Mutations in the VLGR1 Gene Implicate G-Protein Signaling in the Pathogenesis of Usher Syndrome Type II (original) (raw)
Abstract
Usher syndrome type II (USH2) is a genetically heterogeneous autosomal recessive disorder with at least three genetic subtypes (USH2A, USH2B, and USH2C) and is classified phenotypically as congenital hearing loss and progressive retinitis pigmentosa. The VLGR1 (MASS1) gene in the 5q14.3-q21.1 USH2C locus was considered a likely candidate on the basis of its protein motif structure and expressed-sequence-tag representation from both cochlear and retinal subtracted libraries. Denaturing high-performance liquid chromatography and direct sequencing of polymerase-chain-reaction products amplified from 10 genetically independent patients with USH2C and 156 other patients with USH2 identified four isoform-specific VLGR1 mutations (Q2301X, I2906FS, M2931FS, and T6244X) from three families with USH2C, as well as two sporadic cases. All patients with VLGR1 mutations are female, a significant deviation from random expectations. The ligand(s) for the VLGR1 protein is unknown, but on the basis of its potential extracellular and intracellular protein-protein interaction domains and its wide mRNA expression profile, it is probable that VLGR1 serves diverse cellular and signaling processes. VLGR1 mutations have been previously identified in both humans and mice and are associated with a reflex-seizure phenotype in both species. The identification of additional VLGR1 mutations to test whether a phenotype/genotype correlation exists, akin to that shown for other Usher syndrome disease genes, is warranted.
Usher syndrome is a clinically and genetically heterogeneous recessive disease with three clinical subtypes. The subtypes are differentiated on the basis of the severity of hearing loss and presence or absence of vestibular abnormalities. Patients with Usher syndrome type I (USH1) present with profound congenital hearing loss, vestibular areflexia (in most cases), and a progressive retinal degeneration, with impaired night vision and decreased visual fields diagnosed as retinitis pigmentosa (RP). Usher syndrome type II (USH2 [MIM 276900]) have a moderate-to-severe sensorineural hearing loss that is stable in most cases, normal vestibular function, and RP, whereas patients with type III (USH3) present a moderate sensorineural hearing loss with progression to acquired deafness, progressive vestibular dysfunction, and RP. Currently, 11 loci have been identified for Usher syndrome (Kimberling et al. 1990, 1992; Kaplan et al. 1992; Smith et al. 1992; Sankila et al. 1995; Wayne et al. 1996, 1997; Chaib et al. 1997; Hmani et al. 1999; Pieke-Dahl et al. 2000; Mustapha et al. 2002); families that do not link to the known loci indicate additional genes exist. Of the 11 loci, 7 genes have been found with Usher syndrome mutations (Weil et al. 1995, 2003; Eudy et al. 1998; Bitner-Glindzicz et al. 2000; Verpy et al. 2000; Ahmed et al. 2001; Alagramam et al. 2001; Bolz et al. 2001; Bork et al. 2001; Joensuu et al. 2001).
Previously, we had localized the USH2C gene to a 20-cM region on chromosome 5q14.3–5q21.1 (Pieke-Dahl et al. 2000). Reference mRNAs in this linkage interval were obtained from the UCSC Genome Browser Web site and were prioritized for candidate-gene mutation screening. The very large G-coupled receptor gene (VLGR1) was considered a likely USH2C candidate (Burgess 2001; Staub et al. 2002), with EST representation in both human fetal retina and cochlea (obtained from the National Eye Institute Gene Bank Web site) subtracted libraries. Use of the conserved domain architecture retrieval (CDART) tool on VLGR1 (Geer et al. 2002) revealed a motif architecture similar to the cadherin superfamily of integral membrane proteins. This superfamily includes CDH23 and PCDH15, which are the genes mutated in USH1D (MIM 601067) and USH1F (MIM 602083), respectively. In addition, VLGR1 and the USH2A gene product usherin exhibit a high degree of similarity in their respective pentraxin (PTX) homology domains. On the basis of this evidence, we sought to implicate the VLGR1 gene in the pathogenesis of USH2.
Denaturing high-performance liquid chromatography (DHPLC) and direct sequencing were used to detect mutations in PCR-amplified genomic DNA from 10 probands with USH2C (MIM 605472) for 90 VLGR1 exons. The 10 affected probands were members of families consistent with genetic linkage to 5q14-21 (Pieke-Dahl et al. 2000). We had previously obtained informed consent and had extracted DNA from peripheral whole blood of the study subjects according to standard conditions (Puregene). Patients with a clinical diagnosis of USH2 were ascertained through physician referral and self-referral. The study was conducted with the approval of the Boys Town National Research Hospital (BTNRH) institutional review board and met all ethical requirements and Health Insurance Portability and Accountability Act (HIPAA) standards. DNAs from an additional 152 unrelated patients with USH2 without an identifiable USH2A mutation on chromosome 1q41 were used to screen for specific VLGR1 mutations identified from either the mutation screen of the 10 probands with USH2C or from those VLGR1 mutations reported elsewhere (Nakayama et al. 2002).
Genomic VLGR1 PCR primer pairs were designed and synthesized commercially (sequences available upon request [Integrated DNA Technologies]). PCR used 20 ng genomic DNA, 0.2 μM primer, 2.5 mM MgCl2, 200 μM dNTP, in 1 × PCR buffer with 0.5 units AmpliTaq Gold (PE Biosystems) in 25 μl total volume. The initial PCR cycle was 95°C for 8 min and 30 s, followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final step of 70°C for 10 min. BAC clones spanning the VLGR1 genomic interval (CTD-2034A10, exons 1–4; RP11-29K14, exons 2–42; RP11-62E10, exons 18–73; RP11-3B6, exons 51–83; CTD-2001K4, exons 84 and 85; CTD-2266L18, exons 86 and 87; RP11-414H23, exons 88–90) were identified and used as a control template to evaluate PCR-product heteroduplexes that were separated by DHPLC; these clones were also used in direct PCR sequence analysis.
