Identification of 51 Novel Exons of the Usher Syndrome Type 2A (USH2A) Gene That Encode Multiple Conserved Functional Domains and That Are Mutated in Patients with Usher Syndrome Type II (original) (raw)
Abstract
The USH2A gene is mutated in patients with Usher syndrome type IIa, which is the most common subtype of Usher syndrome and is characterized by hearing loss and retinitis pigmentosa. Since mutation analysis by DNA sequencing of exons 1–21 revealed only ∼63% of the expected USH2A mutations, we searched for so-far-uncharacterized exons of the gene. We identified 51 novel exons at the 3′ end of the gene, and we obtained indications for alternative splicing. The putative protein encoded by the longest open reading frame harbors, in addition to the known functional domains, two laminin G and 28 fibronectin type III repeats, as well as a transmembrane region followed by an intracellular domain with a PDZ-binding domain at its C-terminal end. Semiquantitative expression profile analysis suggested a low level of expression for both the long and the short isoform(s) and partial overlap in spatial and temporal expression patterns. Mutation analysis in 12 unrelated patients with Usher syndrome, each with one mutation in exons 1–21, revealed three different truncating mutations in four patients and two missense mutations in one patient. The presence of pathogenic mutations in the novel exons indicates that at least one of the putative long isoforms of the USH2A protein plays a role in both hearing and vision.
Usher syndrome is the most common cause of deaf-blindness (Marazita et al. 1993). Patients present with sensorineural hearing loss and retinitis pigmentosa (RP), with or without vestibular impairment. On the basis of the severity and progression of the hearing loss, the age at onset of RP, and the presence or absence of vestibular impairment, three clinical subtypes are distinguished that are genetically heterogeneous (Smith et al. 1994; Ahmed et al. 2003 [and references therein]). Also, a minority of the patients are described as “atypical,” because their phenotype does not fall within one of the three types (Otterstedde et al. 2001).
The USH2A gene is mutated in patients with Usher syndrome type IIa (MIM 276901), in some patients with atypical Usher syndrome, and in a small number of patients with autosomal recessive RP (Eudy et al. 1998; Liu et al. 1999; Adato et al. 2000; Rivolta et al. 2000; Weston et al. 2000; Bernal et al. 2003). The gene was described as consisting of 21 exons that span ∼250 kb of genomic DNA at chromosome 1q41 (Eudy et al. 1998; Weston et al. 2000) (University of California Santa Cruz [UCSC] Genome Browser, July 2003 freeze [UCSC Genome Bioinformatics Web site]). The encoded protein was predicted to consist of 1,546 amino acids containing an N-terminal signal sequence and conserved functional domains that are commonly seen in extracellular proteins or in extracellular domains of proteins and that are involved in protein-protein interactions, including interactions with other components of the extracellular matrix. The USH2A protein, also called usherin, was shown to interact with type IV collagen and to be part of basement membranes in cochlea, retina, and several other tissues (Eudy et al. 1998; Weston et al. 2000; Bhattacharya et al. 2002, 2004; Pearsall et al. 2002).
Usher syndrome type II seems to be the most common type of Usher syndrome (Eudy et al. 1998). In epidemiologic studies, the ratio between Usher syndrome type I and II varies from 2:3 to 1:2, although there is a bias toward the western European population in the studies (Piazza et al. 1986; Hope et al. 1997; Rosenberg et al. 1997; Spandau and Rohrschneider 2002). In certain populations, such as the Finnish population, the Ashkenazi Jews, or the Acadian population in southwestern Louisiana, specific subtypes of Usher syndrome can be relatively frequent because of founder effects (Keats et al. 1994; Pakarinen et al. 1995; Ness et al. 2003). On the basis of mutation analysis, the USH2A gene is thought to be causative in 74%–90% of cases with Usher syndrome type II (Dreyer et al. 2000; Weston et al. 2000; Pennings et al., in press), and thus Usher syndrome type IIa is the most common subtype of Usher syndrome.
By sequence analysis of all 21 known exons and exon-intron boundaries in patients with Usher syndrome type II, we could detect only ∼63% of the mutant USH2A alleles (Pennings et al., in press). Although no alternative splicing or alternative sites for polyadenylation were reported, northern blots showed transcripts of several sizes, both shorter and longer than the known cDNAs (Eudy et al. 1998; Huang et al. 2002). On the basis of these results and exon predictions presented in the UCSC Genome Browser (April 2003 freeze [UCSC Genome Bioinformatics Web site]), we searched for as-yet-uncharacterized regions of the gene.
We developed three sets of primers in exons (29F, 30R; 30F, 33R; 63F, 64R) predicted to be 3′ of exon 21 with the Primer3 program (Rozen and Skaletsky 2000) (table A [online only]). RT-PCR was performed on RNA derived from the retinoblastoma cell line Y79 (American Type Culture Collection), human fetal cochlea, and human adult retina. The RNA was isolated by use of the method described by Chirgwin et al. (1979) and treated with DNase I (Invitrogen). Synthesis of cDNA was performed with random hexanucleotides as primers, as described by Luijendijk et al. (2003). After nested PCR, specific bands were seen on agarose gel (data not shown). PCR fragments were purified from gel using the QIAquick Gel Extraction kit (Qiagen) and sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing version 2.0 kit (Applied Biosystems) and the ABI PRISM 3730 DNA analyzer (Applied Biosystems). This revealed that at least part of the predicted exons are transcribed. Moreover, we could amplify a fragment with forward primers in the known exon 20 and reverse primers in the novel exon 22, strongly suggesting that the USH2A gene is much larger at the 3′ end than previously reported.
