Matching Strategies for Genetic Association Studies in Structured Populations (original) (raw)
Abstract
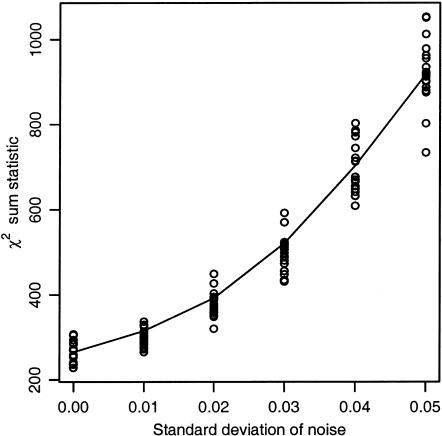
Association studies in populations that are genetically heterogeneous can yield large numbers of spurious associations if population subgroups are unequally represented among cases and controls. This problem is particularly acute for studies involving pooled genotyping of very large numbers of single-nucleotide–polymorphism (SNP) markers, because most methods for analysis of association in structured populations require individual genotyping data. In this study, we present several strategies for matching case and control pools to have similar genetic compositions, based on ancestry information inferred from genotype data for ∼300 SNPs tiled on an oligonucleotide-based genotyping array. We also discuss methods for measuring the impact of population stratification on an association study. Results for an admixed population and a phenotype strongly confounded with ancestry show that these simple matching strategies can effectively mitigate the impact of population stratification.
Introduction
Genomewide association studies provide a powerful approach to implicate DNA variants (and, by extension, the genomic regions they represent) in the predisposition to complex diseases and in the genetic underpinnings of drug efficacy and adverse reactions. The success of these studies relies on the accurate measurement or estimation of allele-frequency differences between case and control subjects. When searching for small genetic effects in large association studies, systematic differences in ancestry between the cases and controls are likely to produce many statistically significant but spurious associations (e.g., Knowler et al. 1988; Lander and Schork 1994). Such differences are expected to be found when genetically distinct population subgroups have a different prevalence of the target phenotype.
The use of family-based association study designs mitigates the impact of systematic ancestry differences (population stratification) but can lead to an increased burden in the recruitment of subjects and in genotyping (Cardon and Palmer 2003). Self-reported ancestry is also useful in matching case and control subjects to reduce the prevalence of spurious associations. Population structure can be empirically determined by individually genotyping all potential cases and controls across a set of unlinked marker loci (Pritchard and Rosenberg 1999). When individual genotypes are known, analysis methods can correct the association test statistic for unmatched groups by use of the inferred population structure (Pritchard et al. 2000_b_; Reich and Goldstein 2001; Satten et al. 2001; Thornsberry et al. 2001; Hoggart et al. 2003).
In association studies using DNA pooled from many individuals, significant causal disease (or pharmacogenetic) associations would be indistinguishable from associations due to ancestry differences between cases and controls. Thus, genetic-ancestry matching prior to DNA pooling is essential. By use of inferred population-structure data, DNA pools can be constructed that are matched to have similar genetic composition, to minimize the likelihood of spurious associations due to population stratification. Allele-frequency estimates in the matched DNA pools should then give a more reliable indication of causal disease association. See the work of Sham et al. (2002) for a recent review of DNA pooling methodologies and implications for association studies.
In genomewide association studies, it is necessary to test at least hundreds of thousands of SNP markers because of the generally limited extent of linkage disequilibrium in the human genome (Risch and Merikangas 1996; Kruglyak 1999; Risch 2000; Patil et al. 2001). We are currently testing >1.5 million SNP markers in association studies, using pooled genotyping with multiple measurements of allele frequency in each of two pools as an efficient screen to enrich for SNPs with significant allele-frequency differences. The SNPs with the greatest apparent allele-frequency differences in the pooled data are then selected for individual genotyping. The pooled genotyping step reduces the number of SNPs that must be individually genotyped to confirm allele-frequency differences between case and control groups. In this context, spurious associations due to population structure force us either to examine more SNPs by individual genotyping or, if that is impractical, to sacrifice power to detect causal associations.
In this study, we describe the use of unlinked SNP markers to detect and correct for population stratification in case and control subjects in an admixed population prior to pooled genotyping for association testing. Using a phenotype that is strongly confounded with ancestry, we show that several strategies for matching case and control groups are successful at eliminating significant stratification. We also discuss methods for measuring the impact of stratification on a pooled genotyping experiment.
Methods
Subject Collection
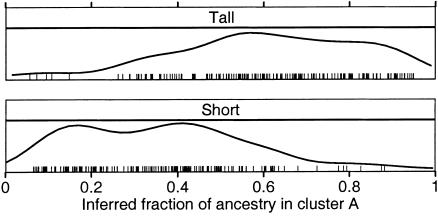
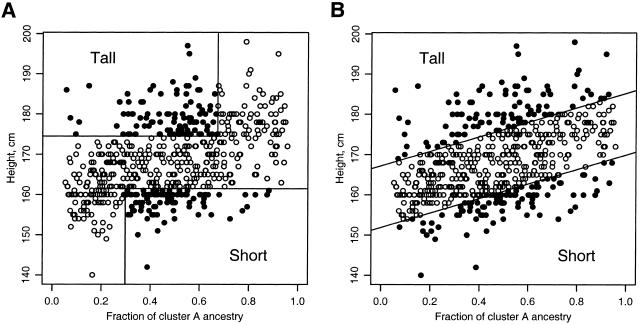
Subjects were chronic alcoholics, some with alcoholic liver disease, recruited in Mexico City under full informed consent. The international institutional review board of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (INCMNSZ), which is registered with the Office of Human Research Protection, approved the human patient sample-collection protocol. Subjects were measured for height in cm at the time of blood-sample collection. The three self-reported ethnicities in this population were “Caucasian,” those of primarily Spanish European ancestry; “Otomi” Indians, from the Pachuca region in Mexico; and “Mestizo,” a mix of Spanish European and Mexican Indian ancestry. A total of 824 Mestizo males were examined to determine the distribution of height. The definitions of “tall” and “short” were chosen to include the upper and lower 25% of the observed distribution. This yielded a minimum height of 174 cm for the “tall” group and a maximum height of 162 cm for the “short” group.
SNP Selection
From a genomewide collection of SNPs discovered by Perlegen Sciences in a globally diverse panel of individuals (Patil et al. 2001), we selected a set of 312 that were roughly equally spaced across the autosomes and were expected to behave well in oligonucleotide array–based genotyping. SNPs were selected to be at least 150 bp from the nearest common repetitive element, as identified by the RepeatMasker 2 program (available on the RepeatMasker Web site), and the 25-bp sequence containing the SNP (± 12 bases of context) was required to be unique in the human genome, according to then-current National Center for Biotechnology Information (NCBI) Build 29 (available on the NCBI Web site). We also required that in Perlegen’s previously collected SNP discovery data, the SNPs have a high rate of high-confidence genotype calls and an allele frequency close to 0.5. A combination of these quality metrics was used to numerically score each candidate SNP. We then selected the highest-scoring candidates from a series of 2-Mb windows spaced at 9-Mb intervals across each NCBI Build 29 chromosome.
Primer Design
PCR primer pairs for each SNP were selected using the program Oligo, version 6.57 (Molecular Biology Insights). We selected primers having a Tm of 59°C–66°C, a length of 18–22 bases, a PCR product size of 50–200 bases, and 3′-end ΔG of between −5.5 and −9.8 kcal/mol. We also required that each primer be at least 5 bases from its target SNP. Primer sequences containing repetitive sequences, as determined by the RepeatMasker 2 program, were excluded. Only primer sequences determined to be unique (P<10-4) in the genome (NCBI Build 29) by use of the BLAST program (available on the NCBI BLAST Web site) (Altschul et al. 1990) were selected.
Genotyping Oligonucleotide Array Design
Genotyping arrays of 25-bp oligonucleotides were designed as four sets of 20 features (80 features per SNP), corresponding to forward and reverse strand tilings for sequences complementary to each of two SNP alleles. A set of 20 features consisted of five sets of 4 features where the location of the SNP within the oligonucleotide varied from position 11 to position 15. A set of 4 features consisted of sequences where A, C, T, or G was substituted at position 13. Thus, each set of four features provided one perfect match to the sequence of the corresponding SNP allele and three features with a single-base mismatch for that allele. Mismatch probes were used to measure background and, by comparison with the signal for the perfect match probes, to detect the presence or absence of a specific PCR product in a sample. Light-directed chemical synthesis of the appropriate oligonucleotides was carried out by Affymetrix (Fodor et al. 1991).
Hybridization Sample Preparation
For analysis of the 312 stratification SNPs, DNA was amplified by PCR in 12-μl volume containing 13 primer pairs at 0.4 mM of each primer, 10 ng of individual genomic DNA, 2 U Titanium Taq (Clontech), 0.5 mM deoxynucleotide triphosphates, 10 mM Tris-HCl (pH 9.1), 3 mM MgCl2, and additives. Thermocycling was performed on a 9700 cycler (Perkin-Elmer), with initial denaturation at 96°C for 5 min, followed by 10 cycles of 96°C for 30 s, 58°C minus 0.5°C/cycle for 30 s, 65°C for 1 min, then 40 cycles of 96°C for 10 s, 53°C for 30 s, and 65°C for 60 s, and, finally, an extension at 65°C for 7 min. PCR products were pooled together and labeled with 0.7 μM biotin-16-ddUTP/dUTP (Roche) with 25 units of terminal deoxynucleotidyl transferase (Roche), by incubating at 37°C for 90 min, after which the reaction was stopped by heat-inactivation at 99°C for 10 min.
Hybridization of Samples to High-Density Oligonucleotide Arrays
Labeled DNA samples were incubated in hybridization buffer (3 M tetramethylammonium chloride, 10 mM Tris-HCl [pH 7.8], 0.01% Triton X-100, 100 μg/ml herring sperm DNA, and 50 pM control oligomer) at 99°C for 10 min and hybridized to a chip overnight at 50°C on a rotisserie at 25 rpm. Chips were washed twice in 1 × MES buffer (0.1 M 2-[N-morpholine]ethane sulfonic acid [pH 6.7], 1 M NaCl, and 0.01% Triton X-100), and incubated with 5 μg/ml streptavidin (Sigma-Aldrich) and 2.5 mg/ml acetylated bovine serum albumin (Sigma-Aldrich) in 1 × MES for 15 min on a rotisserie at room temperature (RT). After two washes with 1 × MES at 35°C, chips were incubated with antibody solution (1.25 μg/ml biotinylated antistreptavidin antibody [Vector Laboratories] and 2.5 mg/ml BSA in 1 × MES) for 15 min on a rotisserie at RT, followed by another two washes with 1 × MES at 35°C. Then, chips were stained with 1 μg/ml streptavidin-Cy-chrome conjugate (Molecular Probes) and 2.5 mg/ml BSA for 15 min on a rotisserie at RT, followed by two washes with 1 × MES at 35°C. Chips were incubated for 30 min at 37°C in 0.2 × SSPET (30 mM NaCl, 2 mM NaH2 PO4, 0.2 mM EDTA [pH 7.4], 0.01% Triton X-100), followed by a wash with 1 × MES at RT. Hybridization of the labeled sample to the chip was detected using a confocal laser scanner (Perlegen) (Patil et al. 2001).
SNP Genotyping
For each SNP, we measured ratios of the mean intensity of perfect-match features for one allele to the sum of mean intensities for both alleles. In principle, these ratios should take on values near 1.0, 0.5, or 0.0 for AA, AB, or BB genotypes. We discarded data if, for both alleles, <9 out of 10 perfect-match features were brighter than their corresponding mismatch features. We used an expectation-maximization algorithm and a normal mixture model to assign intensity ratios to clusters.
For the stratification analyses, we only used data for SNPs that showed consistently good genotyping results (table 1). We excluded SNPs that had a pass rate of <80% on the basis of the perfect-match/mismatch comparison. We also excluded SNPs for which fewer than three genotype clusters could be identified, as well as those that had >20 ambiguous cluster assignments. Many SNPs showed moderate departures from Hardy-Weinberg equilibrium, which would be expected in a heterogeneous population. We excluded only those SNPs showing extreme deviations that could be traced back to convergence failures of the clustering algorithm. For the 275 SNPs passing these criteria, the overall call rate was 98.4%. In a set of 24 individuals genotyped in triplicate for these SNPs, we had a concordance of 99.8%. The 275 SNPs and all individual genotype data used in this study have been submitted to dbSNP (ss12673803–ss12674077) (available on the dbSNP Web site). SNP positions in NCBI Build 33 are also shown in table A (online only).
Table 1.
Quality-Control Checks for SNP Genotyping Results
| Data-Quality-Filter Criterion | No. of SNPs Passing | % Passing |
|---|---|---|
| Pass rate >80% | 309 | 99% |
| Three genotype clusters identified | 308 | 99% |
| <20 ambiguous calls | 305 | 98% |
| _P_>.00001 for Hardy-Weinberg equilibrium | 303 | 97% |
| Maximum cluster width | 282 | 90% |
| All criteria | 275 | 88% |
Table A.