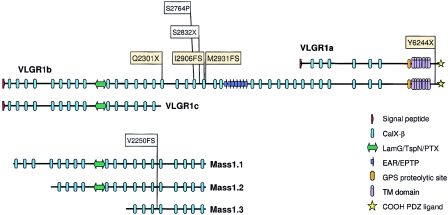
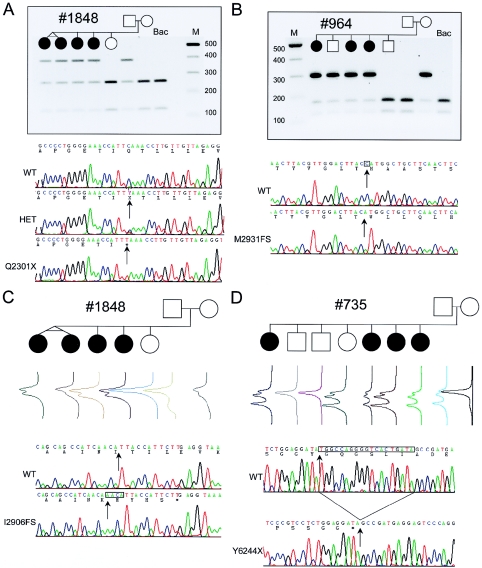
Three human VLGR1 mRNA isoforms are expressed (VLGR1a, VLGR1b, and VLGR1c), of which only VLGR1b and VLGR1c isoforms are expressed in mouse (McMillan et al. 2002) (fig. 2). Of 72 nucleotide changes discovered in VLGR1 (see table A [online only]), 4 mutations would prematurely terminate VLGR1a and VLGR1b protein translation (fig. 1). The VLGR1 6901C→T (Q2301X) and 8716-17insAACA (I2906FS) mutations were found in repulsion exclusively in the members of family 1848 who were affected with USH2C (fig. 1_a_ and 1_c_). Family 964 (with USH2C) was found to segregate a maternal VLGR1 8790delC (M2931FS) mutation (fig. 1_b_). A paternal 19-bp deletion (18732–18750del19bp; T6244X) segregates appropriately with the phenotype in family 735 (fig. 1_d_). Families 735 and 964 have been shown elsewhere to genetically link to the 5q14.3-q21 region, with all affected siblings sharing family-specific 5q 14.3-q21 haplotypes (Pieke-Dahl et al. 2000). No other potential VLGR1 variant was found associated with the maternal allele of family 735; however, a paternal IVS75-15G→A mutation absent in 190 control chromosomes was found segregating with the phenotype in family 964 (data not shown). It remains to be determined whether and how IVS75–15G→A affects VLGR1 expression. Screening for USH2C mutations in 152 patients with USH2 revealed the Q2301X mutation in two other unrelated patients with sporadic USH2. Table 1 presents a clinical summary of all patients with USH2C who harbor VLGR1 mutations. A total of five independent probands with USH2 and eight affected siblings were found to bear pathological VLGR1 mutations. Two probands have pathological mutations identified in both alleles; three occult mutant alleles either have escaped detection by the methodology employed, lie in unscreened regions of the gene, or have been detected but not appreciated as disease alleles. None of the four VLGR1 mutations was observed in 190 control chromosomes. These results establish VLGR1 as the USH2C gene and implicate the protein, which is the largest cell surface receptor known (McMillan et al. 2002), in the pathogenesis of USH2.
Figure 2.
Domain structure of VLGR1/MASS1 isoforms. Conceptual translation of VLGR1b mRNA reveals a protein with a large ectodomain with 35 Calx-β domains, 1 LamG/TspN/PTX homology domain (Beckmann et al. 1998), 7 EAR/EPTP repeats forming a putative β-propeller folding domain (Scheel et al. 2002; Staub et al. 2002), a GPS, a B-family 7TM domain, a putative intracellular tail with multiple potential serine phosphorylation sites, and a putative PDZ-binding COOH-terminal end (McMillan et al. 2002). The PTX domains of VLGR1 and the USH2A protein usherin are most similar to one another, with a BLASTP score of 2e-09. The locations of human USH2C (yellow) and febrile-seizure (FS; gray)–associated mutations are shown on VLGR1a, VLGR1b, and VLGR1c isoforms. The mouse audiogenic reflex seizure model Frings and the age-related hearing loss model BUB/BnJ are both homozygous for the V2250FS mutation and are noted on Mass1.1, Mass1.2, and Mass1.3 isoforms (Skradski et al. 2001).
Table A.
VLGR1 DNA Variants, Including Pathological USH2C Mutations[Note]
| Frequency in Present Study | |||||||
|---|---|---|---|---|---|---|---|
| Exon | Mutation | Codon | USH2C | USH2Af | Controls | SNP Database No. | Reference |
| 4 | 380T→G | L127R | 1/20 | ||||
| 7 | 746G→A | R249Ka | 1/20 | 0/188 | |||
| 9 | IVS9+54C→T | 4/20 | |||||
| 10 | IVS10+53delA | 4/20 | |||||
| 12_13 | IVS11-19G→T | 4/20 | 1344030 | Nakayama et al. 2002 | |||
| 12_13 | IVS12+8C→T | 3/20 | 2366773 | Nakayama et al. 2002 | |||
| 15 | IVS14-34C→G | 4/20 | Nakayama et al. 2002 | ||||
| 17 | 3141A→G | A1047A | 3/20 | 950692 | Nakayama et al. 2002 | ||
| 17 | 3279G→T | L1093F | 5/20 | 2366777 | Nakayama et al. 2002 | ||
| 22 | IVS22+87T→A | 6/20 | 1028191 | ||||
| Ah | IVS22 +234A→G | 6/20 | 1028192 | ||||
| Ah | IVS22+244A→G | 1/20 | |||||
| 24 | 5304G→A | E1768E | 1/20 | 1/190 | |||
| Bh | IVS25+780C→T | 3/20 | |||||
| 28 | IVS27-23T→C | 5/20 | Nakayama et al. 2002 | ||||
| 28 | 5780C→T | T1927M | 1/20 | ||||
| 28 | 5851A→G | I1951V | 5/20 | Nakayama et al. 2002 | |||
| 28 | 5953A→G | N1985D | 3/20 | Nakayama et al. 2002 | |||
| 28 | 5960C→T | P1987L | 5/20 | Nakayama et al. 2002 | |||