Table A.
Primers for USH2A RT-PCR and Nested PCR
| Exon | Forward Primer 1 | Forward Primer 2 |
|---|---|---|
| 20 | GTTGGTTGTGTGACCAGTGC | TTGTGTGACCAGTGCTTCGG |
| 29 | GATAGCCTCGCATGAAGGAG | TCGAGACGTACAACAGGAG |
| 30 | CCTGGGATGAACCTGTTGTC | CTCTGCCAGTGCTGAATTTG |
| 33 | TTCCCCAGTTCTGACTGTCC | ACTGTCCTGGATTCTAGAAC |
| 40 | TTTCCAATCCTTCTGCATCG | GGTTGGTTGCCTCCAATG |
| 41 | TCCCAAGTCCAGAGCTGTTC | CGAGGAGTCATTCCATGAGG |
| 44 | TTTTACACCGAGCCGAGAAG | AGAGGAGCCAATCTCACTGC |
| 46 | TGCTTCCTCCAGAGGTTGTC | CCCAAATGGTGTTGTCACTG |
| 49 | ACAGGAGGCACAACCAAATC | TGCTCTGGGTATTACGCTAG |
| 53 | AACAGCTATGGGCGAGGAC | CAAAGCTGTGAGAGCCAGAAC |
| 57 | TTGGAGTCTGCCAGAAAAGC | GAGTCGTAATGGAAACTTGC |
| 59 | TAACACCTCTGGCCTTCTCC | TCATCAATCCACCCTTCTGG |
| 61 | GCTGGCATTGAAGAGGAGTC | ACCCTGAGGCCTTTCACAC |
| 63 | CCACCAGCATCACAACTCTG | GTCAGCCCTCCAGATCTTTG |
| 64 | CTGCAGCCAAATGGAAAAG | CAGCCTAGAAAATCCAATCCAG |
| 66 | GGAGTAGAGGCCTGCACCTG | CAAAGGACCGACAGCTGAAC |
| 70 | TCTACAGCGAGCTGTGGTTC | GCGAGCTGTGGTTCATAGTG |
| Reverse Primer 1 | Reverse Primer 2 | |
| 21 3′ UTR | CGGGGTGCATACTGAGAATC | ATAGCATGAGGGCATTTTGC |
| 22 | TCCCTTCTTCAACTGAAGTGC | CTCTTCCTGATTGCCAGGTG |
| 29 | ACGTCCTCGACTCCAATCAC | CTCCTTCATGCGAGGCTATC |
| 33 | CCCAGGAACTGTTTGTACAGC | CTGGTGGGGTGTAAATACTGC |
| 40 | CTATGTGCACTGCCAAATCC | TTAACGATGCAGAAGGATTGG |
| 41 | TCTGGACTTGGGATCCCTTC | TCTGGGACTCTGGTGAAAGG |
| 44 | TTGGCTCCTCTCTCTGGAAG | TTCTCGGCTCGGTGTAAAAC |
| 46 | ACACCATTTGGGTTTGAAGG | TGACAACCTCTGGAGGAAGC |
| 49 | GGTTGTGCCTCCTGTATTCG | TCTTCACAACAGCGATGTCC |
| 53 | AGGGGGACTCACTCCTTGAG | AGCTGTTCCAGGCAGAAATC |
| 57 | ACCCAGGAAAAGCAAGTTTC | GGCTTTTCTGGCAGACTCC |
| 63 | AGGGGCTTCATCTGTCCAC | GGAACCCGTCACTGAAGATG |
| 64 | CCTGGAGCCATTTGTACCTC | TGCATCAAAGGTGCAATCTC |
| 65 | GGCTCTGAGACCTTCTGGTG | TCTGCACCATGTCCAGCTAC |
| 70 | GACAGAAAAATGGCCAACAAG | CCACAGCTCGCTGTAGAACTC |
| 71 | TTGCGGATACTCACAGGTGTC | ACTCACAGGTGTCCCAGACC |
| 72 | CTGCCAAACAGAACCAAGTG | TCCAGGGTTACGTCTTCTGG |
To further investigate which of the predicted exons are indeed transcribed and to find exons that are not predicted by gene-prediction programs, forward and reverse primers were designed (table A [online only]) for a large number of predicted exons, and RT-PCR and subsequent nested PCR were performed on RNA isolated from retinoblastoma cells of the Y79 cell line. Fragments were isolated from agarose gels and sequenced. On the basis of the obtained results, primers were chosen to amplify larger, overlapping cDNA fragments encompassing exons 17–29, 20–33, 30–44, 41–53, 49–63, 61–65, 63–70, and 66–72 from human retina Marathon-Ready cDNA (Clontech), according to the manufacturer's protocol. Sequence analysis revealed that overlapping cDNA fragments were indeed amplified, and 51 novel exons were identified. The coding sequence of exon 21 was shown to be 14 base pairs (bp) shorter in the long transcript(s) when compared with the short transcript that was described elsewhere (Eudy et al. 1998; Weston et al. 2000); the termination codon in exon 21 is therefore excluded from the long transcript(s). To characterize the 3′ end of the gene, a 3′ rapid amplification of cDNA ends (RACE) was performed using the human retina Marathon-Ready cDNA (Clontech), the Advantage cDNA Polymerase kit (Clontech), a nested gene-specific forward primer (table A [online only]), and the Marathon-AP1 primer as the reverse primer, according to the manufacturer's protocol. The resulting PCR product was isolated from agarose gel and directly cloned in the pCR4-TOPO vector using the TOPO TA Cloning kit (Invitrogen). Clones were screened by performing internal PCR reactions from exon 70 to exon 71 and from exon 70 to exon 72. Sequence analysis of one of the positive clones revealed an in-frame termination codon in exon 72 and 2,888 bp of 3′ UTR. The longest ORF extends from exon 2 to exon 72, encoding a putative protein of 5,202 amino acids. The USH2A gene now spans ∼790 kb of genomic DNA, and the 3′ UTR ends only ∼4 kb distal to the KCTD3 gene (potassium channel tetramerization domain containing 3) (UCSC Genome Browser, July 2003 freeze [UCSC Genome Bioinformatics Web site]). The cDNA sequence encompassing the longest ORF has been deposited in GenBank (accession number AY481573). The sequences of the 5′ UTR and of exons 1–20 were derived from the previously reported sequence of the known USH2A gene (GenBank accession number NM_007123).