SNP Positions, Alleles, and Flanking Sequences
| dbSNP ID | Chromosome | Accession No. | Position | Reference Allele | Alternate Allele | Flanking 51-bp Sequence (−25 to +25) |
|---|---|---|---|---|---|---|
| ss12673885 | 1 | NC_000001.4 | 9936378 | C | T | AATAGAATAAAAGGGCCTAGAGTTACGGCATTTCACTTTGTAAAGGTTGCT |
| ss12673848 | 1 | NC_000001.4 | 16347653 | G | A | TATGGCTGACCAGGGGCATCTTTACGCATTGAACTCTCAGGTCACAAGTAT |
| ss12673890 | 1 | NC_000001.4 | 31426127 | A | G | TGACCATCTTGGCCAATTGCTCATCAAGTCCATGAAAGAAGATTGAATTTA |
| ss12673851 | 1 | NC_000001.4 | 39299078 | T | C | TATTCCTGAGTTTGGTACGCTTCAATGTTATATGCGGTTGTTCAGCCAAAC |
| ss12673850 | 1 | NC_000001.4 | 48093199 | G | A | CCCATTCCATGGACAGTAAAATTAAGGTTTGAGATCTCTAGGTATTCCCAC |
| ss12673891 | 1 | NC_000001.4 | 54476998 | G | C | CAAGAAACACCTCCAAGCTGAACCTGAATGTAGCTCCAGATTCTCATGGGA |
| ss12673892 | 1 | NC_000001.4 | 71079585 | G | A | CTCACTGTTCATGTTAACCGTCTGTGCTCTCTACACGGAGTGGAGCCCGTG |
| ss12673887 | 1 | NC_000001.4 | 77499794 | A | T | AAAGAGCCAACATTATATCCAACCAACTCTTGGCTCTAGACAATGAAGGTA |
| ss12673893 | 1 | NC_000001.4 | 85573463 | C | A | GCTGTTAAAGTTTCGTAGGCCGTAACCTGATGCCATGAGGCTCTGATATGT |
| ss12673894 | 1 | NC_000001.4 | 93335361 | G | C | GCTTCGTTCCCCTCTTACTTTTCACGTTACTACTTGCAATTCTCTAGCTCA |
| ss12673895 | 1 | NC_000001.4 | 100468314 | G | T | TTTGTGTTGCAAATGAGTTATAGAGGTGAATCCATGTGGGGCCAGAAAGTA |
| ss12673896 | 1 | NC_000001.4 | 109070518 | A | C | TGAGAAATCTCTTGAAAACCATTCTAAATTCCAGTTCCTATAAAATCAGAC |
| ss12673888 | 1 | NC_000001.4 | 116699736 | G | A | GCAACAGACCTGGCTAGGAGCTACAGATAGTTCCAACCAACCAGCTAGAAA |
| ss12673897 | 1 | NC_000001.4 | 149656841 | C | G | ACACACAGCATTCCAATGGGAGATTCAGGCCTAGAGCATGTCCTGTGGCTC |
| ss12673898 | 1 | NC_000001.4 | 157108893 | A | G | GCCACTGCTCTCAAAGGTACATATTAGGATGAGATACTGTTTACCCAGAGT |
| ss12673847 | 1 | NC_000001.4 | 173882748 | C | T | ACGGTCTGTATTGACTGGCTGCGCACGGACAAGTGTCATCTTGCCACACCT |
| ss12673899 | 1 | NC_000001.4 | 180522150 | C | T | ATAAAATATGACATCATTCTTACCACTGGTGCTGAAAAATGTCACTGATAC |
| ss12673900 | 1 | NC_000001.4 | 189969066 | A | G | AGAAAACCATGTAATTTTTCAGCTTAATTTAACATTGTATCTAGGGCAAGC |
| ss12673901 | 1 | NC_000001.4 | 197042249 | A | G | CAAAGTTAGGAAGGCCAATGAGAATACAATAAAGATTATGGGAGAGTAACA |
| ss12673902 | 1 | NC_000001.4 | 214750406 | G | A | ATAACCTACCATGTTTACCAGGTCCGCCAAGACTATGAAACAAATATATGT |
| ss12673903 | 1 | NC_000001.4 | 221802714 | G | A | GTGTAAGTTGTAATCACACAAAGGCGTAATCATGAAAAGTAGTAAAACATA |
| ss12673849 | 1 | NC_000001.4 | 229272493 | C | T | TTTACACATCAGTGCATTTTTGTATCTAGCACAATTCCTGGCATGTGGCTG |
| ss12673886 | 1 | NT_077961.1 | 186534 | T | C | AACTTGAGACATGTAATTGGGGTTTTGTAACTAGCTACTCACTATATAGCT |
| ss12673884 | 1 | NT_077984.1 | 2495 | G | C | ACAAGCTGATAATTACCATCATTTTGGAATTGTTCAGAACCATGAAGAATA |
| ss12673904 | 2 | NC_000002.5 | 9639015 | G | A | TTTGACACAAGGCAATAACTTCCGCGTAATGAGTACCCAAGAAAGTAGAAC |
| ss12673905 | 2 | NC_000002.5 | 17853426 | C | G | ACAGCAGAGTCAACTGGCTTCAGAACTGATCTTTTCCTCACTAATCCAAGT |
| ss12673906 | 2 | NC_000002.5 | 26279750 | A | G | AACCTTTGAATTTGTATTTGTCTGAATCATAGAATTTAGAATTATAATGGC |
| ss12673907 | 2 | NC_000002.5 | 35537401 | A | G | GCGAATAAAATAACAAGTCACATCAAGAAGTTGTGGCCTGATTTAAAACAA |
| ss12673854 | 2 | NC_000002.5 | 41971450 | A | T | TTCTGAACATACACCCAGGAATGTAATATTGTTCCGTTTTTGCAGAAGCTA |
| ss12673852 | 2 | NC_000002.5 | 50417805 | T | G | TTTAAGATTTGAAGCTTCTCAATAATATGGCTGCTTATATCAAGTTCTATA |
| ss12673908 | 2 | NC_000002.5 | 76828447 | A | G | ATCATGCAATTCAGGCAGGGAACCAATCTTTAGAAACTATACCCAGTTTAG |
| ss12673909 | 2 | NC_000002.5 | 86713991 | G | A | TGAGTTTTTCCTATTCAAGGAACCCGTGTTGATAATAACAGCAACCCCGGC |
| ss12673910 | 2 | NC_000002.5 | 113628511 | A | G | CAATAGCAACGTTTTTGAATCAGAGAAGTGATTTTGAACACACTGTACATA |
| ss12673911 | 2 | NC_000002.5 | 123753048 | A | C | TCTTTCCTCATTGTCTGCGATCTGGAAATAGAGCTTTCAGTTCTCATCACT |
| ss12673912 | 2 | NC_000002.5 | 130675905 | A | G | TAGAATCCATTCATTATTATGTGTGACTGGAGAGGTATATTGCTTAAAAAC |
| ss12673913 | 2 | NC_000002.5 | 140961080 | G | A | TGGCTCTTGACTCAGTAATCACTTTATGTCAAAATGTTTCCTAGTCTCCTT |
| ss12673914 | 2 | NC_000002.5 | 157001034 | T | C | ATCATCCCATTTCACATGAAGAAATTTGCATCTAGTGAGGTTCACTAACTT |
| ss12673915 | 2 | NC_000002.5 | 166705382 | G | A | AGGATGCTTCCTAAATTTCAGCAGAGGGATTATGATGCATTTATAAAGAAA |
| ss12673916 | 2 | NC_000002.5 | 175050139 | C | T | TACCATCATCCTAAATTGCTCCAGCCTGGAAAATTTTAAGTCAAATATCCC |
| ss12673917 | 2 | NC_000002.5 | 183322843 | A | T | AATGTACATTCAAAACAATCGAGGTAGGCTTTAAAGGAGCATTCAAAATCA |
| ss12673918 | 2 | NC_000002.5 | 191081229 | G | A | ATCAATATCACAAAAGACTTGCTATGAACTGTGCTAACTTGGGTATTTTTC |
| ss12673919 | 2 | NC_000002.5 | 199615627 | A | G | ACATGGCTAGCACATTGCTTGGCACATTCTCAGAGGTGAATAGGTCATTTT |
| ss12673853 | 2 | NC_000002.5 | 207935369 | C | T | CAACAGCCTTTCTCCATGAAGTTCCCCTGCAAGAAGCGTGAATCATACATG |
| ss12673920 | 2 | NC_000002.5 | 216170075 | A | G | TAGATTCCATGGTACCATGTTGAGAATTGTGCCTAGCTACTGAGAGTCTTT |
| ss12673921 | 2 | NC_000002.5 | 225720515 | C | G | TTTTAAAAATTATGCAGACCAGAGACTGTCAATTTAAGTCAGATCTGGGGC |
| ss12673922 | 2 | NC_000002.5 | 233086431 | G | A | TTCAGCCAGAGATACATCTTAATGAAGTGCTGACATTTTTCAGAGGATAAA |
| ss12674008 | 3 | NC_000003.5 | 3699847 | C | T | GAATCGACAAAGCTCTCCTGGAAAACGGCCATCTCATGGAAGTGATGGCCT |
| ss12674009 | 3 | NC_000003.5 | 8874886 | T | C | TTGTTTCCTTTAGATAACATCCTGCTGGTTACTGATCATACCTGTTGATGA |
| ss12674010 | 3 | NC_000003.5 | 13413791 | G | A | TATCCCTCAATAGCCCTGGGAACACGCTACTGAGAGCCACATTTTGGGGAT |
| ss12673856 | 3 | NC_000003.5 | 21334999 | G | C | TATACTTTTCATCAAGTGACAAGTTGTTCCCCATAGTAGCCTGCATGAAAC |
| ss12674011 | 3 | NC_000003.5 | 30025863 | T | C | CCTACCTGTGATGAACTTACTGGAATGGGAACTTTTCACTTTACAATTAGC |
| ss12674012 | 3 | NC_000003.5 | 40271257 | T | C | ATTAGGCAGACTTGATACCCTTATATGGCAGAACTTTAGAGCAACCACATT |
| ss12673855 | 3 | NC_000003.5 | 47208180 | T | A | AGTGTGGTTTTGCCTGTTGGGAAACTCTTCAGTCACACTTTTCCAAAAGTC |
| ss12674013 | 3 | NC_000003.5 | 56092272 | G | A | TTTGGCTTAAAAGGGGTACAATTAGGTCTTACTCATGCTGATTAAGGCAAA |
| ss12674014 | 3 | NC_000003.5 | 70589259 | C | T | CCGTTTGGGTTCAGCCTTACAGAGCCGTGATTTTGGCTACATCCTTTAGAA |
| ss12673857 | 3 | NC_000003.5 | 84902112 | A | G | ACGGTATAGTGCAAATCCTGACGGTAGGTTCTACAATTATGCTAATAGATT |
| ss12674015 | 3 | NC_000003.5 | 99066776 | T | C | GTGACCACTGACTTTTCAAGAGGTGTGAGGACAAGGCCAGATGACCATAGA |
| ss12674016 | 3 | NC_000003.5 | 106351294 | A | G | TCCAGGGAAAATACATTCCTGGCTGATCGTAGACAAGGGATATTGCCTGAA |
| ss12674017 | 3 | NC_000003.5 | 114254346 | A | G | GTCATTTAGACATGAATCAGAAGGCAAAATGTTTGGGCCTGACTAGAAAGA |
| ss12674018 | 3 | NC_000003.5 | 122316930 | G | C | TCCTTTCTCTGCCCATTTTTTCACTGTTGTTCCACGTTACTTTTCTTAATG |
| ss12674029 | 3 | NC_000003.5 | 136194178 | A | G | GTGGCTACAGATTAAAGGGTCAGTCATTGAAACATTGCTGGGATCACTGCT |
| ss12674030 | 3 | NC_000003.5 | 144332113 | T | C | GAACTCCTACTATGCGTTGGTTACTTGCCGGGCACTGGGAAACAAAGATGA |
| ss12674033 | 3 | NC_000003.5 | 159534573 | G | A | GAGAATTATTTCATGACTTTAGAGCGTAGTATTTTAGCATTTCAGCAGTAG |
| ss12674034 | 3 | NC_000003.5 | 166984897 | C | T | AAGTACAAAATTGTAGGTGGCAAATCGATACCCTGAGGGCTGCATATTTTA |
| ss12674035 | 3 | NC_000003.5 | 174673711 | C | A | ATATGCAGGGCACTGATTTTTGGATCGGAAAGGAGACTAAATTTTCCCCCT |
| ss12674036 | 3 | NC_000003.5 | 182314045 | G | C | AAGGATGGGTAGAGCGAGGACCTATCGGATGTGTAGGCAAAGCTGGTGGCC |
| ss12674037 | 3 | NC_000003.5 | 190743817 | G | A | TCACAGATAAATTCATGGCGCTTTAGCACAAGTATGGAACTTCACATTATA |
| ss12674052 | 4 | NC_000004.5 | 733928 | T | C | CATCCACTCCTCTTTCTAGTCTCCTTCCCGGGCGGTCAGGGAGTGTTGTCT |
| ss12674053 | 4 | NC_000004.5 | 8720247 | G | C | GTCTCCGGGTTGTTCTGAAACCATCGCAGGAATGATGCACACTGTGGCTGA |
| ss12673846 | 4 | NC_000004.5 | 17350708 | A | C | AACTTTTGAACCAGCTGAGAATTTCAAGTGTGATCGGTTTAGATTGGGATG |
| ss12673845 | 4 | NC_000004.5 | 24834655 | G | A | GACCATACTAGATGTCTCCCTCCCAGTGCAAGAAACCAACAGGCAGAAAAC |
| ss12674061 | 4 | NC_000004.5 | 33801012 | C | T | TTTAATCGGTTTTTATGTAAAAGCACGTGTCATTATGAACAAGGAATACAG |
| ss12674059 | 4 | NC_000004.5 | 41840213 | T | C | TTTTGAGAAGACATCACATTTTCTTCCAAAGGCAAGTACCCATTGACTTAG |
| ss12674060 | 4 | NC_000004.5 | 48828164 | T | G | CCCTGGATGGTGTTTACTGAACAAATTATCATCTCAGGCAGTATAACAATT |
| ss12674062 | 4 | NC_000004.5 | 58397869 | C | T | ACTGCAATGAGATTGAATAATGCCTCTAGGGTCTGAATAATACGTGTTGTC |
| ss12674065 | 4 | NC_000004.5 | 66672503 | T | C | AGGTAGGAGAGACCACTTTACGGCTTTCACCATTCACATTCCTCTTGGAGT |
| ss12674066 | 4 | NC_000004.5 | 77117873 | A | G | TTAATGGAGCCTTCTTTGAAATCTTGTAGTTCTGCTGCATGTTAATTGGCC |
| ss12674057 | 4 | NC_000004.5 | 85230848 | T | C | CCATTAAGACAGGTTCCTCACTGTATTGCTTATTCTCTTTCCAGTCTTTTT |
| ss12674058 | 4 | NC_000004.5 | 92088698 | T | C | CATGCTTGGTGATGAGCCTGGCATATGAAACAATTTCCTACAGTAATAAAA |
| ss12674054 | 4 | NC_000004.5 | 100188103 | G | A | AAGCTGCATTATAGAACACAATAACGAAACAGATGATGGTCTGTATACATG |
| ss12674055 | 4 | NC_000004.5 | 108912304 | A | G | AAGAAAATGCATTTGCAGGTTTACAAGAAGCAACCCAGAAATTCAATGGTA |
| ss12674056 | 4 | NC_000004.5 | 115777312 | C | T | GTATTAGGACTTTCACTGATACCGACATTTACTTTTAGGATTTCTAGGATT |
| ss12674067 | 4 | NC_000004.5 | 124057410 | G | A | ATTAAAACAGTATTAGATAGCATGCGGCTTCAAGAAGACAGCTCAGAAGAA |
| ss12674068 | 4 | NC_000004.5 | 132889303 | G | A | GCACCTAGCATGCAATAAGCCGCCAATGAACATCGCTGAGTTCTCAATTAT |
| ss12674073 | 4 | NC_000004.5 | 141354227 | G | A | CCTCCTGACTTCCCACTAATCAAGTGCACTAGCCCTCCTGGATATAAGCTG |
| ss12674075 | 4 | NC_000004.5 | 149160357 | C | T | GACGGAACTCCTGCATTCCAGAGTTCAGTGGTAAAAAGAACACTTTATGTA |
| ss12674063 | 4 | NC_000004.5 | 158093442 | A | G | AAATTATAGCCTGTTCAAATCATCCACAGACTTGTTCAAGAATAGGTAAAG |
| ss12674064 | 4 | NC_000004.5 | 166155853 | T | C | AGGAACTCACCACACATGAGCTATGTGTCCAGAAAACATTAACAGCTGCCT |
| ss12674076 | 4 | NC_000004.5 | 175609800 | T | A | GTGTTTGTTCATTCTGGGCAAATCATCCATTAACACTCAGAAGACTTTTAA |
| ss12674077 | 4 | NC_000004.5 | 182884117 | A | T | ACTGATGTTGTCTGAACCATGATTGAGGTCCTCCAAAAGTTATTCCCTTTA |
| ss12674027 | 5 | NC_000005.4 | 2026753 | C | T | CTACTCCAGCACCATTACCTCCTTACGCAAATGACCACAACTGAATTAACT |
| ss12673871 | 5 | NC_000005.4 | 7856314 | G | A | CCAGCCAGAACAAGAATCGTGGGGCGTTTTGGCAGCCTAGTCTAATCATTT |
| ss12673868 | 5 | NC_000005.4 | 16716847 | T | C | TAAAGTAGGGTGCACACTACAGTAATACCTCTAGCCCACAACTCCCAATCA |
| ss12673869 | 5 | NC_000005.4 | 30043011 | C | A | CATTGTAAAATATTCAATGCAGGAGCATAATCATGTTTGCCTTCTGATCTG |
| ss12674028 | 5 | NC_000005.4 | 41185313 | C | A | GATTGGTTGTTTTGGAACTGGAAAGCAACTTCCATCAGAACAATATTACAT |
| ss12673803 | 5 | NC_000005.4 | 50605403 | C | T | AGTGTTGAAAATGTGTCAGGCACTTCAGGGCATTGACCTATTAATCCTCCC |
| ss12673870 | 5 | NC_000005.4 | 59427503 | C | T | GACACTGAGAGAAGGGCCGCAATAACTCTCATGGCTCCTTTCCACTTTCTA |
| ss12674044 | 5 | NC_000005.4 | 67386238 | C | T | CTGCCTTGTTTACTGTGCTTTCTGACGCCTACCTCCAAGGTCAAAGGGGGA |
| ss12674045 | 5 | NC_000005.4 | 74960399 | G | A | CCTGGTTCTCTTCAGGATGGAATGAGCGCCCTCCACTTTGCCACTCAGAGC |
| ss12674041 | 5 | NC_000005.4 | 82273510 | G | A | ATTTGCTCTGGTAGAAATCATGTCCGCCCTTTTCCAGTTATCTTAGGTGAG |
| ss12674042 | 5 | NC_000005.4 | 91333028 | C | T | TGCTTGCATGAAATACCTATTCGCCCTCCATGAAACAGTTTGGAAAATGTT |