| 28 | 6012G→T | L2004F | 3/20 | 20/170 | Nakayama et al. 2002 | ||
| 30 | 6695A→G | Y2232C | 6/20 | 36/144 | Nakayama et al. 2002 | ||
| 31 | 6901C→T | Q2301X | 1/20 | 3/282 | 0/190 | ||
| 32 | IVS31-10G→A | 8/20 | Nakayama et al. 2002 | ||||
| 32 | c.7034A→G | N2345S | 8/20 | 2366926 | Nakayama et al. 2002 | ||
| 33 | 7135G→A | G2379A | 0/20 | 1/190 | |||
| 33 | 7155G→T | L2385L | 0/20 | 1/190 | |||
| 33 | 7176C→T | S2392Sb | 1/20 | 0/190 | |||
| 33 | 7179C→T | D2393D | 0/20 | 2/190 | |||
| 33 | 7206G→A | E2402E | 5/20 | 69/190 | Nakayama et al. 2002 | ||
| 33 | 7751G→A | S2584N | 5/20 | 1878878 | Nakayama et al. 2002 | ||
| 33 | IVS33+27A→C | 7/20 | |||||
| 34 | IVS34+82T→C | 6/20 | |||||
| 36 | IVS35-60G→A | 0/20 | 1/190 | ||||
| 36 | 8291C→T | S2764L | 1/20 | 6/190 | Nakayama et al. 2002 | ||
| 37 | 8495C→A | S2832X | 0/20 | 0/276 | Nakayama et al. 2002 | ||
| 37 | 8407G→A | A2803T | 1/20 | 1/190 | |||
| 37 | 8538T→G | L2846L | 4/20 | 21/190 | Nakayama et al. 2002 | ||
| 38 | IVS37-60G→T | 6/20 | 1160121 | ||||
| 38 | IVS37-13C→T | 0/20 | 1/28 g | Nakayama et al. 2002 | |||
| 38 | 8716-17insAACA | I2906FS | 1/20 | 0/276 | 0/190 | ||
| 38 | IVS38+10insT | 6/20 | |||||
| 39 | 8790delC | M2931FS | 1/20 | 0/280 | 0/190 | ||
| 39 | ISV39+50A→→C | 1/20 | 6/190 | ||||
| 41 | IVS41+57C→T | 2/20 | |||||
| 45 | 9650C→T | A3217V | 1/20 | 3/190 | |||
| 45 | 9743G→A | G3248D | 4/20 | 21/190 | |||
| 47 | IVS46-35C→A | 6/20 | |||||
| 47 | 9927T→G | P3309P | 8/20 | ||||
| 48 | IVS48+26G→T | 6/20 | |||||
| 49 | 10411G→A | E3471K | 3/20 | 2366928 | |||
| 51 | 10577T→C | M3526T | 1/20 | 3/182 | |||
| 51 | IVS51+9A→G | 0/20 | 2/182 | ||||
| 56 | IVS55-3insC | 8/20 | 90/190 | ||||
| 56 | 11599G→A | E3867K | 8/20 | 75/190 | |||
| 56 | 11682C→T | P3894P | 8/20 | 92/190 | 2438349 | ||
| 59 | 12269C→A | T4090Nc | 1/20 | 0/166 | |||
| 64 | IVS63-36A→T | 2/20 | 19/190 | ||||
| 64 | IVS63-31C→T | 2/20 | 6/190 | ||||
| 67 | 13590C→T | P4530P | 2/20 | 3/158 | |||
| 67 | 13599A→G | T4533T | 3/20 | 29/158 | |||
| 74 | 15987C→T | Y5329Yd | 1/20 | 0/190 | |||
| 74 | 16031G→A | G5344E | 0/20 | 4/190 | 2438374 | ||
| 76 | IVS75-15G→Ae | 1/20 | 0/190 | ||||
| 76 | 16248C→T | V5416V | 0/20 | 3/190 | |||
| 76 | 16312A→G | T5438G | 0/20 | 1/190 | |||
| 82 | 17626A→G | I5876V | 5/20 | 2247870 | |||
| 88 | IVS87 complexi | 3/20 | 34/190 | ||||
| Ch | IVS24a+1G→A | 6/20 | 50/192 | ||||
| 89 | IVS88-7T→C | 3/20 | 12/192 | ||||
| 89 | 18732-18750delj | Y6244X | 1/20 | 0/282 | 0/190 | ||
| 89 | 18741G→A | G6247G | 0/20 | 23/192 | |||
| 89 | ISV89+44C→G | 0/20 | 1/40 g |
Figure 1.
Segregation of VLGR1 mutations Q2301X, M2931FS, I2906FS, and T6244X in families 735, 964, and 1848 (with USH2C). A, Paternal inheritance of Q2301X in family 1848 is shown by agarose-gel electrophoresis of exon 31 PCR product _Xmn_I digests. Q2301X mutation is a 6901 C→T transition, 33 bp on the 3′ end of the alternate exon 31 splice donor, affecting only the VLGR1b mRNA isoform (fig. 2_a_). Shown here are sequence electropherograms from the heterozygous family 1848 proband (HET), an apparent Q2301X homozygote, and wild-type (WT) BAC clone RP11-29K14, with sequence and amino acid translation differences (arrows). The Q2301X homozygote was the result of a brother-sister incestuous union, whereas another singleton case was a Q2301X/occult heterozygote (data not shown; table 1). B, Maternal inheritance of M2931FS in family 964 is shown by agarose-gel electrophoresis of exon 39 PCR product _Nco_I digests. Sequence electropherograms of the WT and the cloned M2931FS allele show 8790delC in exon 39 (arrows). This deletion causes a 10-codon frameshift. ending with a TAG stop encoded by the last 3 bases of exon 39, affecting VLGR1b. C, Maternal inheritance of I2906FS in family 1848 by DHPLC of exon 38 PCR products. Sequence comparison of the cloned I2906FS mutation shows an 8716–17insAACA (arrows) causing a frameshift of 5 codons and ending with a TGA stop 1 bp short of the 3′ end of exon 38, affecting VLGR1b. D, Paternal inheritance of Y6244X detected by DHPLC of exon 89 PCR products. A 19-bp deletion brings a TAG stop codon immediately in-frame. Y6244X removes 63 amino acids from the COOH end of VLGR1a and VLGR1b. The putative maternal and paternal mutations in family 735 and family 964 have not been identified.
Table 1 .