Besides the alternative splicing in exon 21, RT-PCR on human retina Marathon-Ready cDNA provided evidence for alternative splicing that leads to the skipping of exon 42, exons 50–52, and exon 62. The alternative splicing of exon 62 is also seen in the retinoblastoma cells. Only the skipping of exon 42 leads to interruption of the reading frame. Additional studies are needed to elucidate the functional significance of the detected alternative splicing and to fully characterize all of the alternative transcripts and their spatial and temporal distribution.
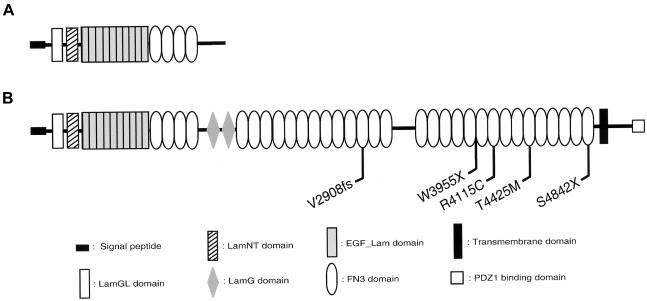
The putative protein of the 5,202 amino acid residues encoded by the longest ORF has a predicted molecular weight of 576 kD. When the predicted amino acid sequence is analyzed with the Pfam software (Pfam Home Page) (Bateman et al. 2002), a protein appears that consists of the previously described combination of a domain that is homologous to a thrombospondin N-terminal-like domain (TSPN), a laminin G–like domain (LamGL), and a pentaxin (PTX) domain, in addition to a laminin N-terminal (LamNT) domain, 10 laminin-type EGF-like (LE) repeats, and four fibronectin type III (FN3) repeats (Eudy et al. 1998; Weston et al. 2000) (fig. 1_A_), as well as two laminin G (LamG) and 28 FN3 repeats (fig. 1_B_). Between FN3 repeats 13 and 14 there is a partial FN3 domain and a stretch of ∼350 amino acids with no significant homology to any known domain (fig. 1_B_). The TMHMM software predicts with high significance the presence of an outside-in transmembrane region (residues 5,041–5,063), followed by an intracellular domain of 139 amino acids with a type I PDZ-binding domain (DTHL) (Sheng and Sala 2001) at its C-terminal end (fig. 1_B_). This indicates that the longest putative USH2A protein is a transmembrane protein. Skipping of exon 42 is predicted to result in an isoform of 2,742 amino acids that contains two LamGL and eight FN3 repeats, in addition to the USH2A protein domains that are already known to exist. Skipping of exons 50–52 or exon 62 does not interrupt the reading frame but can be predicted to lead to protein isoforms without the residues 3,247–3,462 or 4,023–4,098, respectively. Residues 3,247–3,462 encode most of the region between FN3 repeat 13 and 14. Deletion of amino acids 4,023–4,098 removes the C-terminal part of an FN3 repeat and the N-terminal part of the following one, leading to an isoform that is one FN3 repeat shorter.
Figure 1.
Schematic representation of the USH2A protein structure. A, The previously described short isoform. B, The putative long isoform corresponding to the longest ORF. Novel mutations in the long isoform are depicted.
