissues, the S-COMT form predominates, accounting for ∼95% of total COMT activity (Jeffery and Roth 1984; Grossman et al. 1985). However, the MB-COMT form is the more prevalent species in brain (Tenhunen et al. 1994). Studies prior to that of Shifman et al. (2002) that investigated genetic association between schizophrenia and the COMT gene have yielded inconsistent results. These have largely focused on an SNP that produces a valine→methionine (Val/Met) substitution at codons 108 and 158 in the S-COMT and MB-COMT transcripts, respectively (Lachman et al. 1996). The Val/Met polymorphism is reported to have a functional effect on enzyme activity, which is associated with altered thermostability of the COMT protein (Lotta et al. 1995; Lachman et al. 1996). Although the majority of case-control studies have failed to find evidence for association between this polymorphism and schizophrenia (e.g., Daniels et al. 1996; Liou et al. 2001; but see Ohmori et al. [1998]), preferential transmission of the reportedly high activity (Val) allele has been reported in several studies in which the transmission/disequilibrium test (TDT) was employed (e.g., Li et al. 1996; Kunugi et al. 1997; Egan et al. 2001).
The study of Shifman et al. (2002) is the largest case-control analysis in schizophrenia that has been reported to date, with >700 patients and 4,000 control subjects. The authors tested not only the Val/Met polymorphism, but also several other SNPs across the COMT gene. It is interesting that the evidence for association between schizophrenia and the Val/Met polymorphism was modest, but, when it was analyzed as part of a haplotype that included two other noncoding SNPs, extremely high levels of statistical significance were achieved. Moreover, the two additional polymorphisms included in the haplotype—one in the first intron of the MB-COMT transcript (rs737865) and one purported to lie near the 3′ UTR (rs165599)—were themselves highly significantly associated. These data suggest that if the COMT locus is indeed involved in susceptibility to schizophrenia, this cannot be wholly accounted for by the Val/Met polymorphism; therefore, other functional variants are likely to exist.
One possibility is that there exists genetic variation at the COMT locus that affects expression of the gene. Given that COMT degrades dopamine, that most treatments for schizophrenia block dopaminergic transmission (whereas drugs that enhance it are psychotomimetic), and that deletion of 22q11 (including COMT) is associated with schizophrenia, we postulated that the COMT haplotype associated with schizophrenia would also be associated with low COMT expression.
To investigate potential _cis_-acting influences on gene expression, it is possible to use SNPs within an expressed sequence as a tag for mRNA transcribed from each chromosomal allele. It is then possible to apply quantitative methods of allele discrimination to mRNA from individual subjects who are heterozygous for the marker polymorphism to measure relative allelic expression (e.g., Singer-Sam et al. 1992; Cowles et al. 2002; Yan et al. 2002). Each allele serves as an internal control against which expression of the other allele can be measured within each individual mRNA sample, thereby enabling detection of genuine _cis_-acting phenomena that affect expression, while controlling for _trans_-acting confounders.
We have applied this principle to investigate possible _cis_-acting mechanisms that affect expression of the COMT gene in human brain. In particular, we sought to test whether the haplotype previously implicated in schizophrenia susceptibility (Shifman et al. 2002) is also associated with altered COMT expression. To this end, we have assayed mRNA derived from human cerebral cortex using a highly quantitative method of allele discrimination (Norton et al. 2002), capable of determining small percentage differences in allelic expression.
Methods
Samples
Postmortem brain tissue derived from frontal, parietal, or temporal cortex of 60 unrelated, anonymous human adults was obtained from three sources (The MRC London Neurodegenerative Diseases Brain Bank, United Kingdom; The Stanley Medical Research Institute Brain Bank, Bethesda; The Karolinska Institute, Stockholm). Subjects were drawn principally from psychiatric control groups. From each individual, ∼500 mg of tissue was processed for genomic DNA, by use of standard phenol/chloroform procedures, and ∼300–500 mg of tissue was processed for total RNA, by use of the RNAwiz isolation reagent (Ambion). Total RNA was treated with DNAse prior to reverse transcription, which was performed by use of the RETROscript kit (Ambion).