Clinical Summary of Patients with USH2C Harboring VLGR1 Mutations[Note]
| Family No. andIndividual No. | Severity ofHearing Loss | Age atRP Diagnosis(years) | Sex | RP Symptoms | VLGR Mutation(s) |
|---|---|---|---|---|---|
| 735a: | |||||
| 1 | Moderate-severe | 27b | Female | Typical | T6244X/occult |
| 5 | Moderate-severe | 25 | Female | NA | T6244X/occult |
| 6 | Moderate-severe | 20 | Female | NA | T6244X/occult |
| 7c | Moderate-severe | 19 | Female | NA | T6244X/occult |
| 964d: | |||||
| 1 | Moderate-severe | 24 | Female | Typical | M2931FS/occult |
| 3 | Moderate-severe | 21 | Female | Typical | M2931FS/occult |
| 4 | Moderate-severe | 33 | Female | Mild | M2931FS/occult |
| 1127: | |||||
| 1 | Moderate-severe | 42 | Female | Typical | Q2301X/occult |
| 1848a: | |||||
| 1 | Moderate-severe | 32b | Female | NA | Q2301X/I2906FS |
| 2 | NA | 29 | Female | NA | Q2301X/I2906FS |
| 3 | NA | 29 | Female | NA | Q2301X/I2906FS |
| 4 | NA | 21 | Female | NA | Q2301X/I2906FS |
| 2684e: | |||||
| 1 | Moderate-severe | 17 | Female | NA | Q2301X/Q2301X |
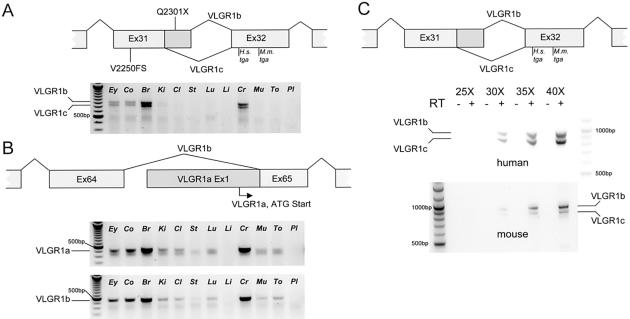
Because the USH2 phenotype manifests cochlear and retinal defects, we wanted to determine if any of the three VLGR1 isoforms were tissue specific in these organs (fig. 3_a_ and 3_b_). We performed semiquantitative RT-PCR by use of total RNA isolated from nine human fetal tissues. Primer pairs for the human isoforms VLGR1a, VLGR1b, and VLGR1c have been described elsewhere (McMillan et al. 2002). Total RNA from three human tissues (fetal brain, fetal liver, and placenta) was purchased from Clontech. For cDNA synthesis, total RNA was treated with DNase I (Invitrogen), according to the manufacturer’s conditions, and random-primed in a volume of 250 μl from 3.1 μg total RNA using 4 pmol random hexanucleotides, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 0.01% (v/w) gelatin, 5 mM MgCl2, 1 mM DTT, 95 U RNAguard (Pharmacia), and 2,500 U MMLV reverse transcriptase (Invitrogen). Semiquantitative RT-PCRs were performed by sampling 50 μl RT-PCR reactions at the 25th, 30th, 35th, and 40th cycle. PCR reactions included 87 ng of randomly primed cDNA, 11.2 pmol of each primer, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 0.01% gelatin, 2.5–3 mM MgCl2, 160 mM of each dNTP, and 1.75 U Taq DNA polymerase (Invitrogen) under the following conditions: 92°C for 4 min, followed by 40 cycles of 95°C for 30 s, 50°C / 55°C for 30 s, and 72°C for 1 min, with a final step at 72°C for 10 min. RT-PCR results show expression of all three VLGR1 isoforms from fetal retina and cochlea, confirming the EST expression profile of VLGR1. High relative expression in fetal brain and spinal cord confirms the neuronal expression of VLGR1 (McMillan et al. 2002) (fig. 3_a_ and 3_b_). Although there appears to be a consistent difference in the amount of VLGR1a, VLGR1b, and VLGR1c RT-PCR products amplified from different tissues, no apparent tissue-specific differences in the relative abundance between the three isoforms was seen in any of the fetal tissues tested (fig. 3_a_ and 3_b_). In addition, we find the relative amount of the VLGR1a and VLGR1b isoforms to be roughly equal as judged by RT-PCR, which is in contrast to an approximately fourfold difference in expression of the VLGR1b isoform over the VLGR1a isoform previously observed in human fetal kidney, brain, and lung (McMillan et al. 2002). One explanation for this disparity could be the way the RT-PCR of the VLGR1a and VLGR1b isoforms was performed, as the study by McMillan et al. (2002) amplified VLGR1a and VLGR1b in multiplex, whereas we chose to amplify them in separate reactions because of a significant difference in the TMs of the sense primers.
Figure 3.
RT-PCR of VLGR1 mRNA splice forms from human and mouse fetal tissue. Human fetal tissues from left to right: eye (Ey), cochlea (Co), brain (Br), kidney (Ki), colon (Cl), stomach (St), lung (Lu), liver (Li), spinal cord (Cr), muscle (Mu), tongue (To), and adult placenta (Pl). A, The relative locations of the human Q2301X and the Frings and BUB/BnJ mouse V2250FS mutations to the alternative donor site that differentiates the VLGR1b and VLGR1c splice isoforms. These isoforms differ by 83 bp because of the alternative use of a 5′ splice-donor site in exon 31 (Skradski et al. 2001; McMillan et al. 2002). Expression from eye and cochlear tissue is less intense at 30 cycles of RT-PCR than from brain and spinal cord. At 40 cycles, all tissues but liver and placenta show VLGR1 expression of similar relative intensities for VLGR1c, compared with VLGR1b (fig. 2_b_; data not shown). B, The location of the VLGR1a 5′ start site, relative to the exon 65 3′ splice acceptor of VLGR1b. RT-PCR results at 30 cycles for VLGR1a and VLGR1b splice forms show a similar tissue distribution to that in figure 2_a_, as well as similar relative intensities. All RNAs were positive for D-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (data not shown). C, A comparison of VLGRb and VLGR1c concentration in human and mouse reveals a species-specific difference in the relative abundance of these isoforms, which were amplified from fetal brain mRNA, indicating a difference in the use of the alternate exon 31 donor sites between species.