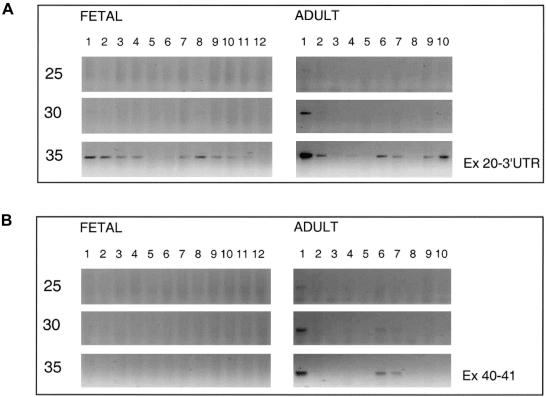
Expression was studied for both the short and the long isoforms in fetal and adult tissues. A semiquantitative RT-PCR was performed, as described by Luijendijk et al. (2003), and 10 μl aliquots were taken after 25, 30, and 35 PCR cycles. For the short isoform, primers were designed in exon 20 and in the 3′ UTR of exon 21 (table A [online only]). In fetal tissues, bands were visible on agarose gel only after 35 PCR cycles and were strongest in eye, cochlea, and heart (fig. 2_A_). Weak bands were seen in brain, CNS, intestine, skeleton, tongue, kidney, and lung. No expression was detectable in stomach and liver. In adult tissues, the highest expression was seen in neural retina, already visible after 25 cycles (fig. 2_A_). In testis and retinal pigment epithelium (RPE)–choroid, expression was seen after 30 cycles. In brain, heart, kidney, and liver, expression was seen only after 35 cycles. The expression in RPE-choroid might be due to contamination with cells of the neural retina since RPE-choroid is extremely difficult to isolate without any remains of neural retina. Expression of the long isoform(s) was tested using primers in exons 40 and 41 (fig. 2_B_) and primers in exons 59 and 60 (data not shown). The two primer combinations revealed the same expression pattern. No expression was detected in fetal tissues after 35 PCR cycles (fig. 2_B_). Expression of the long isoform was, as was seen for the short transcript, highest in adult neural retina and was visible after 25 PCR cycles. After 30 cycles, expression was detectable in heart and kidney. We can conclude that, in fetal tissues and in adult tissues other than retina, USH2A transcripts are not abundant; this is especially true for the transcripts encoding the long isoform(s). For testis, RT-PCR suggests differential expression, since a clear fragment was only seen for the short transcript. Although not detected in expression profiling, there is very low expression of the long isoform(s) in fetal cochlea, since several fragments could be amplified by nested PCR. Comparison of our data with previous data on USH2A expression is difficult, since previous studies do not discriminate between the long and the short isoforms. However, the highest expression of USH2A in retina is in agreement with the results of Huang et al. (2002), who analyzed USH2A transcripts in adult mouse and rat tissues, and with the results of Eudy et al. (1998), who studied some fetal human tissues, including eye.
Figure 2.
Expression profile analysis by semiquantitative RT-PCR analysis of USH2A. A, RT-PCR specific for the transcript encoding the short isoform. B, RT-PCR specific for transcript(s) encoding the long isoform(s). Samples were taken after 25, 30, and 35 cycles. The RNA used for RT-PCR analyses was derived from the following tissues: fetal (1) eye, (2) cochlea, (3) brain, (4) CNS, (5) stomach, (6) intestine, (7) skeleton, (8) heart, (9) tongue, (10) kidney, (11) lung, and (12) liver; adult (1) retina, (2) RPE-choroid, (3) cornea, (4) brain, (5) skeletal muscle, (6) heart, (7) kidney, (8) lung, (9) liver, and (10) testis. Expression of the short isoform was detected in several fetal and adult tissues, whereas expression of the long isoform was only detected in adult retina (predominantly), heart, and kidney.
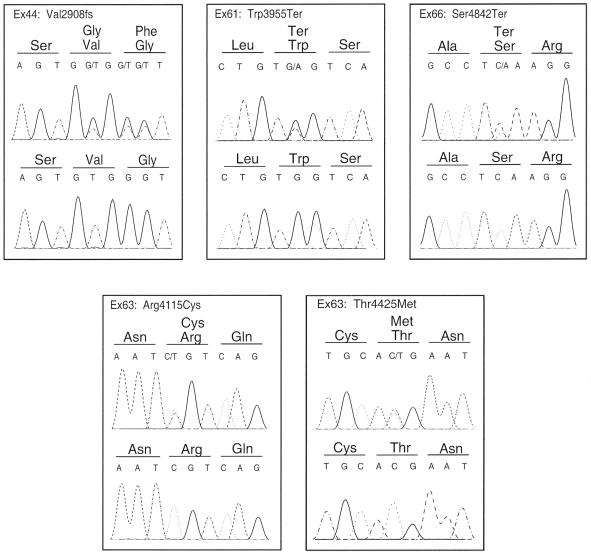
To address the question of whether the putative long USH2A protein is essential for normal functioning of the cochlea and the retina, we sequenced the protein-coding parts and flanking intron sequences of exons 22–72, as described above, in 2 unrelated familial subjects and 10 isolated Dutch patients with USH2. In these patients, only one pathogenic mutation was detected after sequencing exons 1–21 (Pennings et al., in press). Primer sequences are given in table B (online only). The study was approved by the local ethics committee, and informed consent was obtained from all patients and healthy family members. In four patients, a truncating mutation was detected in the novel 3′ region of the gene (fig. 3). The c.11864G→A substitution in exon 61 introduces a termination codon (Trp3955Ter) in patients 20608 and 20662. Patient 20608 was already known to carry the c.949C→A (Arg317Arg) mutation, which was predicted to create an additional 5′ splice site in exon 5. Patient 20662 carries the Cys419Phe (c.1256G→T) mutation, in addition to the Trp3955Ter mutation. A nonsense mutation, c.14525C→A (Ser4842Ter) in exon 66, was found in patient 20625, who was heterozygous for the 2299delG (Glu767fs) mutation. In patient 20599, who was also previously found to be heterozygous for the 2299delG (Glu767fs) mutation, a deletion of a TG dinucleotide in exon 44 was predicted to cause a frameshift (Val2908fs). Two missense mutations in exon 63, Arg4115Cys and Thr4425Met, were found on the same allele in patient 20609, who also carries the Cys419Phe mutation (fig. 3). None of the five mutations was found in 94 ethnically matched control samples.
