Nonmuscle Myosin Heavy-Chain Gene MYH14 Is Expressed in Cochlea and Mutated in Patients Affected by Autosomal Dominant Hearing Impairment (DFNA4) (original) (raw)
Abstract
Myosins have been implicated in various motile processes, including organelle translocation, ion-channel gating, and cytoskeleton reorganization. Different members of the myosin superfamily are responsible for syndromic and nonsyndromic hearing impairment in both humans and mice. MYH14 encodes one of the heavy chains of the class II nonmuscle myosins, and it is localized within the autosomal dominant hearing impairment (DFNA4) critical region. After demonstrating that MYH14 is highly expressed in mouse cochlea, we performed a mutational screening in a large series of 300 hearing-impaired patients from Italy, Spain, and Belgium and in a German kindred linked to DFNA4. This study allowed us to identify a nonsense and two missense mutations in large pedigrees, linked to DFNA4, as well as a de novo allele in a sporadic case. Absence of these mutations in healthy individuals was tested in 200 control individuals. These findings clearly demonstrate the role of MYH14 in causing autosomal dominant hearing loss and further confirm the crucial role of the myosin superfamily in auditive functions.
Hereditary hearing loss is caused, in >50% of cases, by mutations in single genes, including myosins, connexins, transcription factors, potassium channels, and other cellular components that play an important function in ear cells (see Hereditary Hearing Loss Homepage). Different members of the myosin superfamily are responsible for syndromic and nonsyndromic hearing impairment transmitted as an autosomal dominant or recessive trait. Phylogenetic analyses have identified 18 distinct classes of myosins: 17 classes represent unconventional myosins, and class II includes conventional muscle and nonmuscle myosins (Berg et al. 2001). Among unconventional myosins, mutations in the MYO7A gene are responsible for type 1B Usher syndrome (MIM 276903), a syndromic deafness combined with retinitis pigmentosa and vestibular abnormalities. MYO7A is also implicated in two nonsyndromic forms of dominant and recessive hearing loss, DFNA11 and DFNB2, as well as in a strain of deaf mice called “shaker-1” (Liu et al. 1997; Weil et al. 1997). The MYO6 gene is mutated in human DFNA22 and in the Snell’s waltzer mouse (Avraham et al. 1995; Melchionda et al. 2001). The MYO15A gene is mutated in human DFNB3 and in mouse shaker-2 (Probst et al. 1998; Wang et al. 1998). Most recently, it has been shown that normal hearing in humans requires myosin IIIA (MYO3A) and myosin IA (MYO1A), which are implicated in two nonsyndromic forms of hearing impairment, DFNB30 and DFNA48, respectively (Walsh et al. 2002; Donaudy et al. 2003).
The conventional myosins of class II consist of 15 genes, including 6 skeletal-muscle myosin heavy chains, 2 cardiac myosin heavy chains, a smooth-muscle myosin heavy chain, and 3 nonmuscle myosin heavy chains II (NMHC IIA, NMHC IIB, and NMHC IIC) (Berg et al. 2001). The three nonmuscle myosins are encoded by the MYH9, MYH10, and MYH14 genes located on chromosomes 22q11.2, 17p13.3, and 19q13.3, respectively (Simons et al. 1991; Leal et al. 2003). Among nonmuscle myosins, MYH9 was the only gene known to be responsible for a human pathology, including four autosomal dominant diseases reported elsewhere as May Hegglin anomaly and as Sebastian, Fechtner, and Epstein syndromes (Seri et al. 2003). Patients with these diseases have congenital macrothrombocytopenia and aggregates of NMHC IIA in neutrophils. During life, they can develop deafness, nephritis, and cataract. The same gene is also mutated in a family affected by nonsyndromic hearing impairment, DFNA17 (MIM 603622) (Lalwani et al. 2000).
It is interesting that MYH14 is located within a candidate region in which a nonsyndromic autosomal dominant form of hearing impairment, DFNA4, has been mapped independently in two unrelated families (Chen et al. 1995; Mirghomizadeh et al. 2002). The two candidate regions partly overlap, defining a critical region of ∼4 Mb between markers D19S412 and D19S246. Therefore, the MYH14 gene was considered a strong candidate for hearing loss. First, we ascertained its expression in cochlea, using a specific anti-NMHC IIC antibody. We then performed mutational screening of the gene in a large series of 300 sporadic or familial hearing-impaired patients from Italy, Belgium, and Spain. The identification of one nonsense and three missense mutations clearly indicates a fundamental contribution of MYH14 to autosomal dominant hearing loss.