The VLGR1b and VLGR1c isoforms differ by 83 bp as a result of the alternative exon 31 donor splicing (Skradski et al. 2001; McMillan et al. 2002). A mouse Vlgr1 mutation in exon 31 (7009delG; V2250FS ) is responsible for the monogenic audiogenic seizure susceptible (MASS) phenotype (Skradski et al. 2001). The _Vlgr1_7009delG (Vlgr1 Mass1) mouse allele mutates both mouse Vlgr1b and Vlgr1c splice isoforms, whereas the human exon 31 VLGR1 mutation (Q2301X), in addition to the other three USH2C mutations, affects VLGR1b (figs. 2 and 3). To determine if species-specific differences in the VLGR1b and VLGR1c isoforms might help to explain the VLGR1 mutant phenotypes, RT-PCR of VLGR1b and VLGR1c isoforms from mouse and human fetal RNA was performed. For RT-PCR of the VLGR1b and VLGR1c isoforms, random-primed total mRNA from human and mouse fetal brain RNA (Clonetech) was reverse transcribed with Superscript III (Invitrogen), according to standard conditions, and PCR with AmpliTaq Gold (PE Biosystems) was as described for genomic PCR amplification, except that the amount of MgCl2 was 3.0 mM. Amplification primers for the mouse Vlgr1b and Vlgr1c isoforms were as follows: sense 5′-CGAACTGGAGGAACATTTGCAG-3′ and antisense 5′-GCAGACACGTTCACCCAAGTTC-3′. Cloning and sequencing of VLGR1b and VLGR1c RT-PCR products from both human and mouse confirm the evolutionarily conservation of the alternative exon 31 splice donor sites in both species (data not shown). It is not surprising that, although the VLGR1b messages of human and mouse share 81% sequence identity overall, the last 113 bp of exon 31 are 100% conserved at the DNA level, indicating that the sequence context of the mRNA in this region is critically important, perhaps defining a critical cis element necessary for alternative mRNA processing. A comparison of RT-PCR products from fetal human and mouse brain revealed a difference in the relative abundance of VLGR1b and VLGR1c isoforms, indicating a species-specific difference in mRNA processing through the alternative exon 31 donor sites (fig. 3_c_). These results suggest that isoform-specific mutations in VLGR1, either alone or in combination with a specific difference in mRNA processing, may underlie the manifestation of phenotype in these species.
In situ hybridization of mouse embryo sections had shown Vlgr1 expression concentrated in the ventricular zone where neural progenitor cells reside during embryonic neurogenesis, suggesting that the Vlgr1b and Vlgr1c isoforms play a role in the development of the murine CNS (McMillan et al. 2002). This idea is strengthened by the association of the MASS phenotype with the Vlgr1 mutant allele (Skradski et al. 2001). The Vlgr1 mouse ortholog includes in its open reading frame three additional and distinct mRNAs previously characterized as isoforms of the mouse Mass1 gene (Mass1.1, Mass1.2, and Mass1.3) on mouse chromosome 13 (Skradski et al. 2001) (fig. 2). The Frings mouse has not been reported to have deficient hearing or vision; however, no histological or electrophysiological data have been published that explicitly rule out such possibilities. In addition to Frings, the BUB/BnJ inbred mouse was also found to be a Vlgr1 Mass1 homozygote, and it exhibits the MASS phenotype until a progressive age-related hearing loss (AHL) phenotype manifests in this strain by 3 wk of age (Skradski et al. 2001). BUB/BnJ is one of several inbred mouse strains exhibiting AHL (Zheng et al. 1999). The variable severity and progression of hearing loss in many AHL mouse strains, including BUB/BnJ, is influenced by a major modifier locus on mouse chromosome 10 (ahl, also the modifier-of-deaf-waddler locus, mdfw). Both ahl and mdfw are caused by the _Cdh23_753G→A splicing mutation, a synonymous transition of the last nucleotide of exon 7 (Noben-Trauth et al. 2003). It remains to be determined if the early onset of hearing loss in BUB/BnJ is influenced by the Vlgr1 Mass1 mutation, as has been shown for other AHL “accelerating alleles” in combination with _Cdh23_753A (Noben-Trauth et al. 2003). It should be noted that BUB/BnJ is also homozygous for rd1, a mutation in the Pde6b gene that results in early photoreceptor-cell apoptosis because of a lack of cGMP phosphodiesterase activity. The fact that BUB/BnJ carries homozygous mutations in Pde6b (rd1), Cdh23 (ahl, mdfw), and Vlgr1 (Mass1) suggests caution should be used in interpreting the phenotypic effects of these genes in this mouse strain.
The VLGR1 protein belongs to a 33-member subgroup of the large N-terminal family B (LNB) seven-transmembrane (7TM) receptors. Of the 33 LNB 7TM members, 32 are orphan receptors and only a few have demonstrated specific G-protein signaling (Foord et al. 2002). All LNB 7TM members, including VLGR1, have a G-protein–coupled proteolysis site (GPS); cleavage at this site is thought to be a requirement for the proper expression of this class of 7TM receptors on the cell surface (Krasnoperov et al. 2002). VLGR1 may function, like other LNB 7TM members, as a natural protein chimera, with an extracellular cell-adhesion subunit and a canonical G-protein–coupled receptor subunit that are linked by noncovalent interactions (Krasnoperov et al. 2002). The possible function of VLGR1 might be best explored by trying to predict the functions of the two putative subunits separately.
The C-terminal residues of VLGR1 correspond to the consensus motif that is recognized as a ligand for the class I subfamily of PDZ domains (PDZ-binding interface [PBI] [Nikkila et al. 2000; Hung and Sheng 2002]). Interestingly, the C-termini of cadherin 23, protocadherin 15, harmonin, and SANS, all proteins with mutations that cause the USH1 phenotype, have class I PBIs. An interaction through the 2nd PDZ domain of harmonin and the C-terminal PBI ligand of cadherin 23 has been independently confirmed (Boeda et al. 2002; Siemens et al. 2002). Colocalization of harmonin and SANS in transfected HeLa cells suggests that these proteins also interact, perhaps through a PDZ-PBI interaction (Weil et al. 2003). The fact that all USH1 protein mouse mutants, including the nonmuscle myosin VIIa, which also has a protein interaction with harmonin, phenotypically present with defects in hair cell stereocilia development suggests that these proteins function as a macromolecular complex necessary to shape and maintain the stereocilia as an organized cohesive unit (Boeda et al. 2002; Siemens et al. 2002; Weil et al. 2003). It remains to be determined whether one or more of the three PDZ domains of harmonin—or other PDZ domain–bearing protein—have a high-affinity interaction with the PBI of VLGR1.