| ss12674046 | 5 | NC_000005.4 | 100229818 | T | C | AAAAGGTTGATTCTGAGATTTCCTATGATGGAAGTGTGGGAAAAAGTTAGG |
| ss12674047 | 5 | NC_000005.4 | 108102517 | T | C | CAGCTTATCTGGCCTGTTTCCTTTCTGGTTGGTTCGTTAGCTCCAGTCATG |
| ss12674072 | 5 | NC_000005.4 | 115453797 | T | C | AGGCTCTTCCCTGCGCAGTACAAGGTTTCCCTAGTAGTTTGGTTTGCCCAA |
| ss12674048 | 5 | NC_000005.4 | 130864809 | C | T | TTCAGTTTTTGCCTTTTAAGGCTCTCGTCCACTTTTAGCATGCTATTCTGT |
| ss12673867 | 5 | NC_000005.4 | 139501013 | C | G | TAAGCTGGCTGGTAAAGCCTCCACGGGACAACAGTTTCACTTTGCTTCGGG |
| ss12674049 | 5 | NC_000005.4 | 147071503 | A | G | TCGACAGTTATGAGAGACCTCAAAGATTCACAACATGAAGCCCTTTGTAAC |
| ss12674050 | 5 | NC_000005.4 | 163553409 | T | C | AATACTGGATGGAAAACATTTACAATGGGATAATAATAGAGCTAGAAACAG |
| ss12673872 | 5 | NC_000005.4 | 171593857 | T | C | GAACAAAAGAAGTCAATCAAGGGCATCAGTGACAATATTAACACCCAGAAT |
| ss12674051 | 5 | NC_000005.4 | 177958304 | G | A | GGAATGTGCAAAGGCCACGTCAGCAGATGGTTAGGTGCAATTTCACGCCTC |
| ss12673923 | 6 | NC_000006.5 | 1504798 | C | T | TGCTAAGTCTTCATTACAGGTTTCACTTTTTTATCGTCTATGACCACTATG |
| ss12673823 | 6 | NC_000006.5 | 8572931 | A | T | TTCTTATAGAATCAACCTTACTATGATCCTAAACTTTTGTTCTCAGAAACA |
| ss12673819 | 6 | NC_000006.5 | 15865771 | A | C | GTGGCTCGCGTGATATGAAAGGCCAAAGCATAGAGTTTCGTGAGGAAGAAG |
| ss12673924 | 6 | NC_000006.5 | 25789873 | C | T | CGAAAGCTATGAGCATTATGAATTCCTTCGTCACTGATATCTTTGAGCGTA |
| ss12673925 | 6 | NC_000006.5 | 39933409 | C | A | ATGTCCTTCTCCTGAACCACAGAAACGTGCTCTGCCTTAAGCACCTGTAAC |
| ss12673926 | 6 | NC_000006.5 | 41245155 | C | T | GTGAGACGCTGACTTTAGAAATAGCCGGTGATTACAGATTTAATTCATGTT |
| ss12673927 | 6 | NC_000006.5 | 57634762 | G | C | CCCAGGACCATTCCAGAGCTTATTCGTTCTACCTTGTTCTTCCTTGGGATG |
| ss12673820 | 6 | NC_000006.5 | 66890421 | A | G | CCCCAGGTACTTTGCATGTCTCACAACATTACGAATGGATAACTGAATCTC |
| ss12673928 | 6 | NC_000006.5 | 75359591 | T | C | TTTTCTTACTTTCTGCATAGTAATCTTTCATTCAGCACAGGACTTGAAAAC |
| ss12673929 | 6 | NC_000006.5 | 83061795 | C | T | TAATTCTATGTGGTAGCTACAGTTACCGATTCCGCTTATACAAAGTAATTG |
| ss12673930 | 6 | NC_000006.5 | 92114257 | C | T | GAAATGTGAATTTAGTATTTGTCAACTAATGCTGTTAAGTTAGAGACCTGT |
| ss12673818 | 6 | NC_000006.5 | 109008107 | T | G | GCATGAACTTGAGCACCTGAGTCCCTTGAATGCTGCTAAGGATAGGATGGA |
| ss12673825 | 6 | NC_000006.5 | 117082712 | T | C | TTCATACTTCTGTGCAATAGCTAATTGAGTTCCTGATTTAATGAATGATCT |
| ss12673931 | 6 | NC_000006.5 | 125000422 | T | A | AGAGCATATTGGTTACTTTGATTAATGGCTGATGATATTAAAACAGCATAG |
| ss12673822 | 6 | NC_000006.5 | 133738141 | C | G | TGCAAGTTTTGATCTAAATTGGCACCGACAAATTTTAAAACTATAGCCATT |
| ss12673824 | 6 | NC_000006.5 | 152028449 | G | A | TAAAAGCAGCCATGTCCAATTAGCAGTAAGTGCCATGCACCTGCAGTTACT |
| ss12673932 | 6 | NC_000006.5 | 160529746 | T | G | AATCCGTACATAGCTTTTGTTCATTGGATAATCGGGTGTAATATATGCAAA |
| ss12673821 | 6 | NC_000006.5 | 169356087 | T | C | GGATGTCCCTAAATCACGTTGTAACTGAGCAGACATTCACAGGGAAAACTT |
| ss12673843 | 7 | NC_000007.7 | 2525072 | A | G | ACTAACATCTTTCAAGTTTTTGGATAGACAATACATGCACAGAGTACCAAA |
| ss12673833 | 7 | NC_000007.7 | 10586231 | A | G | AGTCCATGTTGTCAATTCAGACCACACTTAGGGAATCAGACTCTCCAGGGA |
| ss12673834 | 7 | NC_000007.7 | 19501292 | T | C | ATTGATAGGTGCTGTCCACAAAGGTTTGGAATATAAAACCAGCACTGCTCT |
| ss12673835 | 7 | NC_000007.7 | 27111483 | G | T | ATTTTTTCACCTCTTGTGATATTCCGCCAAAGTAAACAATAGAGGTATTAC |
| ss12673839 | 7 | NC_000007.7 | 36197746 | G | T | TTGCTTTAATTACTCTGTACCTCATGTACTTGTAGTCTTTCTCACTATAAA |
| ss12673836 | 7 | NC_000007.7 | 44314883 | A | G | CCTTCCCATGTAACCTTCGGCTCTGAATGCACCTGAGTTTACCTAGCAAGC |
| ss12673841 | 7 | NC_000007.7 | 68684292 | A | G | TTGGGGCCAGGGCTCTGCACCTGGAAAGGCTTTATAACGTGAGATTCTCAA |
| ss12673831 | 7 | NC_000007.7 | 81030028 | C | T | GCAGTTGGGTATCTCAAGTGCCTGCCACAAGTAAATAGTTGTAAAAGCAAG |
| ss12673837 | 7 | NC_000007.7 | 98149562 | C | T | AAATGTAATACTCCACTCGAGCATGCGGCATTATTTAATCACTGATAGTTC |
| ss12673838 | 7 | NC_000007.7 | 107294194 | C | T | ATGAAAGGTATTAATCAGTCATTTCCGGCTCTTTATGTACAAGTGGTTCAT |
| ss12673832 | 7 | NC_000007.7 | 113711846 | A | G | AAACAAATGTGTTTTTGGAAACTAGATATGGTTTGGCTGCCTTCGAAATCT |
| ss12673933 | 7 | NC_000007.7 | 124117659 | C | T | AACTCTAGCTGCCATGTGATACTTACGAATTCCACCAGTATTTATTGGTTT |
| ss12673842 | 7 | NC_000007.7 | 140143478 | C | T | GTTTAACAGTAGAGTCCATTTTGTTCTCACTCAGCTGTTCTAGTTGAAGCA |
| ss12673840 | 7 | NC_000007.7 | 148344112 | G | A | TTCTGGGCCCAACTACAGTACAGACGTTGATGAGACCAACTCTGACTTTGG |
| ss12673963 | 8 | NC_000008.5 | 6543128 | C | T | AGATAATATTTAAAAAGTTTCATTCCGGGAGGCTTGGAACTATAGAGATAG |
| ss12673964 | 8 | NC_000008.5 | 14891730 | G | C | TTCCCATTATGTTCCACTTCTAATAGCTTTCACAAGACTGTCATAAACCAC |
| ss12673965 | 8 | NC_000008.5 | 23192709 | T | A | GAAAACGAGTCATCGTAAACTGAGCTGACCTGTACCCTACGCTGGAGAAAT |
| ss12673966 | 8 | NC_000008.5 | 31938636 | T | C | AATGTGCCAGGCACTGTGTTAAACTTCCAGATGGCAGTGAGAAACAAACTC |
| ss12673967 | 8 | NC_000008.5 | 39612530 | G | C | TATGAGTCTGGGCCAGCTGGAAACAGGTCTGGGATCTTCCAAGAAAGTCCT |
| ss12673968 | 8 | NC_000008.5 | 49331802 | G | A | ATCACAGCTGCCTGTTAACCAGCCTGAATGCAAAAAGTGAAAAAGCATTGC |
| ss12673969 | 8 | NC_000008.5 | 56073823 | C | T | TGAAGGAAGACGTAACAGCCAGAGTTCCTGTAAGAGCAAGAGAGGGTGGCT |
| ss12673970 | 8 | NC_000008.5 | 63475926 | C | T | CAAATATATTTCTGGCATACATCTTCCTTAACCTACATTATCCTCCTACTG |
| ss12673971 | 8 | NC_000008.5 | 71825336 | C | G | TTTCAGAAACCTAGGTCCAAAAGTCCTGCTAGGTATCTGGTATCTGGGATT |
| ss12673972 | 8 | NC_000008.5 | 80590545 | C | T | CGATAAGGTACTGCTTTAAGTTATTCTGAGGTCTTGCCTTTCTATAGACCC |
| ss12673973 | 8 | NC_000008.5 | 87441087 | A | T | TTTGGGAATTAGAATGCGTAGGTTAAGGTCCTAGTTCAATAAGTTAATGCC |
| ss12673974 | 8 | NC_000008.5 | 96224852 | C | A | TGAATCTTGGACTGTGCTACTTTGACTGTGAAAATACATTGACACTTGTGC |
| ss12673975 | 8 | NC_000008.5 | 103925518 | T | C | GTCCAGGTGACAACTCAGGAAAGAATTGCCACTTCGAAGCCGGAACACAAA |
| ss12673976 | 8 | NC_000008.5 | 113781010 | C | T | ATGACCAATTATTATTTGATGTGACCGATAGCTCCAGAACCTAAACAAATG |
| ss12673977 | 8 | NC_000008.5 | 120241683 | C | T | TATCTTCCTAAAGCAGAGCCAAAAACGTTGCTCTTCCAACTAAACATTTTC |
| ss12673978 | 8 | NC_000008.5 | 129227046 | A | G | AAGAAAAAGTTGACATTGTGATTACATATCAGTAGCATGACAAATTACATC |
| ss12673979 | 8 | NC_000008.5 | 135579063 | G | A | GTTCCAAGTGCACACCCTTTCTACTGTACTATCACAGCCTCTTGTTTCCCT |
| ss12674019 | 9 | NC_000009.5 | 9383975 | T | C | ACTTTCCTGATAGCTAGTGCTTTCATGATGCCCTTAGTGTCTACTGCCACG |
| ss12673873 | 9 | NC_000009.5 | 17663047 | C | T | GATTTCTTGCCCTGTTACCCTTACACGTGGCTGTTTGCCATGGTCTGTCAA |
| ss12673876 | 9 | NC_000009.5 | 26070402 | T | A | TTATAGGCATAATTTCTAACTCTCATTTAAGTGAGGCAGTTATCAATGTTG |
| ss12674020 | 9 | NC_000009.5 | 33195409 | T | C | GATGTATTAGCTGAGGGCCCAAAGTTGGGTAATGTGAGAAACCAGGACTCT |
| ss12673877 | 9 | NC_000009.5 | 70506757 | T | C | ACTAATAATTCCAGCCAATGTTTAGTGGAGATATTTCTTCTGACATTCTAA |
| ss12674021 | 9 | NC_000009.5 | 77568465 | C | T | TTAGTAAAGCCATTGTTCAAGCCATCGATATTAGGTTGTCAAATGTCTCTT |
| ss12674022 | 9 | NC_000009.5 | 86491072 | T | C | TCACAGTGTCCCTGTGTGATGCTCTTTTTTGACCCACACACTGTATAGGTC |
| ss12673874 | 9 | NC_000009.5 | 91240102 | C | T | TGGTGCCTGTGCAAAGAGTGGAACCCCAAAGAACACTGGGTGGTCAACACA |
| ss12673875 | 9 | NC_000009.5 | 101820299 | A | G | GCAGAGTTATATTTTGAAATATTGCAGTATTAGAAAAGCACATTATATATG |
| ss12674031 | 9 | NC_000009.5 | 110194153 | G | A | TGATGTGAGGATTTGAAACTTAGGCGGAATAGTAAGTACCAGGCATGGGCC |
| ss12674032 | 9 | NC_000009.5 | 118689216 | A | G | TCTGTACAAAGTGTATCATGGGACCATCCTATAAGGTTAAGCTTTCTCATT |
| ss12674023 | 9 | NC_000009.5 | 126715684 | A | G | CTCTGCATAAACTTGGAGAGAGGCCATTTCCTAATCAGAGGTCACAACTAG |
| ss12673997 | 10 | NC_000010.4 | 509164 | G | C | GATGTGAATCCACCTGTCACATATTGATTACATTCAGGCAATAACAGGGTG |
| ss12673998 | 10 | NC_000010.4 | 9764292 | G | A | TCACCTACATATGAGCAGCCTATCCGTCAGGCCAATGCTTAAGGTACCCCC |
| ss12673999 | 10 | NC_000010.4 | 17081249 | C | A | TTTCATTACCATTGTAATCTAGCCACAACAATGGTTGCTTTTTAAAACTAG |
| ss12674000 | 10 | NC_000010.4 | 25905020 | C | T | CAGGCAAGATCTCGTTTGTAAATTTCGTGGATTGAAAGTGAGGGACTAAGT |
| ss12674001 | 10 | NC_000010.4 | 44291617 | G | A | CTCCACAGCTGTTCCCAGGAATTTCGAAAGGGAGCACACCCTTGACTTGGT |
| ss12674002 | 10 | NC_000010.4 | 53583207 | G | C | TAGCTACTGCTCTTATTGAGGTTGTGTTTCTCTACTCCTCTGTAACATCGT |
| ss12674003 | 10 | NC_000010.4 | 61618552 | G | A | CTGATTTGCCTGTTAAAAGGCAGTAGGAAGGCAGTCCACCTGCTGTTTGCT |
| ss12673861 | 10 | NC_000010.4 | 68562600 | C | A | CCTTAACCATCACTTCTGCTGGAAACTTAGGGTGATCACCTTTTCCTAGAA |
| ss12674004 | 10 | NC_000010.4 | 77221697 | G | T | CAGCTTGGATTATTTTCCCCTGTCAGTTTAGCAATCAACAGCAATAAAAAC |
| ss12673859 | 10 | NC_000010.4 | 83404531 | C | T | CTGATTCATTGGTTCCTATATGGTGCCCCAAATTCTTAAGTCCTAATGCTC |
| ss12674005 | 10 | NC_000010.4 | 92160155 | C | A | GCTGAGGTTAGAAGCCTCCTTTCAACCCTGGTGAGAAGAGGTTGTACAGCG |
| ss12674006 | 10 | NC_000010.4 | 101448303 | A | G | GAGGCTAGATTCTGAAATGTTCCCAAGTCCAGCCATGAGGCCAAGGGAATC |
| ss12673860 | 10 | NC_000010.4 | 110319618 | T | A | TCACTTTTTCTGGTTTTAGCGAGGGTTCATTCGTTCATTCTAGCAGACAAA |
| ss12673858 | 10 | NC_000010.4 | 117679716 | G | A | ATTATGAAATCCATTCTCGAGTGGCGATTTTTTATGATGTTGTGTTATCAC |
| ss12674007 | 10 | NC_000010.4 | 124558750 | T | A | CGTGCAAGCCTAGTGAAACCAACCATGGGTCTCTCATCTGCTTTTACAGGA |
| ss12673947 | 11 | NC_000011.4 | 9978639 | A | G | CCTCTTCCACACTATTTTGGTAAACAGGACCAGCATTTATTCAGTCGCCTA |
| ss12673948 | 11 | NC_000011.4 | 19526219 | T | G | AACTTCTGTAATTTCCAATTCATGATGAAAGCCTAAGTAAAAATATCTGAC |
| ss12673949 | 11 | NC_000011.4 | 26302239 | T | C | ATTAATTCATTAGGAGCTTTTCCCATGTATGATCTGACACATTTCTGCCTT |
| ss12673950 | 11 | NC_000011.4 | 34941032 | G | C | AAATGTGTTTGATCTAGATCTCTTAGCAGTTTAATCCTGCATTCATAACCA |
| ss12673951 | 11 | NC_000011.4 | 50463164 | T | C | TTGAGGTTTTTGGCATCATTGGACATCATGAAATATGTAAATAAGATGGCA |
| ss12673952 | 11 | NC_000011.4 | 58525253 | T | C | AATTAAAAACAGGATGAGGAAAATTTGGTACATTCATTTGTATGCTTCAAT |
| ss12673953 | 11 | NC_000011.4 | 73007371 | A | G | GCTCTGTAAACCTCACAAACGCTCAATCTTTTTAGTCAATCAATCCTTTGC |
| ss12673954 | 11 | NC_000011.4 | 79267467 | T | C | AAAATGAAACTACACCTAATATCTATGAAGCCAATTGTACGTAGTAAAGAT |
| ss12673955 | 11 | NC_000011.4 | 88615466 | A | G | AACAATTCAAAAATCAGGGATCATAGCACTGACAAAAGCTCTAAAGTAATA |
| ss12673956 | 11 | NC_000011.4 | 95783160 | G | A | GTTTGTAGAACACACTAAGATGCTGAGAAGACTGCAGGTAAAGAGTTCTGC |
| ss12673957 | 11 | NC_000011.4 | 102987050 | A | G | AAATGGGTAAAGATTGCACGGGAGCAGTTACAACATTTCTACTTTTGTCCT |
| ss12673844 | 11 | NC_000011.4 | 110769906 | G | A | GTTTTCATCAGTTTTGTGGTCATACGTTTCTGATATGCTTCATTAATTGTT |
| ss12673958 | 11 | NC_000011.4 | 119349702 | T | C | AAATCTTCAATTTTGAAACCAAGTTTGTACTCTTGGCTGTAGAACCCCAAT |
| ss12673879 | 12 | NC_000012.5 | 9734612 | C | T | AGAGAGACCCTTCAAATACTGCTTACGTAACTTAAGAGTCAGCAATACTTG |
| ss12673980 | 12 | NC_000012.5 | 25449289 | C | T | GCTCTAGATTACCCATATAAAGTGGCTGGTTTTAGGCCTATGGCTTTTATT |
| ss12673981 | 12 | NC_000012.5 | 33771690 | G | A | CACATAGGCGATGTGGCTTCCAAGAGTCCCCTGGTCAGAGTAAGCCATGAT |
| ss12673982 | 12 | NC_000012.5 | 41545985 | C | G | GAAAAAGCAAACATTTTCATTGATAGAAGGGTGAGCCATCTTTGCCTTACT |
| ss12673983 | 12 | NC_000012.5 | 48787992 | T | C | CATACATCTCTTCAAAGCAGCAAGTTTGGCCATCTAGAACCACAATGGAAA |
| ss12673984 | 12 | NC_000012.5 | 64435176 | A | G | TCTTGCTGGGATGTCTAGACGTGGTAAAAGGTTTATCTGCTGTGCAATGGA |