Genotyping
Primers (sequences available on request) were designed by use of the Primer3 program. PCR was performed in a total reaction volume of 12 μl that contained 24-ng genomic DNA template, 1× PCR buffer, dNTPs at 0.1 mM, primers at 0.12–0.24 mM, and 0.3-U “Hot Star” Taq polymerase (Qiagen). Genotyping was performed by use of primer extension and the SNaPshot Multiplex Kit (Applied Biosystems).
Allele Expression Assay
Genomic DNA from all subjects was initially genotyped to identify heterozygotes for the marker polymorphisms. The corresponding cDNA samples were initially assayed twice as two separate RT reactions alongside the corresponding genomic DNA samples. This analysis was always replicated on a second occasion. Where the stratified analyses were significant in each of the above two experiments, a third analysis was performed that contained two separate RT reactions for each individual, each amplified twice, alongside a genomic control group. For simplicity, the data presented below represent the average for all assays. For assays using SNPs rs4633 and rs165599, samples were amplified by use of primers that were based on single exonic sequence, capable of amplifying either cDNA or genomic DNA. RNA samples did not yield detectable levels of product in the absence of an RT step. The same analytic conditions were used for genomic DNA and cDNA for these principal analyses to enable us to use the average of the ratios observed from genomic DNA (as a perfect 1:1 ratio of the two alleles) to correct allelic ratios obtained from cDNA analyses for any inequalities in allelic representation specific to each assay (Hoogendoorn et al. 2000). Since we have previously shown that allelic representation in individual samples can be influenced by polymorphisms under the primers (unpublished data), prior to applying the general correction factor to the cDNA, we ensured that the relative allelic representations in all genomic samples were equivalent. Because rs165688 (encoding 108/158 Val/Met) is too near an intron-exon boundary to allow the design of common genomic and cDNA primers, it was genotyped from genomic DNA by use of primers that were based on adjacent intronic sequence, whereas cDNA was assayed at this SNP by use of “cDNA-specific” primers that spanned intron-exon boundaries.
Allelic representation was measured by use of primer extension and SNaPshot chemistry (Applied Biosystems). Amplified samples were incubated with 1 U shrimp alkaline phosphatase (Amersham) and 2 U exonuclease I (Amersham) for 45 min at 37°C and, then, for 15 min at 85°C, prior to primer extension. Primer extension reactions were performed in a total volume of 10 μl, which contained 2 μl treated PCR product, 4.5 μl SNaPshot kit, 2.5 μl water, and 0.3–0.5 pMol extension primer. Primer extension thermocycling conditions consisted of an initial step of 95°C for 2 min, followed by 25 cycles of 95°C for 5 s, 43°C for 5 s, and 60°C for 5 s. Following primer extension, reaction products were treated with 0.5 U shrimp alkaline phosphatase (Amersham) for 45 min at 37°C and, then, for 15 min at 85°C. Aliquots of 1 μl SNaPshot reaction product were combined with 9 μl Hi-Di formamide and loaded onto a 3100 DNA sequencer (Applied Biosystems). Products were electrophoresed on a 36-cm capillary array at 60°C and data processed by use of Genescan Analysis version 3.7 software (Applied Biosystems). Peak heights of allele-specific extended primers were determined by use of Genotyper version 2.5 software (Applied Biosystems). The ratio of cDNA peak heights, corrected, where possible, by use of the average genomic ratio from all heterozygous samples, was used to calculate relative expression of the two alleles in each individual sample.
Statistical Analysis
Predicted haplotype frequencies were calculated by use of EH plus (Zhao et al. 2000). D′ and _r_2 measures of linkage disequilibrium (LD) were calculated by use of the program ldmax within the GOLD software package. Group comparisons were analyzed by t test (two tailed) or, where there was more than one group, by one-way ANOVA, by use of Minitab Statistical Software (Minitab release 13.32).