The very large extracellular portion of VLGR1b has a total of 35 CalX-β modules (fig. 2), named for the homology shared between the regulatory domains of Na+/Ca2+ exchanger proteins and the cytoplasmic portion of integrin-β4 (Schwarz and Benzer 1997; McMillan et al. 2002). Portions of the extracellular domain of VLGR1a that include a subset of the CalX-β domains bind Ca2+ and other cations (Nikkila et al. 2000). Because of the extensive CalX-β modules, one might postulate that the molecule acts as an extracellular Ca2+ sink, as an extracellular Ca2+ monitor sensitive to the regulation of intra- and extracellular Ca2+ trafficking, or in Ca2+-dependent cell adhesion. Disruption of normal Ca2+ metabolism is known to affect both hearing and vision. For example, mutations in the plasma membrane Ca2+ ATPase pump Atp2b2 (PMCA2) cause the deaf waddler (dfw) phenotype in mice (Kozel et al. 1998; Street et al. 1998). It has been postulated that the murine ahl gene (_mdfw, Cdh23_753A) acts epistatically to cause an age-dependent hearing loss through an interaction between the Ca2+-extruding PMCA2 channel and the calcium-dependent homotypic cell-adhesion properties of the cadherin 23 protein (Noben-Trauth et al. 1997, 2003). Furthermore, a functional interaction has been postulated between PMCA2 and the sodium bicarbonate cotransporter, NBC3, at the photoreceptor synapse, as the loss of H+ buffering by NBC3 causes retinal dystrophy (and hearing loss) in NBC3 knock-out mice (Bok et al. 2003). The possibility that calcium homeostasis is involved in USH2C is especially intriguing in light of the observation that the Ca2+-channel–blocking drug, diltiazem, is protective against noise-induced hearing loss (Maurer et al. 1998; Heinrich et al. 1999). However, there is conflicting evidence about whether diltiazem is neuroprotective for the retina, since none of the animal models tested had RP resulting from a calcium metabolism defect (Frasson et al. 1999; Pasantes-Morales et al. 2002; Pawlyk et al. 2002).
The large VLGR1b ectodomain has two recognized potential protein-interaction domains, LamG/TspN/PTX (Beckmann et al. 1998) and EAR/EPTP (Scheel et al. 2002; Staub et al. 2002) (fig. 2). It has been suggested that the multiple Ca2+-binding CalX-β modules may mediate the protein-protein interactions of these two domains (McMillan et al. 2002). The EAR/EPTP motif occurs as a repeated set of domains that together are postulated to form a seven-bladed-propeller structure that, in other proteins, acts as a highly specific receptor (Pons et al. 2003). Probable ligands for the EAR/EPTP domain of VLGR1 would be extracellular or cell-adhesion proteins. Usherin, the product of the USH2A gene, is a known extracellular basement-membrane protein (Bhattacharya et al. 2002). When usherin is defective, the phenotype is remarkably similar to that seen in patients with inactivated VLGR1. Immunolocalization of usherin to the perinurium of the spiral ganglion cells of the cochlea (Bhattacharya et al. 2002) and the likely neuronal localization of the VLGR1 receptor indicates that VLGR1 and usherin may colocalize in specific subcellular compartments. Furthermore, the significant homology between the LamG/TspN/PTX motifs of VLGR1 (fig. 2) and usherin indicates that the two proteins may share an affinity for a common binding partner, reinforcing the hypothesis of an interaction between these proteins. Whether usherin is the extracellular ligand or another yet-to-be-identified USH2 protein remains to established.
Because VLGR1 is expressed as at least three different isoforms in the human, questions arise as to which of the isoforms is responsible for USH2C and, as a corollary, whether mutations involving other isoforms cause a different phenotype, especially a seizure disorder. The USH2C mutations we have observed all involve isoform VLGR1b, but not isoform VLGR1c. The Frings mutation affects both VLGR1b and VLGR1c. We specifically asked all of the patients with USH2C in our study about seizures of any type and received uniformly negative responses. It is plausible to hypothesize that the Frings seizure disorder is primarily due to mutations affecting the VLGR1c isoform. Whether a functional protein is encoded by VLGR1c will be important in helping to determine whether the disparity in phenotype between human and mouse VLGR1 mutants are governed by species-specific differences in VLGR1 function, the inactivation of a subset of VLGR1 isoforms, or a combination of the two. The fact that the VLGR1 S2832X and S2764P mutations (isoform VLGR1b) segregate in families with both febrile and afebrile seizures (FEB4 [MIM 604352]) suggests a causal relationship between human seizures and VLGR1. But the relative low frequency of FEB4-associated mutations, the lack of a significant association between VLGR1 SNPs and FEB4, and the lack of any self-reported seizure-related phenotype in our USH2C patients suggest that VLGR1b is not a significant contributor to any seizure disorder (Nakayama et al. 2002).
All 13 patients with USH2C and VLGR1 mutations are female, a highly significant deviation from expectation (P<.002, Fisher’s exact test). The biologic significance, if any, of this observation can only be speculated on. Although some selection bias could conceivably play a role, it is difficult to understand why the shift in sex ratio is so extreme. A likely explanation is that males are being excluded from the sample: the male phenotype might be more severe, possibly lethal; it could be associated with some other hearing-/vision-loss syndrome; or the phenotype could be very mild or nonexistent. Nonetheless, the possibility that an endocrine-based component could influence the Usher syndrome phenotype as a modifier of VLGR1 expression and/or protein function is intriguing. Mutations in VLGR1 do not seem to be responsible for a large proportion of the USH2 cases in our sample, despite the fact that the gene is large and one might have expected otherwise. The possibility of male lethality could explain the apparently low contribution of USH2C mutations to the total USH2 mutational load.