| ss12673985 | 12 | NC_000012.5 | 78087554 | C | T | AGCTCAAGTGTGAGTCAGGCAATTACGAGTACTAGGAGGCAGGACCATCAT |
| ss12673986 | 12 | NC_000012.5 | 85983132 | T | C | CCTGTCTCATTCAAGTTGTATAGTATGAAATAGCATTATTGGAAGTTTTCT |
| ss12673987 | 12 | NC_000012.5 | 94257563 | A | G | TTACAAATCTGGAGATAACCAAATCATTTTTCGGATTTAAGTGAAGACACT |
| ss12673988 | 12 | NC_000012.5 | 102304251 | G | A | TTTCCAGTATAGCAAACTTAACTGCGTTCTCAAATAGTGCATTATGAACAT |
| ss12673880 | 12 | NC_000012.5 | 109687937 | A | G | ATTATCATTCTCAGATTTGATCCTTATAAATTCCATAGCTAAGACCCCTTG |
| ss12673989 | 12 | NC_000012.5 | 120061185 | C | T | CAAAGGCACAGAAAACTCAAAGAACCTCCCAAAGGCAACAATACACTCAGC |
| ss12673878 | 12 | NC_000012.5 | 126352865 | T | C | TGCTTTCTTGGAATATCCTCAAATTTGGTCACTCAGGTGACTTTGCTGAAA |
| ss12673990 | 13 | NC_000013.5 | 37569214 | A | C | TTTCACAATTTCTTTCTTGTGTCTCAACATTTTGTATGATTCATGAAAATG |
| ss12673991 | 13 | NC_000013.5 | 46030774 | C | T | CTAGGCAAATATGTATTGGTTCAGACACTATTCGAAATAGGGCTGTTGGCC |
| ss12673992 | 13 | NC_000013.5 | 54733488 | A | G | TGTTGGCGCATTTCAATTGCAGAGAAGTTTTCAAATGATTTTAATTTTTCC |
| ss12673993 | 13 | NC_000013.5 | 61713070 | A | C | TAGATAGGTATTATGGCTAAATGAAACAGTCACATCTACTATTTGTTGAAT |
| ss12673994 | 13 | NC_000013.5 | 71242882 | T | C | ATTTGGGGGATCTTGATTCCACCATTATCTATAGCTCCATCTAGGCTCCAG |
| ss12673881 | 13 | NC_000013.5 | 79078549 | A | G | TGTATTGGAATCCTTAGTGACTCACAGTATACATCCCATTAGATCTGCTGT |
| ss12673995 | 13 | NC_000013.5 | 87134862 | G | T | GTAAAGTATAACGGAGTCTACCATTGTATTGGGTACATGAGAAACAAATAA |
| ss12673996 | 13 | NC_000013.5 | 104455053 | G | T | AGAATATGTTCTGAAGTCTTTTCCTGTTGAATACCATCCAGAATTTTTAAA |
| ss12673809 | 14 | NC_000014.4 | 21935494 | A | G | GCTTGGTTCCAGTACATTATGGTATAAACTTTGGCTGCTGCCTCCTCAGCA |
| ss12673810 | 14 | NC_000014.4 | 30627841 | T | C | TAGAATTCAGGCAATGGCTTAATCATAAGGAACTACATGTGAGCCTAATGT |
| ss12673811 | 14 | NC_000014.4 | 37903865 | T | A | TGGATGGTTGTAGTGCACTGGGTTGTTTCAGGTAGGGATGACAAGGTTTTG |
| ss12673817 | 14 | NC_000014.4 | 47597308 | C | A | ATACACAAACAGGTCAGAAAGCTCCCAATGTAGCAGTTAAACAGTGTTTCC |
| ss12673812 | 14 | NC_000014.4 | 56536816 | A | G | TAGGCAACAGCCAGGTTTGACTGCCAACGATGCTAAGACAAGGAGATGAGG |
| ss12673813 | 14 | NC_000014.4 | 64870703 | C | T | ACATTTGCTGAATTACAAAGTAGTGCAGCTGTACATCAAGGCCAAAAGCTA |
| ss12673814 | 14 | NC_000014.4 | 73249181 | C | T | CATAATCTTGTAGTCTCAGGAGAAGCGGCCCTTCTGATGAGAGCTAATCCT |
| ss12673816 | 14 | NC_000014.4 | 81522156 | C | T | TTCTTTTTGCCTAATTGCAAACTTACGATATTCACAAAGACACAAATCTTA |
| ss12673815 | 14 | NC_000014.4 | 98203350 | G | A | TGTTTGCTATCCTGTGCTTGCCTCCGCTCTATCGGGCGCTGTGCCCCATCT |
| ss12674074 | 15 | NC_000015.4 | 31015317 | G | A | AGGTCCAAAACCTATCGCCTTGATAGAAATATGATATGGAAATCAGTAGGG |
| ss12674070 | 15 | NC_000015.4 | 41408146 | C | T | CCTACTCCATCCTCTACTGCTTCATCGCCCTCTAGTACTTGACTAACCTAC |
| ss12674071 | 15 | NC_000015.4 | 46635808 | T | G | TTAAAACATGAACTTGTTGTGCGTGTCTTGGATAGCAAAAAAAATCCCTCT |
| ss12674069 | 15 | NC_000015.4 | 64802044 | C | T | GAAACCTGGGCCAGGGATACATTTTCGCAGGTCCCGCAGACACTGCTAAGC |
| ss12673942 | 16 | NC_000016.4 | 929722 | C | T | AGATGGGAAGATACTTGTGATTTGACGGGAAGTAAAAAAACTTTGGTTATT |
| ss12673943 | 16 | NC_000016.4 | 8210016 | T | G | TTATAAACCAATCACCATTGAGAGGTTCCCCTTAGCCAGATCCTGGTTTAA |
| ss12673944 | 16 | NC_000016.4 | 16027252 | C | T | CCTATTTTGTACTTCTTATTTTATCCGATTGAATTGTGGTGGAGATAGGAA |
| ss12673882 | 16 | NC_000016.4 | 22941613 | A | G | AGAAAACAATGGAACAGTAACAATCGATCATTATGAGCTATCACCAAGACA |
| ss12673883 | 16 | NC_000016.4 | 53860338 | G | A | ACAACTATGAGATATTTCGTATTTTGAATGCCCCACAAATAAACAGATATT |
| ss12673945 | 16 | NC_000016.4 | 60769659 | A | G | TTAGCCTGTATTCCCATGAAAGATGACTCCAGAAACTTCAGAAGGATTGCT |
| ss12673946 | 16 | NC_000016.4 | 68835084 | T | C | TCCTGCCTTTCTTTACTGACCGTCCTGACGCTTTCAGTGAAGTGTCTCAAA |
| ss12673889 | 16 | NC_000016.4 | 76218899 | G | C | AGTAGCTATAATAACTTTGTCACATCAAACAAGATGAGTAAACTGGAATGT |
| ss12674043 | 16 | NC_000016.4 | 83056636 | G | A | ACAGCCTTATTAACTAACTCATCCCGCAGTTTTCAAAGAGCATGTATTTCT |
| ss12673862 | 17 | NC_000017.5 | 17949595 | T | C | GCTCTACAGAGGTCAGGACACAGCTCGGGGTCACGGCGCAAACCTTCAAGC |
| ss12674024 | 17 | NC_000017.5 | 25674535 | G | C | TTGTCCAGTAAGGCTGTCTCTACCAGGTAACACATGACTGCCAAGTGGGTA |
| ss12673863 | 17 | NC_000017.5 | 33821968 | G | A | TATTTTTATTTATCTCGGTCTTGACGGTCTGAATTACTGTGGCCTCCATGT |
| ss12674025 | 17 | NC_000017.5 | 41821078 | G | A | TCATGGAGGCAATTCCAGACAAAGGGATCAGTGCAAGCAAAGGAAGCGAGG |
| ss12673864 | 17 | NC_000017.5 | 49396518 | T | C | TATTTCCTATAATTCTCCTATTTGTTCCATGGCAGTTATCTAAAAATATAC |
| ss12673865 | 17 | NC_000017.5 | 56570082 | C | A | GAAAGAACCCACGGTTACTGACGGGCTTTAGCCATTACAGTGACACTCAAA |
| ss12673866 | 17 | NC_000017.5 | 65526529 | T | C | AAAGACGGAGGTCATGTTAGAGAGATTGTGAAAAGTAAAAATGTGTCAAAG |
| ss12674026 | 17 | NC_000017.5 | 74688517 | T | C | AACCCTGTACCCTTCTTCCTTGTGGTGCTCTCAGAACCCTTATGCATTACA |
| ss12673959 | 18 | NC_000018.4 | 1287985 | C | G | ATTATGGGACTGCTATCTTAGCCTACTAGAATGGAATCAGCATGGGGATCC |
| ss12674038 | 18 | NC_000018.4 | 8059870 | T | C | AAGAAGTAAGCTGGGATACAGAAAACTCACACCCTCAACACACGATCACTA |
| ss12673960 | 18 | NC_000018.4 | 24641105 | G | C | ATTTAACCTCATTTACTTTGTCCCTGTCATAGAACCTGTACTTGATGGATA |
| ss12673961 | 18 | NC_000018.4 | 32549247 | T | A | TGAAGTAAATACTGTGCATTCTTCAAACTGATTTGGGATCCTTCTGATACT |
| ss12673962 | 18 | NC_000018.4 | 54949539 | G | A | CTCTATTCCCTAAAGCAGGCTAAAGGTTTCACTGAAGTCTTATACTCTGTC |
| ss12674039 | 18 | NC_000018.4 | 63533863 | A | G | TGGAAATGGCTACATTATCATTTGCATAAGCCTCTCATGCAGAATTATCTC |
| ss12674040 | 18 | NC_000018.4 | 70863198 | G | A | TTTGTGCAAACTTCATACACTTCCAAATCTTCTGTAGCTGAGACGAGTGAA |
| ss12673934 | 19 | NC_000019.5 | 206544 | C | T | TAGTCATGAAGTTAATGATAAAAGACGACCCATGCCTTATTTATGTAATAA |
| ss12673935 | 19 | NC_000019.5 | 7106531 | A | G | GTTACTCACCCAACAATCTAATGCCACAAGAAAAAATAACTCGGGAACAGC |
| ss12673936 | 19 | NC_000019.5 | 14842230 | T | C | ATACCTTTCCTCCTGTTATTCCAACTCTGAACACATCAGTTTCCTGGGGGA |
| ss12673937 | 19 | NC_000019.5 | 21737353 | T | C | TTGGGTAAAGGTAAAACTGTGTCCATTACTCTCAGTCATCTTGGTTAGAAT |
| ss12673938 | 19 | NC_000019.5 | 34028148 | G | A | TCCACAGTCAGAAGACACGCTAGACGAAGGGCGTCCATCCAGTCTCAGCCC |
| ss12673939 | 19 | NC_000019.5 | 41616809 | T | G | GAGCCGAGTTCTTTCTTAAACTGCCGATTACATTCCCAATCATCTCTGAAA |
| ss12673940 | 19 | NC_000019.5 | 49030287 | T | C | GCTAAAAAAATGAGACTTGAAAAAATCCAGACTTTTGAAGAGTTTAGGAAA |
| ss12673941 | 19 | NC_000019.5 | 57184140 | C | T | GAGCAAGGTCTGAAGAGGAACAAAACGGTAAGTAATTAATAAAGCCTAAAT |
| ss12673830 | 20 | NC_000020.5 | 1332298 | T | C | GGTGAAACTGTAGCCAAAACTCTTATAAATTCTATGGTGGACATTTGGTGA |
| ss12673829 | 20 | NC_000020.5 | 9620074 | A | C | TTAGGCAACTGTCACGAAAATCATAAGACTCTACGGAAAGAAAAAGACTGT |
| ss12673827 | 20 | NC_000020.5 | 18818060 | A | G | TAGCCAAGAAAATCAAATTTCCACTATCCCGAGAAGGTTAGCTCTGTTGTT |
| ss12673828 | 20 | NC_000020.5 | 38659006 | C | G | TTCTGGCTCTTGGAAAGTCATTGTTCTCAAATGGGATGCCATGATTTGTAG |
| ss12673826 | 20 | NC_000020.5 | 46896999 | A | G | ATACTAAATAAAATATCTTTAAGCAATTTAGCAAGTAGCATCTTTGAAAAT |
| ss12673804 | 21 | NC_000021.3 | 21432625 | A | G | TCTGTAAATTGAAGATGATTACAGTAGTCGTAGTTCCCCAATCTTAAGCTA |
| ss12673805 | 22 | NC_000022.4 | 15767574 | G | T | GGTCGTGCCTCCTGCGGACCTGAGTGACCTCATGGAACAGAGCCAACGACA |
| ss12673806 | 22 | NC_000022.4 | 25256704 | G | A | CAGACAGGAACAAATCAGATGACCAGGAATTGAGAGACTGAACATTTCCTC |
| ss12673807 | 22 | NC_000022.4 | 32926742 | A | G | CCATGTGGACTCGCTACAGAGGTACATGCATAGGTCCAAGATAGGCGTCCC |
| ss12673808 | 22 | NC_000022.4 | 42338946 | C | T | TGTTAGAACCTTCTTTTTCTATAGACGGCCAGCACTGGCATGAAGAGATGC |
Statistical Analysis
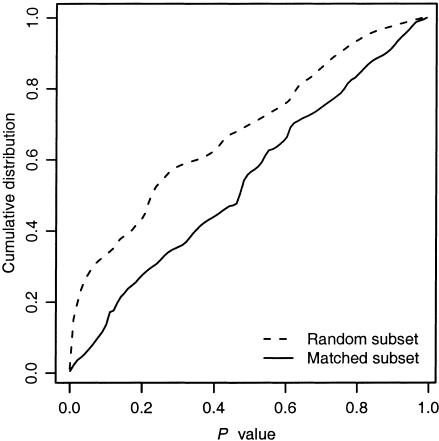
We used the structure program (Pritchard et al. 2000_a_) to identify population subgroups and infer admixture information from SNP genotype data. All runs were 100,000 cycles, after a 20,000-cycle burn-in period. We selected a model with admixture and with correlated allele frequencies; we used the defaults for other settings. We did not use prior information about population membership to direct the clustering. Without this information, the structure program cannot distinguish between solutions with permuted cluster labels; therefore, we manually assigned labels to clusters, for consistency across multiple analyses. Genetic distances (_F_ST) were calculated from _structure_’s allele-frequency estimates, as in the study by Weir (1996). False-discovery rates were calculated using Q-VALUE (available on the Q-VALUE Software Web site) (Storey and Tibshirani 2003). All other statistical analyses were performed with the R package (available on the R Project Web site) (Ihaka and Gentleman 1996).
Results
Assessment of Population Structure
A total of 707 individuals recruited in Mexico City were selected for genotyping. The majority of subjects (655) were of Mestizo (“mixed”) ancestry; small numbers of individuals of self-reported Caucasian (23) and Otomi Indian (29) ancestry were also included. Using high-density oligonucleotide arrays, we genotyped these subjects for 312 uniformly spaced, unlinked SNPs. Of the 312 markers, 275 yielded high-quality genotype data. Many of the SNPs showed larger-than-expected allele-frequency differences between the three subpopulations, measured as an excess of small P values in χ2 tests (table 2). Controlling for false-discovery rate (Storey and Tibshirani 2003), we also counted SNPs having q values < 0.05 and found many significant associations. The q value method accounts for multiple testing, and it indicates the number of SNPs with significant associations such that, on average, only 5% will be false positives.
Table 2.
Association Test Results for Population Subgroups with 275 SNPs
| Number of SNPs with χ2 Test Statistics | |||||
|---|---|---|---|---|---|
| P<.0001 | P<.001 | P<.01 | P<.1 | q<.05 | |
| Expected | 0 | 0 | 2.75 | 27.5 | 0 |
| Caucasian—Mestizo | 2 | 5 | 23 | 85 | 8 |
| Otomi—Mestizo | 0 | 1 | 15 | 50 | 0 |
| Otomi—Caucasian | 3 | 14 | 34 | 105 | 32 |
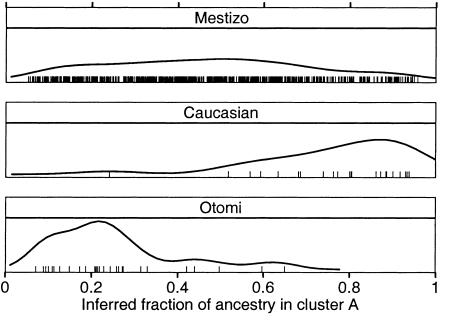
We analyzed this genotype data for population structure using the structure program (Pritchard et al. 2000_a_; available on the Pritchard Lab Web site). This is a model-based method for identifying subpopulations in which, within each subpopulation, all markers are in Hardy-Weinberg and linkage equilibrium. The analysis supported the presence of two genetically distinct population clusters, one of mostly European ancestry (“cluster A”), and one of mostly Indian ancestry (“cluster B”). The estimated cluster-membership proportions for self-reported Caucasian and Otomi Indian samples are well separated; Mestizo samples are uniformly distributed across nearly the full range of values (fig. 1). There was no strong evidence for models with more than two population clusters. On the basis of their estimated allele frequencies, we determined a genetic distance of F _ST_=0.14 between the two clusters. Phenotype information and cluster-membership proportions for each sample are reported in table B (online only).