Results
Genotyping for COMT Polymorphisms
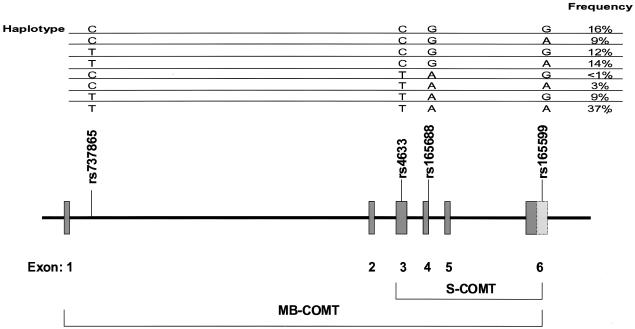
The sample was individually genotyped for SNPs rs737865 and rs165599 (which gave strongest evidence for association in the study of Shifman et al. [2002]), rs165688 (which encoded the Val/Met polymorphism), and rs4633 (a synonymous SNP located within exon 3 of the MB-COMT transcript and exon 1 of the S-COMT transcript). Positions of SNPs and predicted haplotype frequencies are shown in figure 1. Consistent with previous data (Shifman et al. 2002), SNPs rs165688 (Val/Met) and rs4633 were found to be in complete LD (_D_′=1; _r_2=1). SNP rs737865, located within intron 1 of the MB-COMT transcript, also showed strong LD with SNPs rs165688/rs4633 (_D_′=0.84; _r_2=0.24). SNP rs165599, located at the 3′ end of the COMT gene, showed more modest LD with rs165688/rs4633 (_D_′=0.46; _r_2=0.12).
Figure 1.
Genomic structure of the COMT gene, which indicates exon usage of the membrane-bound (MB-COMT) and soluble (S-COMT) forms. Location of assayed SNPs are shown, together with estimated haplotype frequencies.
_Cis_-Acting Differences in COMT Expression
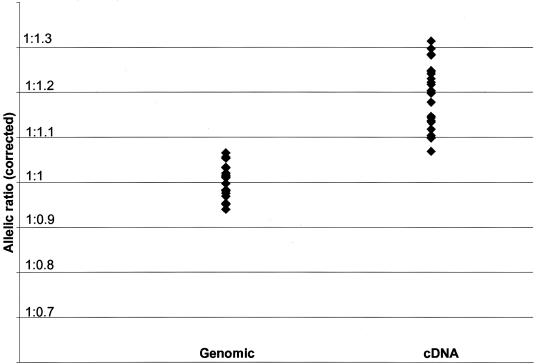
Alleles are not equally represented by the SNaPshot assay, even when present in equimolar concentrations. Therefore, to obtain exact estimates of the relative allelic expression, it is necessary to correct the estimate obtained from cDNA for differential representation of alleles with a correction factor derived from genomic DNA, where the exact ratio of alleles is known to be equimolar. SNP rs165688 (Val/Met) is too near an intron/exon boundary to enable the design of primers that amplify both genomic DNA and cDNA. Our primary analysis was, therefore, based on expressed SNP rs4633, which is in perfect LD with the Val/Met polymorphism. Twenty-three heterozygotes were assayed for allelic differences in expression at SNP rs4633 (fig. 2). The genomic ratios for each individual sample were similar, which indicates that the data are not confounded by unidentified sequence variants that might alter the relative efficiencies of allelic representation between samples (on the basis of two measures from each genomic sample, the maximum deviation from the mean ratio was 6%). The correction factor for inequalities in allelic representation was, therefore, derived from the average genomic ratio obtained from all heterozygotes in each experiment. Analysis of the corrected cDNA peak-height ratios in all three replicate analyses showed that, in all individuals, the C allele was significantly reduced in expression, relative to the T allele. Even though they were conducted on three different occasions, repeat assays showed good reproducibility of individual cDNA allelic ratios, with an average SD of 0.03. When the data from all individuals across all experiments were combined, the expression of the C allele remained significantly reduced (P<.0001), with an average relative reduction of 17% (95% CI 14%–21%). The C allele at rs4633 always lies on the same haplotype as the G allele at rs165688 (encoding valine), which indicates that the valine transcripts are expressed at a slightly lower level than those encoding the 108/158 methionine allele in human brain. In accordance, when SNP rs165688 was used to assay the same mRNA samples, the relative allelic differences in expression correlated significantly with those obtained when assayed using SNP rs4633 (correlation 0.7; P<.001), despite the relatively limited range of ratios observed in each assay.
Figure 2.
Comparison between observed genomic ratios and corrected cDNA ratios assayed at SNP rs4633 (_n_=23). Data are expressed as ratio of C:T alleles. The data for genomic DNA are the averages of two measurements for each individual sample. The data for cDNA are the averages of eight estimates for each individual sample.