Table B.
Intronic Primers for Mutation Analysis
| Exon | Primer Sequence |
|---|---|
| 22 | F: CCATGCGAGTATATTGCTGTG |
| R: GCTGAGGGCAAGTCACATTAC | |
| 23 | F: CAGGAAAGCCAGAATGATGC |
| R: CCCAAAGGCAAATTCAACAG | |
| 24 | F: CAGGCAATGAGGAGAGAGGA |
| R: CCTAAAGGAAATTTTTGGCACA | |
| 25 | F: TGAATCATTAAGAGGCTTGCAG |
| R: TGTGTGCACCATTGGAATAAC | |
| 26 | F: GGTCTTTCTGTCACTTCTGTGC |
| R: TTCAGTTATCAGGCGTGGTG | |
| 27 | F: TGCTTTCAGGGAACTGTTTTG |
| R: GGTGCTGCTTTTAGCCTGAG | |
| 28+29 | F: TGCTGCAGAGGACAAAAATG |
| R: GCTTCAGGGTAATAGTCCTTCC | |
| 30 | F: TGCGGCCATTAAAAGTGAAG |
| R: TGCAGGCTTCCACTACTTTAG | |
| 31 | F: GCAGAAAGGGAGGAAAATGAC |
| R: CAAATTAGGTTGGGGTTTGC | |
| 32 | F: TGATTTATTGGTTTGGGTCTG |
| R: GCATTCTTGATTAAATTTGCAG | |
| 33 | F: TGAAGCATCCTATATGTTATTGC |
| R: CCTCCCCTGATTGAACTCAC | |
| 34 | F: ATTTCCTTTTTGCCCCTCAG |
| R: AGGATGGGAAGAGAGTTTTCAG | |
| 35 | F: TTGGGGAAGTAAAGGTAGCAC |
| R: CCACAATCTCCCCAACTAGAG | |
| 36 | F: AAATCACATCAAGAGTGCTTGC |
| R: CCTGCTTGAAAGGCTAGCTG | |
| 37 | F: TGTGCTTTGATCCTGCTGAC |
| R: AACCAACATCTGTGGCTAAAAG | |
| 38 | F: AATTGGCCAGGTCAACTCAG |
| R: TGTGAGGTGGATGAAAGCAG | |
| 39 | F: CAGGAGTTCAGGAATGAAAATG |
| R: AAGTTTCAGTGGGAAGAAATCC | |
| 40 | F: GAGATCTCATTTGATGGCAGAA |
| R: GGCTCATTTCTTTGCTTTGG | |
| 41 | F: GGCAGCATCCTAGAAAATCC |
| R: GCAGGTCGTGAGGGTCTTG | |
| 42 | F: GCAAAATTCTAGGCCTCGTG |
| R: AAAGCCTCCTTCATTTCCCTAC | |
| 43 | F: ATGCCACAGAACAGCCTGAG |
| R: AGCCGTGACAAAGGCAATAG | |
| 44 | F: TTTTGTAGAGGGGTGGAAGG |
| R: TGTGTACATGGGGGAGGTTC | |
| 45 | F: CATTTCCAAAACAAAGGCTCTC |
| R: TTAGCCTCCACCCCCTTC | |
| 46 | F: TCATCATATCCCACTGGTCAC |
| R: CCCTTCTCTCTTTTCCCTTCC | |
| 47 | F: AGGGAAGGTGGGATTCAGAC |
| R: TGTCATGGCTGAGAGGATACC | |
| 48 | F: CCTCACGCCATGTGTTATTC |
| R: CCTTTCTTTTCCGTGGAGTC | |
| 49 | F: TCGAATGATCCTGGAAAATACA |
| R: TTGTTGAGAGGGAGGTGTTTG | |
| 50 | F: ACCTGTAAGTTGCCATGTGTG |
| R: TTTGGAGGACATGACCTTTTC | |
| 51 | F: ATTTCAGCAACTGCCTGAGC |
| R: AAAGCTTCCCTGAGAACAGC | |
| 52 | F: TGGGAAGCTGCAAAACTG |
| R: GGCCTCAAAGTATGATGGAATG | |
| 53 | F: ACTGGTTGGGGCATGACTG |
| R: AGTCTGCATAACACCCTGACC | |
| 54 | F: ATTGCATTTCTTCCGAACAC |
| R: TCTCTCCTTCCAGCCATAGG | |
| 55 | F: AAAGGGAAATGCTTCTCCAAG |
| R: CCCCCTAACCACAATGACAG | |
| 56 | F: AGCCTCTTATGAGGTTCAGACC |
| R: CAAGCCTGAAGAATGGGAAC | |
| 57 | F: GGGGATGGTGTGACTTTTG |
| R: ATGGCCAATGAATGAGGAAG | |
| 58 | F: GGCAAAGAGTTTGCAATTTGTC |
| R: TTTATCCAGGAGACCGACTATG | |
| 59 | F: CAAACATTTGTTGCCCATTC |
| R: GCCAGACTGTGATTTTTCTGG | |
| 60 | F: TGCAAAAGGACAGGTTAAAA |
| R: GATTCTGCTGTGTTGGAGCA | |
| 61 | F: TGACACCAGGAAGAAACAGC |
| R: TTATCCCCGTGACTACATTGC | |
| 62 | F: TTTGGCCATGAGGTTCAGAG |
| R: TGAAGGGAGTTTTCCCACAG | |
| 63 | F: GGGGGTGAAGGTTAAAAAGG |
| R: TTTAGGTGGATGGAGGTTGG | |
| 64 | F: AATCTCGGCTCACTGCAAG |
| R: AGTGCCTTTTCAAATTGTGC | |
| 65 | F: TGCTTTTGGTTGCCAATTTC |
| R: ACCGTAGAGCAACTGAGAACAG | |
| 66 | F: TGTAGGAGGTGGGATCTTGG |
| R: CTGGGGAGTGCCAGGTAG | |
| 67 | F: GAGCAGTTTCCTGCAAATGG |
| R: TCCCCACAAGAAAATCCTTC | |
| 68 | F: GTTTGGATTGGTTCGGTTTG |
| R: CGTAAAGCTGGGGAACAGAG | |
| 69 | F: CGTCATAACTTGCTTTGGAATC |
| R: CAACACTTGGCACAATTTCTTC | |
| 70 | F: ATCAAATAGCAGGGCCAGAG |
| R: CCTCTCTTGGTCCCCACAC | |
| 71 | F: CGTGTTGTGGGTGCTCATAG |
| R: CCAAGTGCTGGGATTACAGG | |
| 72 | F: TGAGGCTTCTGAGGCTTAG |
| R: CTGCCAAACAGAACCAAGTG |