Figure 1.
Distribution of ancestry for self-reported population subgroups. Density distributions for the inferred fraction of subjects with cluster A ancestry are shown for 655 Mestizo, 23 Caucasian, and 29 Otomi Indian subjects. Each tick mark represents the fractional ancestry of an individual subject.
Table B.
Sample Phenotypes and Inferred Cluster Membership Proportions
| Subject | Ethnicity | Sex | Height (cm) | Cluster A | Cluster B |
|---|---|---|---|---|---|
| 0b7 | Mestizo | Male | .3126 | .6874 | |
| 0b8 | Mestizo | Male | .1795 | .8205 | |
| 0bb | Mestizo | Male | .1457 | .8543 | |
| 0bc | Mestizo | Male | .4727 | .5273 | |
| 0bd | Caucasian | Male | .7723 | .2277 | |
| 0be | Caucasian | Male | .9331 | .0669 | |
| 0bf | Caucasian | Male | .8838 | .1162 | |
| 0bg | Mestizo | Male | .4903 | .5097 | |
| 0bh | Otomi | Male | .4210 | .5790 | |
| 0bi | Mestizo | Male | .3954 | .6046 | |
| 0bj | Mestizo | Female | .3851 | .6149 | |
| 0bk | Mestizo | Male | 162 | .6158 | .3842 |
| 0bl | Mestizo | Male | 170 | .9062 | .0938 |
| 0bm | Caucasian | Male | 168 | .5929 | .4071 |
| 0bn | Mestizo | Male | 166 | .5515 | .4485 |
| 0bo | Mestizo | Male | 162 | .5460 | .4540 |
| 0br | Mestizo | Male | 165 | .6342 | .3658 |
| 0bs | Mestizo | Male | 174 | .8261 | .1739 |
| 0bt | Mestizo | Male | 158 | .3408 | .6592 |
| 0bu | Caucasian | Male | 180 | .9376 | .0624 |
| 0bv | Caucasian | Male | 185 | .8000 | .2000 |
| 0bw | Mestizo | Male | 174 | .7291 | .2709 |
| 0by | Mestizo | Male | 161 | .1883 | .8117 |
| 0bz | Mestizo | Male | 163 | .5132 | .4868 |
| 0c0 | Mestizo | Female | 156 | .4868 | .5132 |
| 0c1 | Mestizo | Male | 165 | .8240 | .1760 |
| 0c2 | Mestizo | Male | 159 | .3869 | .6131 |
| 0c4 | Mestizo | Male | 165 | .2128 | .7872 |
| 0c6 | Mestizo | Male | 169 | .7532 | .2468 |
| 0c7 | Mestizo | Male | 155 | .1457 | .8543 |
| 0c9 | Mestizo | Male | 162 | .4311 | .5689 |
| 0ca | Mestizo | Male | 168 | .6268 | .3732 |
| 0cb | Mestizo | Female | 150 | .6889 | .3111 |
| 0cc | Mestizo | Male | 164 | .6794 | .3206 |
| 0cd | Mestizo | Male | 160 | .1669 | .8331 |
| 0ce | Mestizo | Male | 163 | .5118 | .4882 |
| 0cf | Mestizo | Male | 165 | .4008 | .5992 |
| 0cg | Otomi | Male | 155 | .3292 | .6708 |
| 0ch | Mestizo | Male | 175 | .3374 | .6626 |
| 0ci | Mestizo | Male | 157 | .1288 | .8712 |
| 0cj | Mestizo | Male | 170 | .6687 | .3313 |
| 0ck | Mestizo | Male | 165 | .4926 | .5074 |
| 0cl | Mestizo | Male | 155 | .0853 | .9147 |
| 0co | Mestizo | Female | 141 | .1381 | .8619 |
| 0cp | Mestizo | Male | 165 | .6193 | .3807 |
| 0cq | Caucasian | Male | 179 | .9381 | .0619 |
| 0cr | Mestizo | Male | 160 | .2016 | .7984 |
| 0cs | Mestizo | Male | 168 | .6686 | .3314 |
| 0ct | Mestizo | Male | 177 | .5407 | .4593 |
| 0cu | Mestizo | Male | 170 | .6671 | .3329 |
| 0cv | Mestizo | Male | 174 | .4032 | .5968 |
| 0cw | Mestizo | Male | 170 | .2612 | .7388 |
| 0cx | Mestizo | Male | 179 | .3576 | .6424 |
| 0cy | Mestizo | Male | 170 | .6408 | .3592 |
| 0cz | Mestizo | Male | 157 | .6199 | .3801 |
| 0d0 | Mestizo | Male | 160 | .6221 | .3779 |
| 0d1 | Mestizo | Male | 175 | .6839 | .3161 |
| 0d2 | Mestizo | Male | 173 | .3033 | .6967 |
| 0d3 | Mestizo | Male | 175 | .4385 | .5615 |
| 0d4 | Mestizo | Female | 149 | .2430 | .7570 |
| 0d5 | Mestizo | Male | 170 | .7654 | .2346 |
| 0d7 | Mestizo | Male | 165 | .6873 | .3127 |
| 0d8 | Mestizo | Male | 173 | .2555 | .7445 |
| 0d9 | Caucasian | Male | 171 | .9000 | .1000 |
| 0da | Mestizo | Male | 183 | .3681 | .6319 |
| 0db | Mestizo | Female | 165 | .9016 | .0984 |
| 0dc | Caucasian | Male | 170 | .9298 | .0702 |
| 0dd | Mestizo | Male | 175 | .6059 | .3941 |
| 0df | Caucasian | Male | 181 | .5684 | .4316 |
| 0dh | Caucasian | Male | 172 | .6811 | .3189 |
| 0di | Mestizo | Male | 157 | .7941 | .2059 |
| 0dj | Caucasian | Male | 178 | .7389 | .2611 |
| 0dk | Mestizo | Male | 175 | .9220 | .0780 |
| 0dl | Mestizo | Male | 172 | .4538 | .5462 |
| 0dm | Mestizo | Female | 136 | .2183 | .7817 |
| 0dn | Mestizo | Male | 166 | .3694 | .6306 |
| 0do | Mestizo | Male | 168 | .5476 | .4524 |
| 0dp | Mestizo | Male | 166 | .1653 | .8347 |
| 0dq | Mestizo | Male | 170 | .7309 | .2691 |
| 0dr | Caucasian | Female | 158 | .7624 | .2376 |
| 0ds | Caucasian | Female | 160 | .9170 | .0830 |
| 0dv | Caucasian | Female | 151 | .5181 | .4819 |
| 0dw | Mestizo | Male | 178 | .5863 | .4137 |
| 0dx | Mestizo | Male | 167 | .6549 | .3451 |
| 0dy | Mestizo | Male | 170 | .4953 | .5047 |
| 0dz | Caucasian | Male | 171 | .8717 | .1283 |
| 0e0 | Caucasian | Male | 160 | .2408 | .7592 |
| 0e1 | Mestizo | Male | 172 | .5484 | .4516 |
| 0e2 | Mestizo | Male | 171 | .6700 | .3300 |
| 0e4 | Caucasian | Male | 168 | .6855 | .3145 |
| 0e5 | Mestizo | Male | 167 | .5110 | .4890 |
| 0e6 | Mestizo | Male | 158 | .1879 | .8121 |
| 0e7 | Caucasian | Male | 165 | .6337 | .3663 |
| 0e9 | Caucasian | Male | 173 | .8050 | .1950 |
| 0ea | Caucasian | Male | 178 | .8034 | .1966 |
| 0eb | Mestizo | Male | 161 | .8818 | .1182 |
| 0ed | Mestizo | Male | 153 | .1724 | .8276 |
| 0ee | Mestizo | Male | 179 | .5559 | .4441 |
| 0ef | Mestizo | Male | 170 | .1890 | .8110 |
| 0eh | Mestizo | Male | 170 | .6383 | .3617 |
| 0ei | Mestizo | Male | 158 | .1396 | .8604 |
| 0ek | Mestizo | Male | 161 | .5511 | .4489 |
| 0eo | Mestizo | Male | 155 | .1599 | .8401 |
| 0ep | Mestizo | Female | 153 | .1997 | .8003 |
| 0eq | Mestizo | Female | 154 | .1193 | .8807 |
| 0er | Mestizo | Male | 166 | .1575 | .8425 |
| 0et | Mestizo | Female | 156 | .8033 | .1967 |
| 0eu | Mestizo | Male | 172 | .1903 | .8097 |
| 0ev | Mestizo | Female | 144 | .0896 | .9104 |
| 0ex | Mestizo | Male | 163 | .5826 | .4174 |
| 0f0 | Mestizo | Male | 162 | .1381 | .8619 |
| 0f1 | Mestizo | Male | 160 | .3733 | .6267 |
| 0f2 | Mestizo | Male | 151 | .1719 | .8281 |
| 0f3 | Caucasian | Male | 191 | .8856 | .1144 |
| 0f4 | Mestizo | Male | 164 | .7052 | .2948 |
| 0f5 | Mestizo | Male | 160 | .2973 | .7027 |
| 0f8 | Caucasian | Male | 184 | .8615 | .1385 |
| 0fb | Mestizo | Male | 160 | .0949 | .9051 |
| 0fc | Mestizo | Male | 167 | .2901 | .7099 |
| 0fe | Mestizo | Male | 168 | .1597 | .8403 |
| 0fh | Mestizo | Male | 160 | .2974 | .7026 |
| 0fj | Mestizo | Male | 160 | .0968 | .9032 |
| 0fl | Mestizo | Male | 163 | .3957 | .6043 |
| 0fm | Mestizo | Male | 168 | .0731 | .9269 |
| 0fn | Mestizo | Male | 164 | .0996 | .9004 |
| 0fo | Mestizo | Male | 160 | .6349 | .3651 |
| 0fq | Mestizo | Male | 154 | .4988 | .5012 |
| 0fr | Mestizo | Male | 166 | .2022 | .7978 |
| 0fs | Mestizo | Female | 154 | .4985 | .5015 |
| 0ft | Mestizo | Male | 155 | .4147 | .5853 |
| 0fu | Mestizo | Male | 168 | .9101 | .0899 |
| 0fv | Mestizo | Male | 167 | .8343 | .1657 |
| 0fw | Mestizo | Male | 166 | .4248 | .5752 |
| 0g1 | Mestizo | Female | 118 | .2731 | .7269 |
| 0g2 | Mestizo | Male | 171 | .5132 | .4868 |
| 0g5 | Mestizo | Male | 154 | .7244 | .2756 |
| 0g6 | Mestizo | Male | 171 | .3916 | .6084 |
| 0g7 | Mestizo | Female | 159 | .5678 | .4322 |
| 0g8 | Mestizo | Male | 160 | .3190 | .6810 |
| 0g9 | Mestizo | Male | 165 | .3160 | .6840 |
| 0gb | Mestizo | Female | 144 | .1203 | .8797 |
| 0gc | Mestizo | Male | 185 | .5447 | .4553 |
| 0ge | Mestizo | Male | 162 | .2413 | .7587 |
| 0gh | Mestizo | Male | 165 | .7255 | .2745 |
| 0gj | Mestizo | Male | 167 | .1250 | .8750 |
| 0gk | Mestizo | Male | 165 | .6056 | .3944 |
| 0gl | Mestizo | Male | 162 | .4868 | .5132 |
| 0gm | Mestizo | Male | 160 | .0690 | .9310 |
| 0gn | Mestizo | Male | 161 | .1788 | .8212 |
| 7op | Mestizo | Male | 149 | .2217 | .7783 |
| 7oq | Mestizo | Male | 162 | .0618 | .9382 |
| 7ou | Mestizo | Male | 163 | .4906 | .5094 |
| 7ov | Mestizo | Male | 170 | .3040 | .6960 |
| 7ow | Mestizo | Male | 164 | .1904 | .8096 |
| 7oy | Mestizo | Male | 159 | .6188 | .3812 |
| 7oz | Mestizo | Male | 179 | .7177 | .2823 |
| 7p2 | Mestizo | Male | .5777 | .4223 | |
| 7p3 | Mestizo | Male | 157 | .2603 | .7397 |
| 7p4 | Mestizo | Male | 169 | .4466 | .5534 |
| 7p5 | Mestizo | Male | 168 | .5085 | .4915 |
| 7p7 | Mestizo | Male | 178 | .5731 | .4269 |
| 7p8 | Mestizo | Female | 160 | .8835 | .1165 |
| 7pa | Mestizo | Male | 181 | .9286 | .0714 |
| 7pb | Mestizo | Male | 165 | .6542 | .3458 |
| 7pd | Mestizo | Male | 162 | .2041 | .7959 |
| 7pi | Mestizo | Male | 157 | .5592 | .4408 |
| 7pj | Mestizo | Male | 174 | .4927 | .5073 |
| 7pl | Mestizo | Male | 179 | .5644 | .4356 |
| 7pm | Mestizo | Male | 159 | .4851 | .5149 |
| 7pn | Mestizo | Male | 162 | .4956 | .5044 |
| 7pr | Mestizo | Male | 195 | .5563 | .4437 |
| 7ps | Mestizo | Male | 176 | .5133 | .4867 |
| 7pu | Mestizo | Male | 172 | .4362 | .5638 |
| 7pw | Mestizo | Male | 158 | .0942 | .9058 |
| 7px | Mestizo | Male | 169 | .6493 | .3507 |
| 7pz | Mestizo | Male | 161 | .4779 | .5221 |
| 7q0 | Mestizo | Male | 156 | .7743 | .2257 |
| 7q1 | Mestizo | Male | 162 | .2425 | .7575 |
| 7q2 | Mestizo | Male | 166 | .1697 | .8303 |
| 7q3 | Mestizo | Male | 172 | .5815 | .4185 |
| 7qa | Mestizo | Female | 146 | .4247 | .5753 |
| 7qc | Mestizo | Male | 162 | .3309 | .6691 |
| 7qf | Mestizo | Male | 162 | .4822 | .5178 |
| 7qh | Mestizo | Male | 163 | .1427 | .8573 |
| 7qj | Mestizo | Male | 169 | .3919 | .6081 |
| 7qm | Mestizo | Male | 155 | .6480 | .3520 |
| 7qn | Mestizo | Male | 155 | .4069 | .5931 |
| 7qp | Mestizo | Male | 167 | .6990 | .3010 |
| 7qq | Mestizo | Male | 165 | .7216 | .2784 |
| 7qv | Mestizo | Male | 158 | .5001 | .4999 |
| 7qw | Otomi | Male | 180 | .6493 | .3507 |
| 7qx | Mestizo | Male | 168 | .4843 | .5157 |
| 7qz | Mestizo | Male | 164 | .5880 | .4120 |
| 7r1 | Mestizo | Male | 142 | .3875 | .6125 |
| 7r2 | Mestizo | Male | 162 | .4021 | .5979 |
| 7r3 | Mestizo | Male | 160 | .0884 | .9116 |
| 7r4 | Mestizo | Male | 168 | .5629 | .4371 |
| 7r5 | Mestizo | Male | 154 | .1291 | .8709 |
| 7r6 | Mestizo | Female | 150 | .1211 | .8789 |
| 7ri | Mestizo | Male | 176 | .8348 | .1652 |
| 7rj | Mestizo | Male | 160 | .2914 | .7086 |
| 7rm | Mestizo | Male | 158 | .4687 | .5313 |
| 7rq | Mestizo | Male | 163 | .2027 | .7973 |
| 7rr | Mestizo | Male | 177 | .2619 | .7381 |
| 7rs | Mestizo | Male | 160 | .0780 | .9220 |
| 7ru | Mestizo | Male | 172 | .5849 | .4151 |
| 7rw | Mestizo | Male | 154 | .0928 | .9072 |
| 7ry | Mestizo | Male | 151 | .2050 | .7950 |
| 7rz | Mestizo | Female | 155 | .3809 | .6191 |
| 7s0 | Mestizo | Female | 163 | .5029 | .4971 |
| 7s1 | Mestizo | Male | 178 | .6896 | .3104 |
| 7s3 | Mestizo | Male | 157 | .4147 | .5853 |
| 7s4 | Mestizo | Male | 155 | .3096 | .6904 |
| 7s5 | Mestizo | Male | 156 | .3947 | .6053 |
| 7s6 | Mestizo | Male | 156 | .1174 | .8826 |
| 7s7 | Mestizo | Male | 173 | .6185 | .3815 |
| 7s9 | Mestizo | Male | 160 | .4705 | .5295 |
| 7sb | Mestizo | Male | 168 | .3815 | .6185 |
| 7sd | Mestizo | Male | 160 | .5431 | .4569 |
| 7se | Otomi | Male | 150 | .1733 | .8267 |
| 7sf | Otomi | Male | 168 | .2148 | .7852 |
| 7sg | Otomi | Male | 158 | .1271 | .8729 |
| 7sh | Otomi | Female | 143 | .2110 | .7890 |
| 7si | Otomi | Female | 143 | .0713 | .9287 |
| 7sj | Otomi | Male | 159 | .2113 | .7887 |
| 7sk | Otomi | Female | 153 | .2440 | .7560 |
| 7sl | Otomi | Male | 166 | .1136 | .8864 |
| 7sm | Otomi | Female | 157 | .3143 | .6857 |
| 7sn | Otomi | Female | 150 | .1732 | .8268 |
| 7so | Otomi | Male | 152 | .1007 | .8993 |
| 7sp | Otomi | Male | 166 | .2582 | .7418 |
| 7sr | Otomi | Male | 154 | .2083 | .7917 |
| 7ss | Otomi | Male | 178 | .1095 | .8905 |
| 7st | Otomi | Male | 146 | .2718 | .7282 |