The strongest evidence for association with schizophrenia in the study of Shifman et al. (2002) was yielded by a three-marker haplotype that consisted of a variant in intron 1 of the MB-COMT transcript (rs737865), a polymorphism at the 3′ end of the gene (rs165599), and the valine-encoding allele of the Val108/158Met polymorphism (fig. 1). If reduced expression of COMT is responsible for this association, we would expect the risk haplotype to be the lowest expressed haplotype. We therefore predict that individuals who carry one copy of the risk haplotype and one copy of a nonrisk haplotype will display greater differences in the relative expression of their haplotypes than individuals who carry two copies of nonrisk haplotypes. To test this, the rs4633 data were stratified according to whether subjects carried either zero or one copy of the risk haplotype, as defined by rs4633 and the specific loci included in the stratified analyses (table 1). However, we added an extra level of stringency to our analysis because Shifman and colleagues (2002) also postulate that their data represent the presence of independent risk alleles at these loci. We therefore required that the subjects we examined with one copy of the risk haplotype not carry any of the risk alleles on the other haplotype; that is, they were heterozygous at the particular loci included in each stratified analysis. Conversely, all subjects without a risk haplotype were required to be homozygous at each of the loci included in the stratified analysis, other than the rs16588 polymorphism (owing to the complete LD between it and the test locus), thereby achieving internal control for any influence these loci might have on expression. Although these stratified analyses included subjects who were homozygous at some of the loci, we re-emphasize that all subjects were heterozygous at the test locus and, therefore, each observation is still controlled for the _trans_-acting and tissue-specific variables that could confound such comparisons when based on measurement of the total mRNA level.
Table 1.
Analysis of the Relative Expression of COMT Alleles, as Assayed at SNP rs4633[Note]
| Genotype at Loci | |||||
|---|---|---|---|---|---|
| No. of RiskHaplotypes | rs165688 Val/Met(_D_′=1) | rs737865(_D_′=.84) | rs165599(_D_′=.46) | Mean Reduction of C vs. T Allele % (SD) | P |
| 1 | Heterozygous | … | … | 17 (5) | <.0001 |
| 0 | Heterozygous | Homozygous | … | 14 (4) | … |
| 1 | Heterozygous | Heterozygous | … | 20 (4) | .003 |
| 0 | Heterozygous | … | Homozygous | 16 (5) | … |
| 1 | Heterozygous | … | Heterozygous | 19 (5) | .22 |
| 0 | Heterozygous | Homozygous | Homozygous | 13 (5) | … |
| 1 | Heterozygous | Heterozygous | Heterozygous | 22 (4) | .023 |
Despite small samples sizes, subjects who were heterozygous for SNP rs737865 (_n_=11) showed a significant reduction, in all three experiments (overall _P_=.003), in the expression of the C allele at rs4633 compared with rs737865 homozygotes (_n_=11). The haplotype phases are unambiguous with respect to the rs737865 homozygotes but not the rs737865/rs4633 double heterozygotes. However, given the observed haplotype frequencies, the probability is 0.93 that the haplotypes constructed from rs737865 and the Val/Met polymorphism in a double heterozygote are C-G and T-A. Using the nomenclature of Shifman et al. (2002), this corresponds to the G-G and A-A haplotypes that, in that study, were respectively over- and underrepresented in schizophrenia.
When the same procedure was applied to heterozygosity (_n_=13) and homozygosity (_n_=7) at SNP rs165599, there was only a nonsignificant trend toward lower C-allele expression at rs4633 (_P_=.22). For this analysis, the probability was 0.83 that the haplotypes constructed from the Val/Met and rs165599 loci in double heterozygotes are G-G and A-A, corresponding to the haplotypes over- and underrepresented, respectively, in schizophrenia (Shifman et al. 2002).
The biggest reduction in expression of the C allele at rs4633 was observed in subjects who were heterozygous for both rs737865 and rs165599 (_n_=7), compared with those who were homozygous at both these loci (_n_=5). In individuals who were heterozygous at both loci, the C allele was underexpressed in all three independent experiments, with an overall mean of 22%, compared with a mean reduction of 13% in those rs4633 heterozygotes who were homozygous for SNPs rs737865 and rs165599. Despite the small sample sizes, this difference was significant in each individual assay and where all data were combined (_P_=.023). The probability is 0.84 that, in each treble heterozygote, the phase of the lowest expression haplotype contains the C allele at rs737865 (forward sense, G, according to Shifman et al. [2002]), the G allele at rs16588 (encoding valine), and the G allele at rs165599. This is precisely the haplotype that was overrepresented in patients with schizophrenia in the study of Shifman et al. (2002). Very similar results were obtained for identical analyses we performed without the extra requirement of homozygosity and heterozygosity at rs737865 and rs165599 in subjects with zero and one risk haplotypes, respectively (data not shown).