Figure 3.
Mutations in the novel exons of the USH2A gene. In each panel, the upper sequence pane shows the heterozygous mutation in the patient. The lower pane depicts the wild-type sequence.
DNA samples of additional family members were available for two of the patients, to prove that the identified nucleotide changes were not located on the same allele as the previously detected mutations. For patient 20608, we could show by sequence analysis that the c.949C→A mutation is derived from the mother and the c.11864G→A mutation came from the father (data not shown). In a healthy son of patient 20662, we detected the c.1256G→T mutation but not the c.11864G→A mutation, indicating that these mutations are not part of a complex USH2A allele. To prove the pathogenicity of either of the two amino acid substitutions, or the combination of the two, segregation analysis in family members and functional studies are needed. The presence of pathogenic mutations in the novel exons of the USH2A gene indicates the functional significance of at least one of the long isoforms of the USH2A protein for normal functioning of the cochlea and retina. This is further corroborated by a retina-derived mouse cDNA clone (GenBank accession number AK044734) that encodes a peptide with 69% identity to the putative long human USH2A isoform(s). Also, exons that encode FN3 repeats are predicted in the syntenic region of both mouse and rat (Mouse Genome Browser, February 2003 freeze; Rat Genome Browser, June 2003 freeze [UCSC Genome Bioinformatics Web site]).
Sequencing exons and exon-intron boundaries of other large genes involved in autosomal recessive diseases that display a high allelic heterogeneity, such as the ABCA4 gene, detected ∼80% of the mutations (Shroyer et al. 2001). By sequencing exons 1–21, we identified ∼63% of the mutated USH2A alleles (Pennings et al., in press). From this we can deduce that not more than ∼20% of the mutations in patients with Usher syndrome type IIa are expected to be in the novel exons.
We showed that mutations in the long isoform(s) of USH2A can be causative for Usher syndrome in combination with a mutation in exons 1–21. The question remains whether two alleles with a mutation in the novel exons can also lead to Usher syndrome. Symptoms due to defects in other organs might occur, since USH2A transcripts encoding the long isoform(s) are present in heart and kidney. Also, the USH2A protein was shown—by use of antibodies that are likely to detect both the short and the long isoforms—to be present in several organs in addition to retina and cochlea. (Bhattacharya et al. 2002; Pearsall et al. 2002). Alternatively, this genotype might lead to nonsyndromic hearing loss or RP. It is also interesting to note that the Cys759Phe amino acid substitution was found to be homozygously present in patients with RP and also in nonsymptomatic individuals (Rivolta et al. 2000; Bernal et al. 2003). This might suggest that this mutation is a polymorphism that is linked to a so-far-unknown mutation in one of the novel 3′ exons of the gene in the patients with RP and that two mutations in the long isoform(s) cause nonsyndromic RP. This is in agreement with the very low transcript levels of the long isoform(s) in cochlea.