| 7su | Mestizo | Male | 167 | .3750 | .6250 |
| 7sv | Mestizo | Male | 166 | .6236 | .3764 |
| 7sw | Mestizo | Male | 160 | .1936 | .8064 |
| 7sz | Mestizo | Male | 160 | .6043 | .3957 |
| 7t0 | Mestizo | Male | 161 | .4253 | .5747 |
| 7t2 | Mestizo | Male | 176 | .7161 | .2839 |
| 7t4 | Mestizo | Male | 178 | .8549 | .1451 |
| 7t5 | Mestizo | Male | 180 | .7728 | .2272 |
| 7t7 | Mestizo | Male | 174 | .6574 | .3426 |
| 7t8 | Mestizo | Male | 156 | .1293 | .8707 |
| 7t9 | Mestizo | Male | 170 | .6342 | .3658 |
| 7ta | Mestizo | Male | 168 | .2329 | .7671 |
| 7tb | Mestizo | Male | 166 | .6250 | .3750 |
| 7tc | Mestizo | Male | 169 | .3303 | .6697 |
| 7td | Mestizo | Male | 162 | .1408 | .8592 |
| 7te | Mestizo | Male | 165 | .1545 | .8455 |
| 7tf | Mestizo | Male | 172 | .9084 | .0916 |
| 7tg | Mestizo | Male | 178 | .9062 | .0938 |
| 7th | Mestizo | Male | 181 | .6093 | .3907 |
| 7ti | Mestizo | Male | 180 | .8071 | .1929 |
| 7tk | Mestizo | Male | 158 | .4396 | .5604 |
| 7tl | Mestizo | Male | 156 | .3333 | .6667 |
| 7tn | Mestizo | Male | 160 | .4235 | .5765 |
| 7to | Mestizo | Female | 153 | .6285 | .3715 |
| 7tp | Mestizo | Female | 154 | .4776 | .5224 |
| 7tq | Mestizo | Male | 161 | .1615 | .8385 |
| 7tr | Mestizo | Male | 168 | .1676 | .8324 |
| 7ts | Mestizo | Male | 160 | .1171 | .8829 |
| 7tu | Mestizo | Male | 165 | .4346 | .5654 |
| 7tv | Mestizo | Male | 156 | .1421 | .8579 |
| 7tx | Mestizo | Female | 155 | .5290 | .4710 |
| 7ty | Mestizo | Male | 159 | .2850 | .7150 |
| 7tz | Mestizo | Male | 168 | .2214 | .7786 |
| 7u0 | Mestizo | Female | 152 | .5419 | .4581 |
| 7u3 | Mestizo | Male | 163 | .1941 | .8059 |
| 7u4 | Mestizo | Male | 160 | .4576 | .5424 |
| 7u5 | Mestizo | Male | 161 | .2100 | .7900 |
| 7u6 | Mestizo | Male | 167 | .5917 | .4083 |
| 7u8 | Mestizo | Male | 168 | .9431 | .0569 |
| 7u9 | Mestizo | Male | 165 | .1774 | .8226 |
| 7ua | Mestizo | Male | 163 | .4069 | .5931 |
| 7ub | Mestizo | Male | 172 | .9570 | .0430 |
| 7uc | Mestizo | Male | 190 | .7985 | .2015 |
| 7ue | Mestizo | Male | 172 | .5063 | .4937 |
| 7uf | Mestizo | Male | 165 | .5739 | .4261 |
| 7ug | Mestizo | Male | 157 | .4205 | .5795 |
| 7uh | Mestizo | Male | 170 | .3555 | .6445 |
| 7uk | Mestizo | Male | 160 | .1563 | .8437 |
| 7ul | Mestizo | Male | 168 | .5278 | .4722 |
| 7um | Mestizo | Male | 174 | .7976 | .2024 |
| 7un | Mestizo | Male | 165 | .3994 | .6006 |
| 7up | Mestizo | Male | 160 | .4159 | .5841 |
| 7uq | Mestizo | Male | 165 | .4985 | .5015 |
| 7ur | Mestizo | Male | 158 | .1292 | .8708 |
| 7us | Mestizo | Male | 180 | .8326 | .1674 |
| 7ut | Mestizo | Male | 163 | .9325 | .0675 |
| 7uu | Mestizo | Male | 174 | .6588 | .3412 |
| 7uv | Mestizo | Male | 167 | .6644 | .3356 |
| 7uw | Mestizo | Male | 185 | .9276 | .0724 |
| 7ux | Mestizo | Male | 180 | .3082 | .6918 |
| 7uy | Mestizo | Male | 180 | .6943 | .3057 |
| 7uz | Mestizo | Male | 186 | .8417 | .1583 |
| 7v1 | Mestizo | Male | 172 | .9133 | .0867 |
| 7v2 | Mestizo | Female | 156 | .7400 | .2600 |
| 7v3 | Mestizo | Male | 170 | .1986 | .8014 |
| 7v4 | Mestizo | Male | 170 | .4294 | .5706 |
| 7v5 | Mestizo | Male | 163 | .4120 | .5880 |
| 7v6 | Mestizo | Male | 170 | .3065 | .6935 |
| 7v7 | Mestizo | Male | 173 | .6868 | .3132 |
| 7v8 | Mestizo | Male | 178 | .8400 | .1600 |
| 7v9 | Mestizo | Male | 162 | .2877 | .7123 |
| 7va | Mestizo | Male | 173 | .6125 | .3875 |
| 7vc | Mestizo | Male | 183 | .3608 | .6392 |
| 7vd | Mestizo | Male | 186 | .3975 | .6025 |
| 7ve | Mestizo | Male | 163 | .6361 | .3639 |
| 7vj | Mestizo | Male | 172 | .3938 | .6062 |
| 7vk | Mestizo | Male | 182 | .7810 | .2190 |
| 7vl | Mestizo | Male | 166 | .5863 | .4137 |
| 7vm | Otomi | Male | 162 | .1494 | .8506 |
| 7vo | Mestizo | Male | 164 | .3427 | .6573 |
| 7vp | Mestizo | Male | 163 | .5622 | .4378 |
| 7vq | Mestizo | Male | 186 | .0568 | .9432 |
| 7vw | Otomi | Female | 148 | .1872 | .8128 |
| 7vz | Mestizo | Female | 159 | .2409 | .7591 |
| 7w0 | Mestizo | Male | 170 | .3778 | .6222 |
| 7w2 | Otomi | Female | 153 | .5968 | .4032 |
| 7w3 | Mestizo | Male | 165 | .1099 | .8901 |
| 7w7 | Otomi | Male | 166 | .0945 | .9055 |
| 7w8 | Mestizo | Male | 159 | .1765 | .8235 |
| 7w9 | Mestizo | Male | 160 | .3967 | .6033 |
| 7wa | Mestizo | Male | 162 | .5551 | .4449 |
| 7wc | Mestizo | Male | 180 | .4988 | .5012 |
| 7wn | Mestizo | Male | 180 | .4742 | .5258 |
| 7wp | Mestizo | Male | 178 | .5604 | .4396 |
| 7ws | Mestizo | Male | 166 | .3801 | .6199 |
| 7wt | Mestizo | Male | 172 | .8085 | .1915 |
| 7wu | Mestizo | Female | 157 | .4878 | .5122 |
| 7wv | Mestizo | Male | 177 | .6316 | .3684 |
| 7ww | Mestizo | Male | 180 | .3873 | .6127 |
| 7wx | Mestizo | Female | 156 | .4296 | .5704 |
| 7wy | Mestizo | Male | 175 | .4363 | .5637 |
| 7wz | Mestizo | Male | 163 | .1446 | .8554 |
| 7x0 | Mestizo | Male | 167 | .7575 | .2425 |
| 7x1 | Mestizo | Male | 170 | .2288 | .7712 |
| 7x2 | Mestizo | Female | 162 | .4132 | .5868 |
| 7x3 | Mestizo | Male | 170 | .4404 | .5596 |
| 7x4 | Mestizo | Male | 162 | .6423 | .3577 |
| 7x5 | Mestizo | Male | 160 | .2939 | .7061 |
| 7x6 | Mestizo | Male | 168 | .5462 | .4538 |
| 7x7 | Mestizo | Male | 182 | .5964 | .4036 |
| 7x8 | Mestizo | Female | 162 | .6169 | .3831 |
| 7x9 | Mestizo | Male | 180 | .7221 | .2779 |
| 7xb | Mestizo | Male | 180 | .7376 | .2624 |
| 7xd | Mestizo | Male | 168 | .2357 | .7643 |
| 7xf | Mestizo | Male | 168 | .3490 | .6510 |
| 7xl | Mestizo | Male | 175 | .5976 | .4024 |
| 7xm | Mestizo | Male | 157 | .3636 | .6364 |
| 7xp | Mestizo | Male | 161 | .4115 | .5885 |
| 7xx | Mestizo | Male | 170 | .3199 | .6801 |
| 7xy | Mestizo | Male | 158 | .1743 | .8257 |
| 7y3 | Mestizo | Male | 187 | .5545 | .4455 |
| 7y5 | Mestizo | Male | 178 | .9041 | .0959 |
| 7ya | Mestizo | Male | 173 | .6812 | .3188 |
| 7yb | Mestizo | Male | 175 | .7895 | .2105 |
| 7yc | Mestizo | Male | 183 | .8914 | .1086 |
| 7yd | Mestizo | Male | 178 | .9484 | .0516 |
| 7ye | Mestizo | Male | 175 | .4782 | .5218 |
| 7yf | Mestizo | Male | 171 | .5358 | .4642 |
| 7yh | Mestizo | Male | 180 | .7300 | .2700 |
| 7yi | Mestizo | Male | 162 | .3142 | .6858 |
| 7yj | Mestizo | Male | 176 | .5502 | .4498 |
| 7yl | Mestizo | Male | 164 | .6590 | .3410 |
| 7ym | Mestizo | Female | 158 | .2836 | .7164 |
| 7yn | Mestizo | Male | 170 | .6483 | .3517 |
| 7yo | Mestizo | Male | 165 | .2106 | .7894 |
| 7yv | Mestizo | Male | 154 | .3878 | .6122 |
| 7yw | Mestizo | Male | 158 | .2596 | .7404 |
| 7yy | Otomi | Male | 168 | .4977 | .5023 |
| 7z6 | Otomi | Female | 145 | .2737 | .7263 |
| 7z8 | Mestizo | Male | 168 | .3650 | .6350 |
| 7zd | Mestizo | Male | 180 | .4340 | .5660 |
| 7zf | Mestizo | Male | 162 | .0632 | .9368 |
| 7zg | Mestizo | Male | 163 | .4694 | .5306 |
| 7zi | Mestizo | Male | 162 | .3179 | .6821 |
| 7zl | Mestizo | Male | 187 | .7883 | .2117 |
| 7zm | Mestizo | Male | 182 | .8448 | .1552 |
| 7zo | Mestizo | Male | 158 | .2280 | .7720 |
| 7zp | Mestizo | Male | 160 | .6799 | .3201 |
| 7zq | Mestizo | Male | 176 | .5255 | .4745 |
| 7zs | Mestizo | Male | 185 | .6608 | .3392 |
| 7zy | Mestizo | Male | 175 | .7408 | .2592 |
| 7zz | Mestizo | Male | 175 | .5542 | .4458 |
| 800 | Mestizo | Male | 179 | .0730 | .9270 |
| 804 | Mestizo | Male | 179 | .4978 | .5022 |
| 808 | Mestizo | Male | 180 | .9080 | .0920 |
| 80a | Mestizo | Male | 175 | .3391 | .6609 |
| 80c | Mestizo | Male | 160 | .6212 | .3788 |
| 80g | Mestizo | Male | 178 | .8452 | .1548 |
| 80i | Mestizo | Male | 160 | .6255 | .3745 |
| 80k | Mestizo | Male | 175 | .5195 | .4805 |
| 80p | Mestizo | Male | 185 | .4407 | .5593 |
| 80r | Mestizo | Male | 175 | .6390 | .3610 |
| 80u | Mestizo | Male | 178 | .9401 | .0599 |
| 80x | Mestizo | Male | 162 | .4841 | .5159 |
| cd6 | Mestizo | Female | 160 | .4590 | .5410 |
| cei | Mestizo | Male | 160 | .4917 | .5083 |
| cek | Mestizo | Male | 160 | .2380 | .7620 |
| cem | Mestizo | Male | 160 | .3231 | .6769 |
| cen | Mestizo | Male | 158 | .4957 | .5043 |
| cep | Mestizo | Male | 156 | .3283 | .6717 |
| ceq | Mestizo | Male | 180 | .2894 | .7106 |
| ces | Mestizo | Male | 175 | .6730 | .3270 |
| cev | Mestizo | Male | 160 | .4306 | .5694 |
| cf0 | Mestizo | Female | 158 | .5227 | .4773 |
| cf3 | Mestizo | Female | 160 | .4316 | .5684 |
| cff | Mestizo | Male | 163 | .2125 | .7875 |
| cfm | Mestizo | Female | 160 | .4760 | .5240 |
| cfr | Mestizo | Male | 186 | .5059 | .4941 |
| cfs | Mestizo | Male | 175 | .5130 | .4870 |
| cfu | Mestizo | Male | 155 | .0912 | .9088 |
| cfv | Mestizo | Male | 158 | .3552 | .6448 |
| cfy | Mestizo | Male | 163 | .4233 | .5767 |
| cg1 | Mestizo | Female | 155 | .4061 | .5939 |
| cg2 | Mestizo | Female | 160 | .1271 | .8729 |
| cg3 | Mestizo | Male | 157 | .3245 | .6755 |
| cg4 | Mestizo | Male | 160 | .7861 | .2139 |
| cg6 | Mestizo | Male | 164 | .5470 | .4530 |
| cg8 | Mestizo | Male | 164 | .4259 | .5741 |
| cg9 | Mestizo | Male | 167 | .3282 | .6718 |
| cga | Mestizo | Male | 161 | .2202 | .7798 |
| cgc | Mestizo | Male | 162 | .5532 | .4468 |
| cgf | Mestizo | Male | 164 | .5785 | .4215 |
| cgg | Mestizo | Male | 175 | .6731 | .3269 |
| cgi | Mestizo | Male | 170 | .8933 | .1067 |
| cgj | Mestizo | Male | 180 | .6012 | .3988 |
| cgk | Mestizo | Male | 168 | .7003 | .2997 |
| cgm | Mestizo | Female | 148 | .0753 | .9247 |
| cgo | Mestizo | Male | 154 | .1075 | .8925 |
| cgp | Mestizo | Male | 161 | .3718 | .6282 |
| cgq | Mestizo | Male | 175 | .4953 | .5047 |
| cgr | Mestizo | Male | 160 | .5099 | .4901 |
| cgs | Mestizo | Female | 158 | .1542 | .8458 |
| cgt | Mestizo | Male | 168 | .2268 | .7732 |
| cgu | Mestizo | Male | 160 | .1672 | .8328 |
| cgv | Mestizo | Male | 160 | .8747 | .1253 |
| cgw | Mestizo | Female | 152 | .3015 | .6985 |
| cgx | Mestizo | Male | 162 | .4679 | .5321 |
| cgy | Mestizo | Male | 158 | .4983 | .5017 |
| cgz | Mestizo | Male | 160 | .4002 | .5998 |
| ch1 | Mestizo | Male | 158 | .5599 | .4401 |
| ch2 | Mestizo | Male | 156 | .4946 | .5054 |
| ch5 | Mestizo | Male | 160 | .6014 | .3986 |
| ch6 | Mestizo | Male | 159 | .3855 | .6145 |
| ch7 | Mestizo | Male | 160 | .3793 | .6207 |
| chc | Mestizo | Male | 165 | .4911 | .5089 |
| chd | Mestizo | Male | 162 | .1948 | .8052 |
| che | Otomi | Male | 160 | .0890 | .9110 |
| chf | Mestizo | Male | 168 | .4535 | .5465 |
| chg | Mestizo | Male | 160 | .3978 | .6022 |
| chh | Mestizo | Male | 173 | .1835 | .8165 |
| chi | Mestizo | Male | 160 | .4183 | .5817 |
| chm | Mestizo | Male | 175 | .5505 | .4495 |
| chn | Mestizo | Male | 155 | .5957 | .4043 |
| cho | Mestizo | Male | 170 | .6976 | .3024 |
| chp | Mestizo | Male | 160 | .4180 | .5820 |
| chq | Mestizo | Male | 164 | .3477 | .6523 |
| chr | Mestizo | Male | 160 | .2536 | .7464 |
| chs | Mestizo | Male | 168 | .2898 | .7102 |
| cht | Mestizo | Male | 162 | .5725 | .4275 |
| chu | Mestizo | Male | 162 | .2614 | .7386 |
| ci0 | Mestizo | Male | 184 | .8965 | .1035 |
| ci1 | Mestizo | Female | 165 | .7463 | .2537 |
| ci3 | Mestizo | Male | 168 | .2474 | .7526 |
| ci4 | Mestizo | Female | 153 | .3485 | .6515 |
| ci7 | Mestizo | Male | 172 | .5412 | .4588 |
| ci8 | Mestizo | Male | 158 | .3928 | .6072 |
| ci9 | Mestizo | Male | 165 | .0887 | .9113 |
| cia | Mestizo | Male | 160 | .1353 | .8647 |
| cid | Mestizo | Male | 170 | .5194 | .4806 |
| cie | Mestizo | Male | 170 | .5424 | .4576 |
| cif | Mestizo | Male | 170 | .3959 | .6041 |
| cig | Mestizo | Male | 169 | .5562 | .4438 |
| cih | Mestizo | Female | 150 | .3813 | .6187 |