The genetic analysis of Shifman et al. (2002) suggested sex-specific effects of the associated alleles, both in terms of allele frequency in the general population and in the strength of association with schizophrenia. However, no differences in allelic ratio were observed between males (_n_=12) and females (_n_=11) heterozygous for SNP rs4633 (_P_=.74) or in the subset who were also heterozygous for SNP rs737865 (7M; 4F; _P_=.74). Comparison between rs4633 allelic ratios from cDNA derived from frontal cortex (_n_=15), temporal cortex (_n_=5), and parietal cortex (_n_=3) also showed no significant differences (one-way ANOVA; _F_=0.89, _P_=.42).
SNP rs165599 Is Transcribed in Human Brain and Shows Allelic Differences in Expression
In the study of Shifman and colleagues (2002), SNP rs165599 was the single polymorphism that showed the most significant evidence for association with schizophrenia. This SNP was described as being near the 3′ UTR of COMT, rather than within it, as it is located ∼250 bp downstream from the putative polyadenylation site (Tenhunen et al. 1994). However, certain human EST sequences held in the GenBank database that contain both known COMT cDNA sequence and sequence containing SNP rs165599 (e.g., GI: 24031016; GI: 16174602) suggest that SNP is included in at least some COMT transcripts. Using a forward primer located within known COMT sequence and a reverse that is downstream of SNP rs165599, we were able to amplify and sequence this fragment from cDNA samples, thereby confirming the presence of these transcripts in human brain. Since no product was generated from RNA samples in the absence of reverse transcription, this could not be a result of genomic DNA contamination.
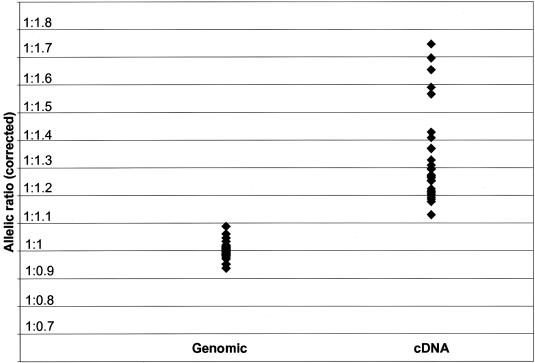
We assayed COMT expression using SNP rs165599 itself as a copy-specific tag, using primers that are both 3′ to the original putative polyadenylation site (Tenhunen et al. 1994), and therefore specific to transcripts containing this polymorphism. A total of 30 individuals heterozygous for SNP rs165599 were assayed (fig. 3). The G allele of SNP rs165599 (the allele that is overrepresented in schizophrenia in the study of Shifman et al. [2002]) showed lower expression relative to the A allele at this locus in all individuals (range 12%–43% relative reduction averaged across independent experiments). The data points from each cDNA sample showed good reproducibility, with an average SD of 0.06. When data from all individuals were combined, this difference in allelic ratio was significant in each individual experiment and in the combined analysis (P<.0001), with the G allele showing an average relative reduction of 24%. Unlike data obtained at SNP rs4633, stratification by other genotypes did not result in differences in allelic expression at this locus (data not shown). As with similar analyses of rs4633 data, no significant difference was observed in allelic ratio at SNP rs165599 between males (_n_=20) and females (_n_=10) (_P_=.76) or between the three tested brain regions of frontal cortex (_n_=20), temporal cortex (_n_=6), and parietal cortex (_n_=4) (one-way ANOVA, _F_=2.29; _P_=.12).
Figure 3.
Comparison between observed genomic ratios and corrected cDNA ratios assayed at SNP rs165599 (_n_=30). Data are expressed as ratio of G:A alleles. The data for genomic DNA are the averages of two measurements for each individual sample. The data for cDNA are the averages of four estimates for each individual sample.