The combination of domains predicted for the USH2A isoforms is not present in any other protein (Pfam Home Page), which might suggest unique functions. The previously known short isoform is an extracellular protein. It has been suggested that LE domains, also present in this short isoform, may interact with collagen IV to stabilize the USH2A protein in the basement membrane (Bhattacharya et al. 2004). The identification of the 3′ exons encoding the putative long USH2A isoform(s)—of which at least some have an intracellular domain—reveals new opportunities to study the function of the USH2A gene. It is interesting that the CDH23 gene is involved in Usher syndrome type I and encodes a protein that is, as are the long putative isoforms of USH2A, composed of a large extracellular region and a transmembrane region followed by an intracellular domain with a PDZ-binding domain at its C-terminal end (Bolz et al. 2001; Bork et al. 2001; Siemens et al. 2002). CDH23 interacts with harmonin through its PDZ-binding domain and is suggested to be part of a protein complex necessary for the development of the hair cell stereocilia in the inner ear (Boëda et al. 2002). Mutations in genes encoding other components of this complex, such as harmonin, myosin VIIa, and sans, also can be causative for Usher syndrome type I (Ahmed et al. 2003 [and references therein]). This illustrates that, besides providing new handles for studying USH2A function, the long isoform(s) of USH2A might guide us to a protein complex with other components that are defective in patients with Usher syndrome type II. So far, two other loci are known for Usher syndrome type II, for which the causative genes have not yet been identified (Hmani et al. 1999; Pieke-Dahl et al. 2000).
Acknowledgments
The authors want to thank the patients and family members, for their participation in this study; Drs. M. Luijendijk and A. den Hollander, for providing cDNA of fetal and adult tissues; and M. van Drie, for technical assistance. The authors are grateful to the Cornea Bank NORI for providing us with corneal material. This work was supported by grant 920-03-222 from ZonMW, the Nijmegen ORL Research Foundation, the Heinsius Houbolt Foundation, the Mgr. J. C. van Overbeek Foundation, the Gelderse Blinden Vereniging, and the Rotterdamse Vereniging Blindenbelangen.
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the “Electronic-Database Information” section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for USH2A short isoform [accession number NM_007123], USH2A long isoform [accession number AY481573], and Ush2a mouse cDNA clone [accession number AK044734])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Usher syndrome type IIa)
- Pfam Home Page, http://www.sanger.ac.uk/Software/Pfam/ (for Pfam software)
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for the Primer3 program)
- TMHMM Server, v. 2.0, http://www.cbs.dtu.dk/services/TMHMM-2.0/
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Adato A, Weston MD, Berry A, Kimberling WJ, Bonne-Tamir A (2000) Three novel mutations and twelve polymorphisms identified in the USH2A gene in Israeli USH2 families. Hum Mutat 15:388 [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Riazuddin S, Wilcox ER (2003) The molecular genetics of Usher syndrome. Clin Genet 63:431–444 10.1034/j.1399-0004.2003.00109.x [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280 10.1093/nar/30.1.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal S, Ayuso C, Antiñolo G, Gimenez A, Borrego S, Trujillo MJ, Marcos I, Calaf M, Del Rio E, Baiget M (2003) Mutations in USH2A in Spanish patients with autosomal recessive retinitis pigmentosa: high prevalence and phenotypic variation. J Med Genet 40:e8 10.1136/jmg.40.1.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya G, Kalluri R, Orten DJ, Kimberling WJ, Cosgrove D (2004) A domain-specific usherin/collagen IV interaction may be required for stable integration into the basement membrane superstructure. J Cell Sci 117:233–242 10.1242/jcs.00850 [DOI] [PubMed] [Google Scholar]
- Bhattacharya G, Miller C, Kimberling WJ, Jablonski MM, Cosgrove D (2002) Localization and expression of usherin: a novel basement membrane protein defective in people with Usher’s syndrome type IIa. Hear Res 163:1–11 10.1016/S0378-5955(01)00344-6 [DOI] [PubMed] [Google Scholar]
- Boëda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners J, Houdusse A, Legrain P, Wolfrum U, Richardson G, Petit C (2002) Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J 24:6689–6699 10.1093/emboj/cdf689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedo Cabrera M, Vila MC, Molina OP, Gal A, Kubisch C (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27:108–112 10.1038/83667 [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, et al (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 68:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294–5299 [DOI] [PubMed] [Google Scholar]
- Dreyer B, Tranebjaerg L, Rosenberg T, Weston MD, Kimberling WJ, Nilssen O (2000) Identification of novel USH2A mutations: implications for the structure of USH2A protein. Eur J Hum Genet 8:500–506 10.1038/sj.ejhg.5200491 [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao SF, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 10.1126/science.280.5370.1753 [DOI] [PubMed] [Google Scholar]