| cii | Mestizo | Male | 168 | .2885 | .7115 |
| cij | Mestizo | Male | 170 | .2932 | .7068 |
| cik | Mestizo | Female | 165 | .3344 | .6656 |
| cil | Mestizo | Male | 170 | .3358 | .6642 |
| cim | Mestizo | Male | 166 | .5217 | .4783 |
| cir | Mestizo | Male | 188 | .4899 | .5101 |
| cis | Mestizo | Male | 162 | .4245 | .5755 |
| ciu | Mestizo | Male | 176 | .4796 | .5204 |
| ciw | Mestizo | Male | 165 | .6046 | .3954 |
| cix | Mestizo | Male | 180 | .6664 | .3336 |
| cj1 | Mestizo | Male | 189 | .5819 | .4181 |
| cj2 | Mestizo | Male | 170 | .4985 | .5015 |
| cj3 | Mestizo | Male | 178 | .4031 | .5969 |
| cj5 | Mestizo | Male | 178 | .7656 | .2344 |
| cj6 | Mestizo | Male | 178 | .7587 | .2413 |
| cj7 | Mestizo | Male | 163 | .7340 | .2660 |
| cj8 | Mestizo | Female | 158 | .5880 | .4120 |
| cja | Mestizo | Male | 171 | .2849 | .7151 |
| cjc | Mestizo | Male | 152 | .2346 | .7654 |
| cjd | Mestizo | Male | 164 | .2892 | .7108 |
| cje | Mestizo | Female | 144 | .0960 | .9040 |
| cjf | Mestizo | Male | 174 | .4539 | .5461 |
| cjk | Mestizo | Male | 162 | .2774 | .7226 |
| cjl | Mestizo | Male | 163 | .5032 | .4968 |
| cjm | Mestizo | Male | 169 | .2475 | .7525 |
| cjp | Mestizo | Male | 162 | .2344 | .7656 |
| cjq | Mestizo | Male | 159 | .5760 | .4240 |
| cjt | Mestizo | Male | 161 | .0901 | .9099 |
| cju | Mestizo | Male | 173 | .8877 | .1123 |
| cjv | Mestizo | Female | 165 | .4959 | .5041 |
| cjw | Mestizo | Male | 185 | .3050 | .6950 |
| cjx | Mestizo | Male | 175 | .3155 | .6845 |
| cjz | Mestizo | Female | 165 | .2790 | .7210 |
| ck0 | Mestizo | Male | 158 | .1991 | .8009 |
| ck1 | Mestizo | Male | 165 | .3378 | .6622 |
| ck2 | Mestizo | Male | 160 | .5444 | .4556 |
| ck4 | Mestizo | Male | 183 | .2729 | .7271 |
| ck9 | Mestizo | Male | 158 | .1429 | .8571 |
| cka | Mestizo | Male | 160 | .1603 | .8397 |
| ckb | Mestizo | Male | 171 | .3592 | .6408 |
| ckc | Mestizo | Male | 164 | .2318 | .7682 |
| ckd | Mestizo | Male | 165 | .2490 | .7510 |
| cke | Otomi | Male | 158 | .2185 | .7815 |
| ckf | Mestizo | Male | 175 | .2882 | .7118 |
| ckg | Mestizo | Male | 170 | .4503 | .5497 |
| cki | Mestizo | Male | 165 | .2414 | .7586 |
| ckj | Mestizo | Male | 172 | .3228 | .6772 |
| ckk | Mestizo | Male | 170 | .7612 | .2388 |
| ckl | Mestizo | Male | 172 | .5816 | .4184 |
| ckm | Mestizo | Male | 165 | .1543 | .8457 |
| cko | Mestizo | Male | 159 | .4716 | .5284 |
| ckq | Mestizo | Male | 167 | .3547 | .6453 |
| ckr | Mestizo | Male | 173 | .6474 | .3526 |
| cks | Mestizo | Male | 170 | .2036 | .7964 |
| ckt | Mestizo | Male | 182 | .6745 | .3255 |
| cku | Mestizo | Male | 180 | .5302 | .4698 |
| ckv | Mestizo | Male | 168 | .2537 | .7463 |
| cky | Mestizo | Male | 160 | .1361 | .8639 |
| cl0 | Mestizo | Male | 163 | .7293 | .2707 |
| cl2 | Mestizo | Male | 170 | .6975 | .3025 |
| cl3 | Mestizo | Male | 160 | .2114 | .7886 |
| cl4 | Mestizo | Female | 150 | .1104 | .8896 |
| cl8 | Mestizo | Male | 164 | .8745 | .1255 |
| cl9 | Mestizo | Male | 160 | .5484 | .4516 |
| cla | Mestizo | Male | 160 | .1149 | .8851 |
| clb | Mestizo | Male | 160 | .0654 | .9346 |
| cld | Mestizo | Male | 160 | .3909 | .6091 |
| cle | Mestizo | Male | .6304 | .3696 | |
| clf | Mestizo | Male | 156 | .3609 | .6391 |
| clg | Mestizo | Male | 162 | .3228 | .6772 |
| clh | Mestizo | Male | 158 | .3679 | .6321 |
| cli | Mestizo | Male | 168 | .2824 | .7176 |
| clj | Mestizo | Male | 160 | .4434 | .5566 |
| cll | Mestizo | Male | 166 | .3035 | .6965 |
| clm | Mestizo | Male | 165 | .6688 | .3312 |
| clo | Mestizo | Female | 173 | .7143 | .2857 |
| clp | Mestizo | Male | 170 | .1587 | .8413 |
| clq | Mestizo | Male | 151 | .4306 | .5694 |
| clu | Otomi | Female | 163 | .2285 | .7715 |
| cly | Mestizo | Male | 170 | .5207 | .4793 |
| clz | Mestizo | Female | 159 | .1337 | .8663 |
| cm0 | Mestizo | Female | 160 | .0803 | .9197 |
| cm1 | Mestizo | Female | 158 | .3935 | .6065 |
| cm2 | Mestizo | Female | 160 | .1687 | .8313 |
| cm3 | Mestizo | Male | 174 | .5492 | .4508 |
| cm4 | Mestizo | Male | 152 | .0764 | .9236 |
| cm5 | Mestizo | Male | 163 | .0531 | .9469 |
| cm6 | Mestizo | Male | 175 | .0951 | .9049 |
| cm8 | Mestizo | Male | 156 | .0731 | .9269 |
| cm9 | Mestizo | Male | 173 | .2500 | .7500 |
| cma | Mestizo | Male | 176 | .8621 | .1379 |
| cmb | Mestizo | Male | 172 | .6413 | .3587 |
| cmc | Mestizo | Male | 153 | .2075 | .7925 |
| cmd | Mestizo | Male | 158 | .4901 | .5099 |
| cme | Mestizo | Female | 148 | .5704 | .4296 |
| cmf | Mestizo | Male | 161 | .3058 | .6942 |
| cmg | Mestizo | Male | 162 | .2842 | .7158 |
| cmi | Mestizo | Male | 165 | .4758 | .5242 |
| cmj | Mestizo | Male | 172 | .2286 | .7714 |
| cmk | Mestizo | Male | 169 | .3634 | .6366 |
| cml | Mestizo | Male | 165 | .1749 | .8251 |
| cmn | Mestizo | Male | 178 | .4953 | .5047 |
| cmo | Mestizo | Male | 160 | .5291 | .4709 |
| cmp | Mestizo | Male | 179 | .5358 | .4642 |
| cms | Mestizo | Male | 173 | .6984 | .3016 |
| cmt | Mestizo | Male | 172 | .2734 | .7266 |
| cmu | Mestizo | Male | 165 | .2056 | .7944 |
| cmv | Mestizo | Male | 172 | .6267 | .3733 |
| cmw | Mestizo | Male | 169 | .0696 | .9304 |
| cn0 | Mestizo | Male | 164 | .3236 | .6764 |
| cn1 | Mestizo | Male | 165 | .8299 | .1701 |
| cn2 | Mestizo | Male | 175 | .7790 | .2210 |
| cn3 | Mestizo | Male | 182 | .8640 | .1360 |
| cn4 | Mestizo | Male | 197 | .5495 | .4505 |
| cn7 | Mestizo | Male | 179 | .8030 | .1970 |
| cn8 | Mestizo | Male | 170 | .4233 | .5767 |
| cn9 | Mestizo | Male | 169 | .4747 | .5253 |
| cnb | Mestizo | Male | 187 | .5456 | .4544 |
| cnc | Mestizo | Male | 170 | .5493 | .4507 |
| cnd | Mestizo | Male | 175 | .6508 | .3492 |
| cne | Mestizo | Male | 178 | .6549 | .3451 |
| cnf | Mestizo | Male | 165 | .3321 | .6679 |
| cnh | Mestizo | Male | 175 | .5416 | .4584 |
| cni | Mestizo | Male | 168 | .2476 | .7524 |
| cnl | Mestizo | Male | 180 | .7485 | .2515 |
| cnm | Mestizo | Male | 187 | .4098 | .5902 |
| cnn | Mestizo | Male | 175 | .3086 | .6914 |
| cno | Mestizo | Male | 178 | .5785 | .4215 |
| cnq | Mestizo | Male | 176 | .7999 | .2001 |
| cnr | Mestizo | Male | 178 | .9065 | .0935 |
| cns | Mestizo | Male | 177 | .7149 | .2851 |
| cnz | Mestizo | Male | 177 | .8719 | .1281 |
| co1 | Mestizo | Male | 185 | .6890 | .3110 |
| co3 | Mestizo | Male | 162 | .7017 | .2983 |
| co6 | Mestizo | Male | 150 | .3495 | .6505 |
| co7 | Mestizo | Male | 182 | .9448 | .0552 |
| co9 | Mestizo | Male | 195 | .9177 | .0823 |
| cod | Mestizo | Male | 176 | .4690 | .5310 |
| coi | Mestizo | Male | 161 | .5186 | .4814 |
| cok | Mestizo | Male | 175 | .8392 | .1608 |
| con | Mestizo | Male | 160 | .6052 | .3948 |
| coo | Mestizo | Male | 175 | .5601 | .4399 |
| cop | Mestizo | Male | 177 | .7855 | .2145 |
| cor | Mestizo | Male | 175 | .6703 | .3297 |
| cou | Mestizo | Male | 170 | .6554 | .3446 |
| cov | Mestizo | Male | 173 | .7484 | .2516 |
| cox | Mestizo | Male | 158 | .5018 | .4982 |
| coy | Mestizo | Male | 172 | .3146 | .6854 |
| coz | Mestizo | Female | 150 | .4496 | .5504 |
| cp1 | Mestizo | Male | 158 | .2756 | .7244 |
| cp2 | Mestizo | Male | 158 | .1475 | .8525 |
| cp4 | Mestizo | Male | 160 | .4598 | .5402 |
| cp5 | Mestizo | Male | 164 | .3057 | .6943 |
| cp6 | Mestizo | Male | 161 | .3064 | .6936 |
| cp7 | Mestizo | Male | 180 | .8891 | .1109 |
| cp8 | Mestizo | Male | 155 | .2484 | .7516 |
| cp9 | Mestizo | Male | 170 | .0968 | .9032 |
| cpc | Mestizo | Male | 160 | .5259 | .4741 |
| cpd | Mestizo | Male | 198 | .7882 | .2118 |
| cpf | Mestizo | Male | 155 | .3186 | .6814 |
| cpi | Mestizo | Male | 178 | .7113 | .2887 |
| cpj | Mestizo | Male | 178 | .1082 | .8918 |
| cpn | Mestizo | Male | 182 | .5435 | .4565 |
| cpr | Mestizo | Male | 176 | .5803 | .4197 |
| cps | Mestizo | Male | 184 | .9105 | .0895 |
| cpv | Otomi | Male | 168 | .4396 | .5604 |
| cpw | Mestizo | Male | 140 | .1643 | .8357 |
| cpz | Otomi | Female | 145 | .2631 | .7369 |
| cq0 | Mestizo | Male | 172 | .5564 | .4436 |
| cq3 | Mestizo | Male | 180 | .4671 | .5329 |
| cq4 | Mestizo | Male | 182 | .6424 | .3576 |
| cq7 | Mestizo | Male | 160 | .3037 | .6963 |
| cq9 | Mestizo | Male | 180 | .3245 | .6755 |
| cqd | Mestizo | Male | 186 | .5922 | .4078 |
| cqe | Mestizo | Male | 160 | .3377 | .6623 |
| cqh | Mestizo | Male | 160 | .3452 | .6548 |
| cqi | Mestizo | Male | 175 | .7030 | .2970 |
| cqk | Mestizo | Male | 179 | .7167 | .2833 |
| cql | Mestizo | Male | 162 | .5204 | .4796 |
| cqm | Mestizo | Male | 160 | .2244 | .7756 |
| cqv | Mestizo | Male | 163 | .4053 | .5947 |
| cqy | Mestizo | Male | 150 | .1871 | .8129 |
| cr0 | Mestizo | Male | 160 | .6213 | .3787 |
| cr1 | Mestizo | Male | 160 | .4885 | .5115 |
| cr2 | Mestizo | Male | 158 | .6159 | .3841 |
| cr3 | Mestizo | Male | 177 | .3927 | .6073 |
| cr6 | Mestizo | Male | 180 | .6227 | .3773 |
| cr9 | Mestizo | Male | 172 | .1045 | .8955 |
| cra | Mestizo | Male | 178 | .4386 | .5614 |
| crb | Mestizo | Male | 163 | .1118 | .8882 |
| crc | Mestizo | Male | 180 | .8329 | .1671 |
| cre | Mestizo | Male | 162 | .6336 | .3664 |
| crg | Mestizo | Male | 177 | .3747 | .6253 |
| cri | Mestizo | Male | 186 | .5990 | .4010 |
| crn | Mestizo | Male | 178 | .4081 | .5919 |
| crq | Mestizo | Male | 177 | .3416 | .6584 |
| crr | Mestizo | Male | 175 | .8971 | .1029 |
| cs1 | Mestizo | Male | 178 | .8305 | .1695 |
| cs6 | Mestizo | Male | 158 | .2088 | .7912 |
| csd | Mestizo | Male | 153 | .4577 | .5423 |
| csf | Mestizo | Male | 183 | .3101 | .6899 |
| csg | Mestizo | Male | 161 | .4497 | .5503 |
| csi | Mestizo | Male | 185 | .8559 | .1441 |
| csr | Mestizo | Male | 175 | .9115 | .0885 |
| css | Mestizo | Male | 167 | .5829 | .4171 |
| cst | Mestizo | Male | 160 | .8267 | .1733 |
| csw | Mestizo | Male | 168 | .6293 | .3707 |
| ct0 | Mestizo | Male | 160 | .4537 | .5463 |
| ct2 | Mestizo | Male | 187 | .1484 | .8516 |
| cta | Mestizo | Male | 184 | .8225 | .1775 |
| cti | Mestizo | Male | 179 | .3820 | .6180 |
| ctk | Mestizo | Male | 185 | .5056 | .4944 |
| ctl | Mestizo | Male | 155 | .4277 | .5723 |
| ctm | Mestizo | Male | 191 | .8045 | .1955 |
| ctn | Mestizo | Male | 175 | .7304 | .2696 |
| ctq | Mestizo | Male | 186 | .6809 | .3191 |
| cts | Mestizo | Male | 177 | .5180 | .4820 |
| ctt | Mestizo | Male | 176 | .9071 | .0929 |
| ctv | Mestizo | Male | 178 | .6137 | .3863 |
| ctx | Mestizo | Male | 176 | .9351 | .0649 |
| cu2 | Mestizo | Male | 187 | .6762 | .3238 |
| cu3 | Mestizo | Male | 168 | .2758 | .7242 |
| cu4 | Mestizo | Male | 170 | .2379 | .7621 |
| cu5 | Mestizo | Male | 180 | .6858 | .3142 |
| cu7 | Mestizo | Male | 175 | .5640 | .4360 |
| cu8 | Mestizo | Male | 181 | .6415 | .3585 |
| cu9 | Mestizo | Male | 183 | .5631 | .4369 |
| cuc | Mestizo | Male | 175 | .6701 | .3299 |
| cuh | Mestizo | Male | 176 | .6290 | .3710 |
| cuk | Mestizo | Male | 180 | .3272 | .6728 |
The admixture model used in the structure program assumes a unimodal distribution of individual admixture proportions. However, we found that our inclusion of small numbers of Caucasian and Otomi samples in the analysis did not significantly perturb the admixture estimates for the Mestizo samples. A separate analysis of just the Mestizo samples, which might be expected to better fit the unimodal admixture model, yielded admixture proportions that had a correlation of 0.9994 with the full analysis (data not shown). Thus, this analysis seems to be robust against some limited misspecification of the admixture model.