Discussion
Using a powerful method for detecting _cis_-acting influences on gene expression (Yan et al. 2002), we have demonstrated altered allelic expression of COMT transcripts in human brain. Moreover, when we stratified the analysis according to the other loci that Shifman and colleagues (2002) used to convincingly implicate COMT in schizophrenia, we demonstrated clear evidence for increased effects on COMT expression at the rs4633 locus, a proxy for the Val/Met polymorphism. Finally, we have shown that the single SNP giving greatest evidence for association with schizophrenia in the previous study is transcribed in human brain and exhibits significant differences in allelic expression, again with lower relative expression of the associated allele.
Our initial analysis of allelic expression was performed by use of a synonymous coding SNP that occurs in all known COMT transcripts (rs4633). This was selected because it is sufficiently distant from the intron-exon boundaries to allow amplification of the single exon, with primers that could be used to amplify the equivalent sequence from genomic DNA. The use of precisely the same analytic conditions for cDNA and genomic DNA is important to enable cDNA ratios to be corrected for inequalities in allele representation (Hoogendoorn et al. 2000). The average allelic ratio derived from genomic heterozygotes is the ideal control for such inequalities, because it constitutes a perfect 1:1 ratio of the two alleles. The direction and extent of any allelic variation in expression in any individual sample can thus be calculated by comparison with the derived 1:1 ratio.
The majority of studies that have sought to test genetic association with the COMT gene have focused on the nonsynonymous SNP rs165688, which produces a valine→methionine (Val/Met) substitution in the encoded protein. The proximity of this SNP to an intron-exon boundary precluded the use of common genomic primers with which to correct observed cDNA ratios. However, the perfect LD between SNP rs165688 and SNP rs4633 allows data from this alternative SNP to be used as a proxy for the extent and direction of allelic differences in its expression. This is borne out by the significant correlation observed between cDNA ratios at each site, despite the relatively limited range of observed ratios. The present data, therefore, indicate that transcripts that contain the G allele at SNP rs165688, encoding 108/158 valine, are expressed in human brain at a slightly lower level than those encoding the methionine allele.
When cDNA ratios derived from analysis of rs4633 heterozygotes were stratified according to genotypes at two other SNPs in the COMT gene (rs737865 and rs165599) that had previously shown association with schizophrenia (Shifman et al. 2002), significant differences in allele ratios were observed. Despite small group sizes, our data show that some of the variance in allelic expression observed at rs4633/rs165688 can be accounted for by genotypes at these two other loci. In particular, we predicted that the relative differences in expression levels would be greatest in individuals who carried one copy of the schizophrenia risk haplotype that consists of the C allele at rs737865 (which corresponds to the G allele in the study of Shifman et al. [2002] forward sense), the G allele at rs165688 (encoding 108/158 valine) and the G allele at rs165599, and one copy of the T-A-A haplotype that was less common in schizophrenics than control subjects in the study of Shifman et al. (2002). Despite the fact that our phase allocation was imperfect, our data are as predicted by the hypothesis. Thus, the schizophrenia-risk haplotype showed significant underexpression in our subjects and also showed a magnitude of underexpression that is greater than can be attributed to the Val/Met locus alone.
The SNP that showed the strongest association with schizophrenia in the study of Shifman et al. (2002) when tested singularly was rs165599. Although the sequence containing SNP rs165599 is slightly downstream of the predicted COMT polyadenylation site (Tenhunen et al. 1994) and is absent from most (but not all) EST sequences in the GenBank database, we were able to amplify it from brain cDNA that was free from genomic contamination, which confirms that it is expressed in human brain. It is, therefore, likely that an alternative, more 3′ polyadenylation site is utilized in some COMT transcripts, in human brain, at least. When SNP rs165599 was used as a tag for expression of transcripts that contained this 3′ sequence, significant differences in allelic expression were again observed. These differences were more pronounced than were seen for rs4633, which suggests that it is associated with larger changes in the transcripts that contain the longer 3′ UTR. Again, the allele at SNP rs165599 that showed lower relative expression (the G allele) was the allele that was overrepresented in schizophrenic patients in the study of Shifman et al. (2002). Since the range of relative allelic expression was observed in one direction, with all assayed heterozygotes showing a relative reduction of the G allele, these data suggest that it is either SNP rs165599 itself responsible for the observed differences or one that is in very strong LD with it.