- Hmani M, Ghorbel A, Boulila-Elgaied A, Ben Zina Z, Kammoun W, Drira M, Chaabouni M, Petit C, Ayadi H (1999) A novel locus for Usher syndrome type II, USH2B maps to chromosome 3 at p23–24.2. Eur J Hum Genet 7:363–367 10.1038/sj.ejhg.5200307 [DOI] [PubMed] [Google Scholar]
- Hope CI, Bundey S, Proops D, Fielder AR (1997) Usher syndrome in the city of Birmingham: prevalence and clinical classification. Br J Ophthalmol 81:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Eudy JD, Uzvolgyi E, Davis JR, Talmadge CB, Pretto D, Weston MD, Lehman JE, Zhou M, Seemayer TA, Ahmad I, Kimberling WJ, Sumegi J (2002) Identification of the mouse and rat orthologs of the gene mutated in Usher syndrome type IIA and the cellular source of USH2A mRNA in retina, a target tissue of the disease. Genomics 80:195–203 10.1006/geno.2002.6823 [DOI] [PubMed] [Google Scholar]
- Keats BJB, Nouri N, Pelias MZ, Deininger PL, Litt M (1994) Tightly linked flanking microsatellite markers for the Usher syndrome type I locus on the short arm of chromosome 11. Am J Hum Genet 54:681–686 [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Hope C, Liang CY, Zou JM, Xu LR, Cole T, Mueller RF, Bundey S, Nance W, Steel KP, Brown SD (1999) A mutation (2314delG) in the Usher syndrome type IIa gene: high prevalence and phenotypic variation. Am J Hum Genet 64:1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijendijk MW, van de Pol TJ, van Duijnhoven G, den Hollander AI, ten Caat J, van Limpt V, Brunner HG, Kremer H, Cremers FP (2003) Cloning, characterization, and mRNA expression analysis of novel human fetal cochlear cDNAs. Genomics 82:480–490 10.1016/S0888-7543(03)00150-2 [DOI] [PubMed] [Google Scholar]
- Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE (1993) Genetic epidemiologic studies of early-onset deafness in the United States school-age population. Am J Med Genet 46:486–491 [DOI] [PubMed] [Google Scholar]
- Ness SL, Ben-Yosef T, Bar-Lev A, Madeo AC, Brewer CC, Avraham KB, Kornreich R, Desnick RJ, Willner JP, Friedman TB, Griffith AJ (2003) Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet 40:767–772 10.1136/jmg.40.10.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterstedde CR, Spandau U, Blankenagel A, Kimberling WJ, Reisser C (2001) A new clinical classification for Usher’s syndrome based on a new subtype of Usher’s syndrome type I. Laryngoscope 111:84–86 10.1097/00005537-200101000-00014 [DOI] [PubMed] [Google Scholar]
- Pakarinen L, Karjalainen S, Smola KO, Laippala P, Kaitalo H (1995) Usher’s syndrome type 3 in Finaland. Laryngoscope 105:613–617 [DOI] [PubMed] [Google Scholar]
- Pearsall N, Bhattacharya G, Wisecarver J, Adams J, Cosgrove D, Kimberling W (2002) Usherin expression is highly conserved in mouse and human tissues. Hear Res 174:55–63 10.1016/S0378-5955(02)00635-4 [DOI] [PubMed] [Google Scholar]
- Pennings RJE, te Brinke H, Weston MD, Claassen A, Orten DJ, Weekamp H, van Aarem A, Huygen PLM, Deutman AF, Hoefsloot LH, Cremers FPM, Cremers CWRJ, Kimberling WJ, Kremer H. USH2A mutation analysis in 79 Dutch families with Usher syndrome type II. Hum Mutat (in press) [DOI] [PubMed] [Google Scholar]
- Piazza L, Fishman GA, Farber M, Derlacki D, Anderson RJ (1986) Visual acuity loss in patients with Usher’s syndrome. Arch Ophthalmol 104:1336–1339 [DOI] [PubMed] [Google Scholar]
- Pieke-Dahl S, Möller CG, Kelley PM, Astuto LM, Cremers CWRJ, Gorin MB, Kimberling WJ (2000) Genetic heterogeneity of Usher syndrome type II: localisation to chromosome 5q. J Med Genet 37:256–262 10.1136/jmg.37.4.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg T, Haim M, Hauch AM, Parving A (1997) The prevalence of Usher syndrome and other retinal dystrophy–hearing impairment associations. Clin Genet 51:314–321 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24:1–29 10.1146/annurev.neuro.24.1.1 [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Wensel TG, Lupski JR (2001) Cosegregation and functional analysis of mutant ABCR (ABCA4) alleles in families that manifest both Stargardt disease and age-related macular degeneration. Hum Mol Genet 10:2671–2678 10.1093/hmg/10.23.2671 [DOI] [PubMed] [Google Scholar]
- Siemens J, Kazmierczak P, Reynolds A, Sticker M, Littlewood-Evans A, Müller U (2002) The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci USA 99:14946–14951 10.1073/pnas.232579599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH, Berlin CI, Hejtmancik JF, Keats BJB, Kimberling WJ, Lewis RA, Moller CG, Pelias MZ, Tranebjaerg L (1994) Clinical diagnosis of the Usher syndromes. Am J Med Genet 50:32–38 [DOI] [PubMed] [Google Scholar]
- Spandau UH, Rohrschneider K (2002) Prevalence and geographical distribution of Usher syndrome in Germany. Graefes Arch Clin Exp Ophthalmol 240:495–498 10.1007/s00417-002-0485-8 [DOI] [PubMed] [Google Scholar]
- Weston MD, Eudy JD, Fujita S, Yao S, Usami S, Cremers C, Greenberg J, Ramesar R, Martini A, Moller C, Smith RJ, Sumegi J, Kimberling WJ, Greenburg J (2000) Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet 66:1199–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]