The discovery of USH2C-associated VLGR1 mutations poses several interesting and important questions regarding the etiology of USH2. Localization and functional studies, in addition to thorough genetic screening for additional VLGR1 mutations, will be needed to elucidate the role of VLGR1 in the development, maintenance, and function of vision and hearing. Furthermore, its capacity as a putative G-coupled receptor makes it a legitimate target in the search for possible therapeutics that may have a broad impact on all types of Usher syndrome.
Acknowledgments
We thank the patients and their families for participating in the research, especially the Ralph and Justine Donnelly family for their patient support of this research. We want to acknowledge Sharon Kuo and Drs. Cynthia Morton, Peter Humphries, and Hannie Kremer for their interest and work in the identification and screening of other USH2C candidate genes. We thank Drs. Frans Cremers, Dana Orten, and Phillip Kelley for their critical reading of this manuscript. This work was supported by grant 5P01 DC01813-08 from the National Institutes of Health–National Institute on Deafness and Other Communication Disorders (NIH-NIDCD) (to W.J.K.), a grant from the Foundation Fighting Blindness (to W.J.K.), a grant from the Morris J. and Betty Kaplun Foundation (to W.J.K), and the European Union commission project grant “GENDEAF Thematic Network” (to C.M.). The work of M.W.J.L. was sponsored by the Netherlands Organization of Scientific Research (NWO) grant 901-04-205.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- National Eye Institute Gene Bank, http://neibank.nei.nih.gov/libs/NbLib0013/NbLib0013.shtml (for human retina) and http://neibank.nei.nih.gov/libs/NbLib0011/NbLib0011.shtml (for human fetal cochlea)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for USH2, USH1D, USH1F, USH2C, and FEB4)
- UCSC Genome Browser, http://genome.cse.ucsc.edu/ (for human VLGR1, AF435925; mouse Vlgr1, AF435926; and VLGR1 genomic DNA, NT_028179)
References
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 69:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Van Camp G, Hageman GS, Woychik RP, Smith RJ (2001) Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet 10:1709–1718 10.1093/hmg/10.16.1709 [DOI] [PubMed] [Google Scholar]
- Beckmann G, Hanke J, Bork P, Reich JG (1998) Merging extracellular domains: fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins. J Mol Biol 275:725–730 10.1006/jmbi.1997.1510 [DOI] [PubMed] [Google Scholar]
- Bhattacharya G, Miller C, Kimberling WJ, Jablonski MM, Cosgrove D (2002) Localization and expression of usherin: a novel basement membrane protein defective in people with Usher’s syndrome type IIa. Hear Res 163:1–11 10.1016/S0378-5955(01)00344-6 [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O’Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 26:56–60 10.1038/79178 [DOI] [PubMed] [Google Scholar]
- Boeda B, El Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners J, Houdusse A, Legrain P, Wolfrum U, Richardson G, Petit C (2002) Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J 21:6689–6699 10.1093/emboj/cdf689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D, Galbraith G, Lopez I, Woodruff M, Nusinowitz S, BeltrandelRio H, Huang W, Zhao S, Geske R, Montgomery C, Van S, I, Friddle C, Platt K, Sparks MJ, Pushkin A, Abuladze N, Ishiyama A, Dukkipati R, Liu W, Kurtz I (2003) Blindness and auditory impairment caused by loss of the sodium bicarbonate cotransporter NBC3. Nat Genet 34:313–319 10.1038/ng1176 [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedo Cabrera M, Vila MC, Molina OP, Gal A, Kubisch C (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27:108–112 10.1038/83667 [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, et al (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 68:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL (2001) Listen carefully: positional cloning of an audiogenic seizure mutation may yield Frings benefits. Neuron 31:507–508 [DOI] [PubMed] [Google Scholar]
- Chaib H, Kaplan J, Gerber S, Vincent C, Ayadi H, Slim R, Munnich A, Weissenbach J, Petit C (1997) A newly identified locus for Usher syndrome type I, USH1E, maps to chromosome 21q21. Hum Mol Genet 6:27–31 10.1093/hmg/6.1.27 [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 10.1126/science.280.5370.1753 [DOI] [PubMed] [Google Scholar]
- Foord SM, Jupe S, Holbrook J (2002) Bioinformatics and type II G-protein-coupled receptors. Biochem Soc Trans 30:473–479 [DOI] [PubMed] [Google Scholar]
- Frasson M, Sahel JA, Fabre M, Simonutti M, Dreyfus H, Picaud S (1999) Retinitis pigmentosa: rod photoreceptor rescue by a calcium-channel blocker in the rd mouse. Nat Med 5:1183–1187 10.1038/13508 [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12:1619–1623 10.1101/gr.278202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich UR, Maurer J, Mann W (1999) Ultrastructural evidence for protection of the outer hair cells of the inner ear during intense noise exposure by application of the organic calcium channel blocker diltiazem. ORL J Otorhinolaryngol Relat Spec 61:321–327 [DOI] [PubMed] [Google Scholar]
- Hmani M, Ghorbel A, Boulila-Elgaied A, Ben Zina Z, Kammoun W, Drira M, Chaabouni M, Petit C, Ayadi H (1999) A novel locus for Usher syndrome type II, USH2B, maps to chromosome 3 at p23-24.2. Eur J Hum Genet 7:363–367 10.1038/sj.ejhg.5200307 [DOI] [PubMed] [Google Scholar]
- Hung AY, Sheng M (2002) PDZ domains: structural modules for protein complex assembly. J Biol Chem 277:5699–5702 10.1074/jbc.R100065200 [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hamalainen R, Yuan B, Johnson C, Tegelberg S, Gasparini P, Zelante L, Pirvola U, Pakarinen L, Lehesjoki AE, de la Chapelle A, Sankila EM (2001) Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet 69:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, Gerber S, Bonneau D, Rozet JM, Delrieu O, Briard ML, Dollfus H, Ghazi I, Dufier JL, Frezal J (1992) A gene for Usher syndrome type I (USH1A) maps to chromosome 14q. Genomics 14:979–987 [DOI] [PubMed] [Google Scholar]