Although many variants may potentially result in differential expression of a gene, it is a striking finding that the allelic differences in COMT expression we have demonstrated are associated with the particular variants and haplotypes that others have implicated in schizophrenia. Moreover, the direction of the effect we have seen is consistent with predictions that arise from the increased incidence of schizophrenia observed in VCFS (Murphy et al. 1999), where interstitial deletions typically encompass the COMT gene (Morrow et al. 1995), and with the classic hyperdopaminergic biochemical hypothesis of schizophrenia (van Rossum 1966; Carlsson 1978). That is, we have shown that the implicated haplotype is associated with reduced expression of the COMT gene in the CNS. However, it is more difficult to reconcile with experiments that suggest that a structural change brought about by the valine-encoding allele—shown to be associated with lower expression in the present analysis—is responsible for higher COMT activity (Lotta et al. 1995; Lachman et al. 1996). One explanation might rest on the differences between the forms of COMT that are expressed in brain and those tissues that have traditionally been used to assay COMT function, such as blood and liver. The COMT protein occurs as two distinct forms: a soluble form found in the cell cytoplasm (S-COMT) and a longer, membrane-bound form (MB-COMT). Whereas, in most assayed tissues, the S-COMT form accounts for ∼95% of total COMT activity (Jeffery and Roth 1984; Grossman et al. 1985), it is the MB-COMT form that is more prevalent in brain (Tenhunen et al. 1994). Since studies suggesting functional consequences of the Val/Met polymorphism have focused on the S-COMT form, these results might not be applicable to human brain. Conversely, lower allelic expression seen in brain might not correlate with that seen in other tissues. For example, the present analysis provides strong evidence for association between lower allelic expression and an SNP within intron 1 of MB-COMT (rs737865). It is quite plausible that this polymorphism has no peripheral effect, since S-COMT is transcribed by use of a separate promoter that is 5′ to exon 3 of the MB-COMT gene (Tenhunen et al. 1994) and which is, therefore, 3′ to the intron 1 polymorphism.
The present analysis does not address whether allelic differences in RNA expression translate to similar differences at the protein level. As with allelic differences in overall COMT mRNA levels observed in this study, these differences may well be rather small, although more pronounced differences in specific transcribed forms are possible.
The present study employed a powerful within-subjects technique that detects _cis_-acting influences on gene expression, while controlling for the confounding effects of _trans_-acting factors. Although this allowed us to obtain strong evidence for functional influences beyond those attributable to the Val/Met polymorphism, we cannot distinguish formally between the variants that are functional per se and those that are in LD with the assayed SNPs. Thus, although our results, together with those of Shifman et al. (2002), suggest that future genetic studies that investigate association with the COMT gene should take account of polymorphisms other than the Val/Met polymorphism, more detailed functional analyses of other SNPs in and around the COMT gene will be required to identify the specific risk variants in schizophrenia.
Acknowledgments
We are grateful to the Department of Clinical Neuroscience at the Karolinska Institute (Stockholm), the Stanley Medical Research Institute (Bethesda), and the MRC London Neurodegenerative Diseases Brain Bank (U.K.) for donating brain tissue. This work was funded by the Medical Research Council (U.K.).