Association of Ancestry with Height
We compared the inferred ancestry information for individuals selected to represent the tallest and shortest 25% of male Mestizo subjects. Of the samples that were genotyped, we identified 164 short and 166 tall individuals. Height is strongly correlated with the inferred proportion of cluster A ancestry (fig. 2), and many spurious allele-frequency differences occur solely as a result of differences in ancestry between the tall and short groups (table 4, all samples). This is an extremely stratified population, and there are multiple SNPs with χ2-test P values of <10−8. This level of significance would exceed genomewide significance thresholds for 1 million independent SNP association tests with conservative adjustment for multiple testing, clearly a problem if these groups were to be used in the type of genomewide association study described above.
Figure 2.
Distribution of ancestry versus height categories. Density distributions for the inferred fraction of subjects with cluster A ancestry are shown for 164 short and 166 tall subjects. Each tick mark represents the fractional ancestry of an individual subject.
Table 4.
Association Test Results for Height in 275 SNPs
| Number of SNPs with χ2 Test Statistics | |||||
|---|---|---|---|---|---|
| Data Set | P<.0001 | P<.001 | P<.01 | P<.1 | q<.05 |
| Expected | 0 | 0 | 2.75 | 27.5 | 0 |
| All samples | 22 | 38 | 69 | 126 | 94 |
| Random subset | 10 | 20 | 44 | 106 | 62 |
| Matched subset | 0 | 0 | 7 | 35 | 0 |
| Leave out 20% | 0 | 0 | 6 | 39 | 0 |
| Linear adjusted | 0 | 0 | 4 | 44 | 0 |
Matching Based on Average Ancestry Estimates
We composed new groups using subsets of the tall and short individuals, so the groups would have the same average proportions of ancestry in clusters A and B, while retaining as many samples as possible. This involved removing tall samples with the highest proportions of cluster A ancestry and short samples with the lowest proportions of cluster A ancestry. We were able to retain 98 tall samples and 98 short samples with this matching strategy. Ancestry proportions before and after matching are shown in table 3. For a direct comparison, 98 samples were also selected at random from the lists of tall and short samples. The random and matched groups were tested for significant allele-frequency differences (table 4, random and matched subsets). Matching removed most evidence for population structure. An overall test for stratification that was based on the sum of χ2 statistics (Pritchard and Rosenberg 1999) for the matched set gave a P value of ∼.005, versus ∼10−71 for the randomly selected set. The distribution of P values for the 275 SNPs is more nearly uniform for the matched groups (fig. 3), and no markers showed significant association after controlling the false-discovery rate.
Table 3.
Average Proportion of Ancestry in Cluster A, for Tall and Short Groups
| No. of Subjectsin | Proportion of Ancestryin Cluster A in | |||
|---|---|---|---|---|
| Data Set | Tall Group | Short Group | Tall Group | Short Group |
| All samples | 166 | 164 | .62 | .36 |
| Matched subset | 98 | 98 | .48 | .48 |
Figure 3.
Cumulative distribution of P values for 275 SNPs, for the random and ancestry-matched subsets of tall and short subjects. In the absence of population structure, the P values should be uniformly distributed, and their cumulative distribution should be a straight line from (0,0) to (1,1). The random subset shows an excess of small P values, whereas the matched subset has a nearly uniform distribution.
In the previous analysis, the SNPs used to test for associations were the same ones used for the stratification analysis. Although the stratification analysis is blind to the phenotype, in principle, this analysis could underestimate the residual population structure expected for other SNPs not included in the stratification analysis. To address this, we split the 275 SNPs into five random subsets of 55. For each subset, we performed a stratification analysis of the other 220 SNPs, matched tall and short groups on the basis of that analysis, and then tested for association in the 20% that had been left out. Then we combined results for all the subsets, yielding a test result for each SNP stratified by use of what was, for that SNP, an independent set of data. Results (table 4, leave-out-20% data set) were essentially the same as for matching on all 275 SNPs, and there were no significant associations.
Matching Based on an Ancestry-Adjusted Phenotype
An alternative approach to eliminating stratification for a quantitative trait is to define groups on the basis of a phenotype that has been adjusted to remove effects of ancestry differences. We performed a linear regression of height against the inferred fraction of cluster A ancestry for the male Mestizo subjects in our study and determined that a 10% increase in cluster A ancestry corresponded, on average, to a 1.8-cm increase in height. We adjusted height by subtracting out this contribution, and we selected the tallest and shortest 98 individuals on the basis of the adjusted phenotype. We did not see any significant associations using these groups (table 4, linear adjusted).
In principle, adjusting for ancestry should yield a cleaner phenotype and a more powerful study design than the simple strategy of matching the mean ancestry of case and control groups. Comparing the distributions of height and inferred ancestry for the two designs (fig. 4), the regression design includes fewer individuals with relatively mild ancestry-adjusted phenotypes and intermediate ancestry coefficients, and more individuals with extreme ancestry-adjusted phenotypes and ancestry coefficients. The regression design may be more challenging to implement, however, if it requires collecting genotype data for additional individuals to accurately determine the relationship between phenotype and ancestry.
Figure 4.
Comparison of a matching strategy with independently determined cutoffs for height and ancestry (A) and a strategy based on a linear regression of height against ancestry (B). The samples retained from tall and short subjects by use of each method are shown as blackened circles, and excluded samples are shown as unblackened circles. The regression method results in inclusion of the tallest and shortest individuals within any narrow window of ancestry values.
Effects of Population Structure on Pooled Genotyping
In many if not most association studies, if the target population is relatively homogeneous, or if there is little confounding between the target phenotype and ancestry, then careful pool matching may not be necessary (e.g., Ardlie et al. 2002). Thus, it is useful to have a way of quantifying the practical impact of population structure on an association study, to decide when corrective action is needed. Significance tests are not appropriate for this purpose because they do not directly measure the magnitude of an effect. One approach is to model population structure as one of various sources of error that lead to an increase in the false-positive rate. If the effect of population structure is determined to be small compared with other known sources of experimental error, then correcting for it will have limited benefit.
We examined the behavior of the sum of χ2 statistics for association tests with data from the tall and short groups matched for average ancestry, as various amounts of random noise were added to allele frequencies in the two groups. Genotypes for each SNP were first permuted to eliminate any residual disequilibrium, so we essentially only preserved overall SNP allele frequencies from the original data. The allele frequencies for each pool were then perturbed by a normally distributed error term, with standard deviation specified in units of allele frequency (fig. 5). The sum statistics for the unpermuted random and matched groups (table 5)—that is, 928 and 338—are comparable to permuted data with additional experimental error of ∼5% and ∼1%, respectively. Additional error on the order of 1% seems tolerable for currently available pooled genotyping technologies, which generally cannot determine allele frequencies with better accuracy than that (Sham et al. 2002). This approach could be combined with estimates of experimental variance components (Barratt et al. 2002) to produce more realistic end-to-end power estimates for pooled genotyping study designs.
Figure 5.
Effect of simulated experimental error on an overall population-structure test statistic. We simulated the effect of experimental error by adding normally distributed noise to allele-frequency estimates in permuted copies of the genotype data for the matched tall and short groups. The overall test statistic is the sum of resulting χ2 statistics for the 275 individual SNPs; this is expected to follow a χ2 distribution, with 275 df. We show results for 20 separate permutations for each value of the noise parameter.
Table 5.
Overall Measures of Population Structure for Height Pools
| Data Set | χ2 Sum | P Value | λa | ESS |
|---|---|---|---|---|
| All samples | 1,380 | 2 × 10−146 | 4.9 | 34 |
| Random subset | 928 | 8 × 10−72 | 3.6 | 27 |
| Matched subset | 338 | 5 × 10−3 | 1.1 | 89 |
| Leave out 20% | 345 | 2 × 10−3 | 1.4 | 70 |
| Linear adjusted | 313 | 6 × 10−2 | 1.3 | 75 |
Genomic control (Devlin and Roeder 1999) provides another approach to estimating the magnitude of the effect of population structure in an association study. In this approach, rather than modeling structure in a population, its effects are measured by the inflation of test statistics for markers that, in aggregate, should not show evidence for association. We estimated the variance-inflation factors (λ) due to population structure for each set of tall and short groups by use of this approach. One interpretation of the variance inflation is as a reduction in effective sample size (ESS), which we estimate here as (N / λ), where N is the original sample size (table 5). Genomic control would effectively maintain a desired type I error rate in the presence of population structure in this example; however, it does so at a substantial cost in the ESS and, hence, power to detect causal associations. Our results show that matching to mitigate the impact of population structure can substantially boost the ESS, despite the reduction in raw sample count.
Discussion
Our results indicate that relatively simple matching strategies can effectively control for population stratification in case-control association studies, for a phenotype with a very large ancestry effect in an admixed population. The genotyping can be efficiently implemented in the laboratory in a high-throughput setting, with a single generic SNP genotyping array carrying around 300 uniformly distributed SNPs that are chosen without regard to their allele frequencies in specific target populations. We have now processed many thousands of these arrays.
Although we chose to use the structure program to infer admixture proportions, other methods are available, including the ADMIXMAP program (available on the Genetic Epidemiology Group Web site) (McKeigue et al. 2000; Hoggart et al. 2003), which may offer significant benefits in some situations. The admixture model in structure suffers from a theoretical deficiency (Pritchard et al. 2000_a_; Hoggart et al. 2003), in that it does not permit specification of prior allele-frequency information for the ancestral populations and thus cannot disambiguate between symmetric modes that differ only in the labels assigned to clusters. Also, interpretation of the admixture coefficients relies on the sampler only exploring one of these symmetric modes. In our analysis, we verified that individual structure runs consistently settled in one (randomly selected) mode, and we could easily determine consistent cluster labels when comparing results across multiple runs. The matching strategies we describe are also invariant under permutations of the cluster labels. Still, it is possible that the structure sampler may have more trouble in situations with more clusters or less clearly separated ones.
In the context of a pooled genotyping screen, absolute control of population structure is probably not required in many cases. It is probably only necessary to ensure that the incremental increase in variance due to population differences between case and control pools is small compared with other sources of variance in the genotyping experiment. In an association study design consisting of an initial screen of many markers by pooled genotyping followed by individual genotyping of candidates, there should be more tolerance for spurious associations in the pooled step. In these cases, a test for population structure on a representative subset of cases and controls may be sufficient to place bounds on the impact of population stratification on the entire study, thus avoiding unnecessary recruitment or individual genotyping effort.
A complete association study would consist of three phases. First, some or all samples would be individually genotyped to ascertain their population structure using our array of ∼300 SNPs. On the basis of those results, and constrained by the form of the phenotype and its ascertainment method, a strategy for mitigating population structure would be selected and validated using the available genotype data. Both of our matching strategies require genotyping some individuals who will end up being excluded from the matched case and control pools. The second phase would consist of pooled genotyping of many SNPs in replicate experiments. In a third phase, candidate SNPs would be selected for individual genotyping on the basis of the pooled data. Samples originally excluded from the pools could be genotyped at this point and could be analyzed using one of the structured association approaches. Genomic control could also be used to adjust significance tests for any residual population structure left in the matched pools.
The matching strategies we discuss here were designed for whole-genome association studies for which we required that a solution could not increase the experimental effort required at the pooled genotyping stage. This constraint (a practical, economic one) severely limited the range of solutions that we could consider. Another approach to controlling for population structure would be to perform a stratified analysis of subpools composed of individuals of similar ancestry. For experimental designs permitting many replicates, this may be a useful strategy for discrete traits that cannot be adjusted to remove ancestry effects. Such a design would allow all individuals to be included in the pooled analysis; however, strata with very unbalanced representation of the trait values would have somewhat lower informativeness for equal experimental effort. The number of strata required to account for most of the variance in ancestry would multiply the experimental effort required for allele-frequency determination, since this would be orthogonal to any replication required to characterize experimental variance.
The strategies we describe can be extended to more complex structured populations. For either admixed populations or populations composed of several unadmixed groups, our approach would be either to match the average genetic contribution of each empirically identified cluster in the case and control groups by excluding samples, or to use multivariate regression to determine an ancestry-adjusted phenotype for each individual on the basis of the individual's inferred cluster-membership proportions. In the absence of admixture, a multiethnic pooled study would be most sensitive for detecting loci that account for phenotypic variation in all of the included populations; such a study would be insensitive for loci accounting for fixed differences between populations.
Admixed populations are attractive targets for association studies because these groups should show more linkage disequilibrium over larger physical distances (Chakraborty and Weiss 1988). If the admixture is between populations with significantly different genetic predispositions to a target phenotype, then heritability of a trait in the admixed population may also be higher than in the more homogeneous ancestral populations. Although linkage-based admixture mapping (McKeigue 1998) can be a more efficient approach for identifying loci that specifically explain phenotypic variance between populations, an association study in an admixed population has the ability to detect loci that explain variance either between or within populations. Pooled allele-frequency differences would not distinguish within- from between-population associations, but these could be resolved later by modeling ancestry effects at associated loci by use of individual genotyping data. The groups used in this study are small, and larger sample sizes would be required for a whole-genome association study of a complex multigenic phenotype. The impact of stratification would be correspondingly larger for more realistic study designs, because although sampling variation in allele frequencies becomes smaller for larger pool sizes, the variance due to population stratification does not. Careful management of population structure is likely to be an important component of future whole-genome association studies.
Acknowledgments
We wish to acknowledge Dr. Raul Bernal Reyes (Instituto Mexicano del Seguro Social, Pachuca), Dr. Armando Diaz Belmont (Hospital General de México), and A. Christian Perez Pruna and Marta Garcia Sandoval (Instituto Nacional de Nutrición Salvador Zubirán), for their invaluable efforts to recruit subjects for this study. We wish to thank Robin Li, Coleen Hacker, Naiping Shen, Claire Marjoribanks, and Albert Yee for excellent technical assistance and overall contribution to this work. We thank Alberto Cevallos and Jesse Hsu, for helping to establish our Mexican collaboration, and Pascual Starink, for assistance with sample tracking. We also thank Kelly Frazer and two anonymous reviewers for helpful comments on the manuscript.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for ss12673803–ss12674077)
- Genetic Epidemiology Group Web Site, http://www.lshtm.ac.uk/eu/genetics/ (for ADMIXMAP software)
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for BLAST search engine)
- NCBI Home Page, http://www.ncbi.nlm.nih.gov/
- Pritchard Lab, http://pritch.bsd.uchicago.edu/ (for the structure program)
- Q-VALUE Software, http://faculty.washington.edu/~jstorey/qvalue/
- R Project for Statistical Computing, http://www.r-project.org/
- RepeatMasker Web Server, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
References
- Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case-control studies. Am J Hum Genet 71:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Barratt BJ, Payne F, Rance HE, Nutland S, Todd JA, Clayton DG (2002) Identification of the sources of error in allele frequency estimations from pooled DNA indicates an optimal experimental design. Ann Hum Genet 66:393–405 10.1046/j.1469-1809.2002.00125.x [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ (2003) Population stratification and spurious allelic association. Lancet 361:598–604 10.1016/S0140-6736(03)12520-2 [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Weiss KM (1988) Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA 85:9119–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 [DOI] [PubMed] [Google Scholar]
- Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D (1991) Light-directed, spatially addressable parallel chemical synthesis. Science 251:767–773 [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding in genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comp Graph Stat 5:299–314 [Google Scholar]
- Knowler WC, Williams RC, Pettitt DJ, Steinberg AG (1988) GM 3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. Am J Hum Genet 43:520–526 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 10.1038/9642 [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork N (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BTN, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SPA, Cox DR (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 10.1126/science.1065573 [DOI] [PubMed] [Google Scholar]
- McKeigue PM (1998) Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations by conditioning on parental admixture. Am J Hum Genet 63:241–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM, Carpenter JR, Parra EJ, Shriver MD (2000) Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet 64:171–186 10.1046/j.1469-1809.2000.6420171.x [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000_a_) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000_b_) Association mapping in structured populations. Am J Hum Genet 67:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB (2001) Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol 20:4–16 [DOI] [PubMed] [Google Scholar]
- Risch N (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 10.1038/35015718 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Satten GA, Flanders WD, Yang Q (2001) Accounting for unmeasured population structure in case-control studies of genetic association using a novel latent-class model. Am J Hum Genet 68:466–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham P, Bader JS, Craig I, O’Donovan M, Owen M (2002) DNA pooling: a tool for large-scale association studies. Nat Rev Genet 3:862–871 10.1038/nrg930 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES 4th (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289. 10.1038/90135 [DOI] [PubMed] [Google Scholar]
- Weir B (1996) Genetic data analysis II. Sinauer, Sunderland, MA [Google Scholar]