- Kimberling WJ, Moller CG, Davenport S, Priluck IA, Beighton PH, Greenberg J, Reardon W, Weston MD, Kenyon JB, Grunkemeyer JA (1992) Linkage of Usher syndrome type I gene (USH1B) to the long arm of chromosome 11. Genomics 14:988–994 [DOI] [PubMed] [Google Scholar]
- Kimberling WJ, Weston MD, Moller C, Davenport SL, Shugart YY, Priluck IA, Martini A, Milani M, Smith RJ (1990) Localization of Usher syndrome type II to chromosome 1q. Genomics 7:245–249 [DOI] [PubMed] [Google Scholar]
- Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, Riddle T, Duffy JJ, Doetschman T, Miller ML, Cardell EL, Shull GE (1998) Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem 273:18693–18696 10.1074/jbc.273.30.18693 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V, Lu Y, Buryanovsky L, Neubert TA, Ichtchenko K, Petrenko AG (2002) Post-translational proteolytic processing of the calcium-independent receptor of α-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein–coupled receptor: role of the G protein–coupled receptor proteolysis site (GPS) motif. J Biol Chem 277:46518–46526 10.1074/jbc.M206415200 [DOI] [PubMed] [Google Scholar]
- Maurer J, Mann WJ, Amedee RG (1998) Calcium channel blockers for prevention of noise trauma in otologic surgery. J La State Med Soc 150:400–405 [PubMed] [Google Scholar]
- McMillan DR, Kayes-Wandover KM, Richardson JA, White PC (2002) Very large G protein–coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J Biol Chem 277:785–792 10.1074/jbc.M108929200 [DOI] [PubMed] [Google Scholar]
- Mustapha M, Chouery E, Torchard-Pagnez D, Nouaille S, Khrais A, Sayegh FN, Megarbane A, Loiselet J, Lathrop M, Petit C, Weil D (2002) A novel locus for Usher syndrome type I, USH1G, maps to chromosome 17q24-25. Hum Genet 110:348–350 10.1007/s00439-002-0690-x [DOI] [PubMed] [Google Scholar]
- Nakayama J, Fu YH, Clark AM, Nakahara S, Hamano K, Iwasaki N, Matsui A, Arinami T, Ptacek LJ (2002) A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann Neurol 52:654–657 10.1002/ana.10347 [DOI] [PubMed] [Google Scholar]
- Nikkila H, McMillan DR, Nunez BS, Pascoe L, Curnow KM, White PC (2000) Sequence similarities between a novel putative G protein-coupled receptor and Na+/Ca2+ exchangers define a cation binding domain. Mol Endocrinol 14:1351–1364 [DOI] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR (2003) Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet 35:21–23 10.1038/ng1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR, Nishina PM (1997) mdfw: a deafness susceptibility locus that interacts with deaf waddler (dfw). Genomics 44:266–272 10.1006/geno.1997.4869 [DOI] [PubMed] [Google Scholar]
- Pasantes-Morales H, Quiroz H, Quesada O (2002) Treatment with taurine, diltiazem, and vitamin E retards the progressive visual field reduction in retinitis pigmentosa: a 3-year follow-up study. Metab Brain Dis 17:183–197 10.1023/A:1019926122125 [DOI] [PubMed] [Google Scholar]
- Pawlyk BS, Li T, Scimeca MS, Sandberg MA, Berson EL (2002) Absence of photoreceptor rescue with D-_cis_-diltiazem in the rd mouse. Invest Ophthalmol Vis Sci 43:1912–1915 [PubMed] [Google Scholar]
- Pieke-Dahl S, Moller CG, Kelley PM, Astuto LM, Cremers CW, Gorin MB, Kimberling WJ (2000) Genetic heterogeneity of Usher syndrome type II: localisation to chromosome 5q. J Med Genet 37:256–262 10.1136/jmg.37.4.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons T, Gomez R, Chinea G, Valencia A (2003) Beta-propellers: associated functions and their role in human diseases. Curr Med Chem 10:505–524 [DOI] [PubMed] [Google Scholar]
- Sankila EM, Pakarinen L, Kaariainen H, Aittomaki K, Karjalainen S, Sistonen P, de la Chapelle A (1995) Assignment of an Usher syndrome type III (USH3) gene to chromosome 3q. Hum Mol Genet 4:93–98 [DOI] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K (2002) A common protein interaction domain links two recently identified epilepsy genes. Hum Mol Genet 11:1757–1762 10.1093/hmg/11.15.1757 [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Benzer S (1997) Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc Natl Acad Sci USA 94:10249–10254 10.1073/pnas.94.19.10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J, Kazmierczak P, Reynolds A, Sticker M, Littlewood-Evans A, Muller U (2002) The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci USA 99:14946–14951 10.1073/pnas.232579599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skradski SL, Clark AM, Jiang H, White HS, Fu YH, Ptacek LJ (2001) A novel gene causing a Mendelian audiogenic mouse epilepsy. Neuron 31:537–544 [DOI] [PubMed] [Google Scholar]
- Smith RJ, Lee EC, Kimberling WJ, Daiger SP, Pelias MZ, Keats BJ, Jay M, Bird A, Reardon W, Guest M (1992) Localization of two genes for Usher syndrome type I to chromosome 11. Genomics 14:995–1002 [DOI] [PubMed] [Google Scholar]
- Staub E, Perez-Tur J, Siebert R, Nobile C, Moschonas NK, Deloukas P, Hinzmann B (2002) The novel EPTP repeat defines a superfamily of proteins implicated in epileptic disorders. Trends Biochem Sci 27:441–444 10.1016/S0968-0004(02)02163-1 [DOI] [PubMed] [Google Scholar]
- Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K (1998) Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deaf waddler mice. Nat Genet 19:390–394 10.1038/1284 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain–containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55 10.1038/79171 [DOI] [PubMed] [Google Scholar]
- Wayne S, Der Kaloustian VM, Schloss M, Polomeno R, Scott DA, Hejtmancik JF, Sheffield VC, Smith RJ (1996) Localization of the Usher syndrome type ID gene (Ush1D) to chromosome 10. Hum Mol Genet 5:1689–1692 10.1093/hmg/5.10.1689 [DOI] [PubMed] [Google Scholar]
- Wayne S, Lowry EB, McLeod DR, Knaus R, Farr C, Smith RJH (1997) Localization of the Usher syndrome type 1F (Ush1F) to chromosome 10. Am J Hum Genet Suppl 61:A300 [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 10.1038/374060a0 [DOI] [PubMed] [Google Scholar]
- Weil D, El Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C (2003) Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet 12:463–471 10.1093/hmg/ddg051 [DOI] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res 130:94–107 10.1016/S0378-5955(99)00003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]