Electronic-Database Information
URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
- GOLD, http://www.sph.umich.edu/csg/abecasis/GOLD/ (for graphical overview of linkage disequilibrium) [DOI] [PubMed]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia, COMT, and VCFS)
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for primer design)
References
- Axelrod J, Tomchick R (1958) Enzymatic O-methylation of epinephrine and other catechols. J Biol Chem 233:702–705 [PubMed] [Google Scholar]
- Carlsson A (1978) Mechanism of action of neuroleptic drugs. In: Lipton MA, DiMascio A. Killam KF (eds) Psychopharmacology: a generation of progress. Raven Press, New York, pp 1057–1070 [Google Scholar]
- Cowles CR, Joel NH, Altshuler D, Lander ES (2002) Detection of regulatory variation in mouse genes. Nat Genet 32:432–437 [DOI] [PubMed] [Google Scholar]
- Daniels JK, Williams NM, Williams J, Jones LA, Cardno AG, Murphy KC, Spurlock G, Riley B, Scambler P, Asherson P, McGuffin P, Owen MJ (1996) No evidence for allelic association between schizophrenia and a polymorphism determining high or low catechol O-methyltransferase activity. Am J Psychiatry 153:268–270 [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 98:6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman MH, Creveling CR, Rybczynski R, Braverman M, Isersky C, Breakefield XO (1985) Soluble and particulate forms of rat catechol-O-methyltransferase distinguished by gel electrophoresis and immune fixation. J Neurochem 44:421–432 [DOI] [PubMed] [Google Scholar]
- Grossman MH, Emanuel BS, Budarf ML (1992) Chromosomal mapping of the human catechol-O-methyltransferase gene to 22q11.1-q11.2. Genomics 12:822–825 [DOI] [PubMed] [Google Scholar]
- Hoogendoorn B, Norton N, Kirov G, Williams N, Hamshere ML, Spurlock G, Austin J, Stephens MK, Buckland PR, Owen MJ, O’Donovan MC (2000) Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum Genet 107:488–493 [DOI] [PubMed] [Google Scholar]
- Jeffery DR, Roth JA (1984) Characterization of membrane-bound and soluble catechol-O-methyltransferase from human frontal cortex. J Neurochem 42:826–832 [DOI] [PubMed] [Google Scholar]
- Kunugi H, Vallada HP, Sham PC, Hoda F, Arranz MJ, Li T, Nanko S, Murray RM, McGuffin P, Owen M, Gill M, Collier DA (1997) Catechol-O-methyltransferase polymorphisms and schizophrenia: a transmission disequilibrium study in multiply affected families. Psychiatr Genet 7:97–101 [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM (1996) Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 6:243–250 [DOI] [PubMed] [Google Scholar]
- Li T, Sham PC, Vallada H, Xie T, Tang X, Murray RM, Liu X, Collier DA (1996) Preferential transmission of the high activity allele of COMT in schizophrenia. Psychiatr Genet 6:131–133 [DOI] [PubMed] [Google Scholar]
- Liou YJ, Tsai SJ, Hong CJ, Wang YC, Lai IC (2001) Association analysis of a functional catechol-O-methyltransferase gene polymorphism in schizophrenic patients in Taiwan. Neuropsychobiology 43:11–14 [DOI] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J (1995) Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry 34:4202–4210 [DOI] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, Collins J, Dunham I, ODonnell H, Scambler P, Shprintzen R, Kucherlapati R (1995) Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet 56:1391–1403 [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ (1999) High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiat 56:940–945 [DOI] [PubMed] [Google Scholar]
- Norton N, Williams NM, Williams HJ, Spurlock G, Kirov G, Morris DW, Hoogendoorn B, Owen MJ, O’Donovan MC (2002) Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum Genet 110:471–478 [DOI] [PubMed] [Google Scholar]
- Ohmori O, Shinkai T, Kojima H, Terao T, Suzuki T, Mita T, Abe K (1998) Association study of a functional catechol-O-methyltransferase gene polymorphism in Japanese schizophrenics. Neurosci Lett 243:109–112 [DOI] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Gottesman II (2002) Schizophrenia. In: McGuffin P, Owen MJ, Gottesman II (eds) Psychiatric genetics & genomics. Oxford University Press, Oxford, UK, pp 247–266 [Google Scholar]
- Pulver AE (2000) Search for schizophrenia susceptibility genes. Biol Psychiat 47:221–230 [DOI] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisanté-Shalom A, Lev-Lehman E, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Beckmann JS, Yakir B, Risch N, Zak NB, Darvasi A (2002) A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet 71:1296–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J, Chapman V, LeBon JM, Riggs AD (1992) Parental imprinting studied by allele-specific primer extension after PCR: paternal X chromosome-linked genes are transcribed prior to preferential paternal X chromosome inactivation. Proc Natl Acad Sci USA 89:10469–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhunen J, Salminen M, Lundstrom K, Kiviluoto T, Savolainen R, Ulmanen I (1994) Genomic organization of the human catechol O-methyltransferase gene and its expression from two distinct promoters. Eur J Biochem 223:1049–1059 [DOI] [PubMed] [Google Scholar]
- van Rossum JM (1966) The significance of dopamine-receptor blockade for the mechanism of action of neuroleptic drugs. Arch Int Pharmacodyn Ther 160:492–494 [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297:1143 [DOI] [PubMed] [Google Scholar]
- Zhao H, Curtis D, Sham PC (2000) Model-free analysis and permutation tests for allelic associations. Hum Hered 50:133–139 [DOI] [PubMed] [Google Scholar]