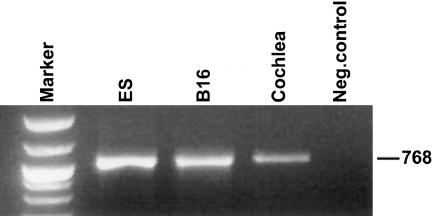
To determine whether the MYH14 gene is expressed in cochlea, we first assayed for its expression in mice. Total RNA from mouse cochlea, from mouse melanoma cell line B16 (Chirivi et al. 1994), and embryonic stem (ES) TVB2 cells, was reverse transcribed to perform RT-PCR. The correct fragment length of 768 bp was amplified from cochlear cDNA (fig. 1).
Figure 1.
RT-PCR of the Myh14 gene in mouse cochlea. Specific oligonucleotides C1F (5′-AGCATTGGAAGAGGAGTCCC-3′) and C2R (5′-GCTCCAGTCGTTCTACAGC-3′), spanning coding exons 26–30, were used to detect the expression of Myh14 in mouse cochlea RNA, as well as in two mouse cell lines, B16 (melanoma) and ES cells. Cochlea RNA was kindly provided by K. Avraham (Tel Aviv). In brief, it has been prepared from inner ears dissected out of the temporal bones of mice at embryonic day (E) 14, E16, E18, and P0. Negative control was performed without cDNA aliquot in the reaction. Marker: 1-kb DNA ladder (BioLabs).
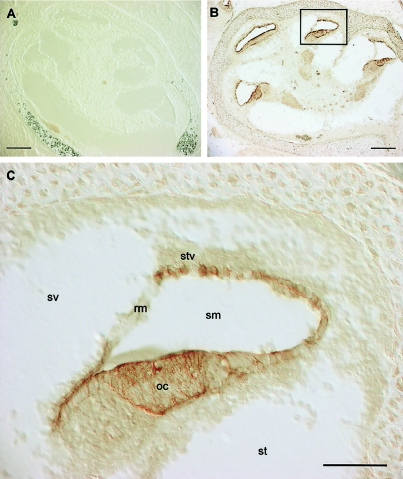
The NMHC IIC protein was detected in mouse cochlear neonatal sections (postnatal day [P0]) by use of polyclonal antibodies raised against mouse C-terminal NMHC IIC (Golomb et al. 2004). High expression was detected in the organ of Corti and the stria vascularis, as well as in cells of the cochlear duct, such as Hensen and Claudius cells, external sulcus cells, and epithelium of the spiral prominence (fig. 2). It is interesting that low or no expression of NMHC IIC was detected in the Reissner membrane and spiral ligament, in contrast with the MYH9 expression in the latter structures in the rat (Lalwani et al. 2000). It is also interesting that no expression was detectable in vestibular epithelia (data not shown), in agreement with phenotypic findings in patients (see below).
Figure 2.
Immunohistochemistry of NMHC IIC in mouse neonatal cochlea. Gelatin tissues at P0 were sectioned (10 μm) and fixed with 4% paraformaldehyde. A, Negative control obtained using only biotin-conjugated immunoglobulin G secondary antibodies. B, Section immunolabeled with anti-NMHC IIC antibodies (Buxton et al. 2003; Golomb et al. 2004), revealing a high expression in the cells surrounding the scala media. The insert is enlarged in panel C. sv = scala vestibuli; sm = scala media; st = scala tympany; rm = Reissner’s membrane; oc = organ of Corti; stv = stria vascularis. No expression in the vestibular epithelia has been detected (data not shown). Scale bars = 20 μm.
An overall number of 300 subjects were analyzed for mutations in the MYH14 gene. Inclusion criteria were: (a) absence of the most common mutations within the GJB2 gene, (b) sensorineural hearing loss, and (c) normal tympanometric evaluation. All subjects underwent a physical examination, including otoscopy; pure-tone audiometry with air and bone conduction performed at 125, 250, 500, 1,000, 2,000, 4,000, and 8,000 Hz with a Beltone 2000 audiometer; and tympanometry. The series includes subjects with a variable degree of hearing loss, ranging from mild to profound, and with a variable age at onset, ranging from congenital to late onset. Familial records were available, in most cases. The majority of patients came from central and southern Italy (200), whereas 48 and 52 were from Spain and Belgium, respectively. A large German family (Tü-N) that made possible the mapping of the DFNA4 locus to a 2.7-Mb map segment in 19q13.33 was also included in the screening (M.P., personal communication). After receipt of informed consent, peripheral blood was obtained from all subjects, and DNA was isolated from blood leukocytes according to standard methods.
An overall number of 40 primer pairs were designed to amplify the 40 coding exons of the MYH14 gene, including the splice sites (PCR primer sequences and conditions available on request). All amplicons were screened by DHPLC on a WAVE Nucleic Acid Fragment Analysis System HSM (Transgenomic), according to supplier protocols. DHPLC data analysis was based on a subjective comparison of sample and reference chromatograms. PCR products showing an abnormal chromatographic profile were directly sequenced on an automated sequencer (ABI 3100 [Perkin Elmer]).
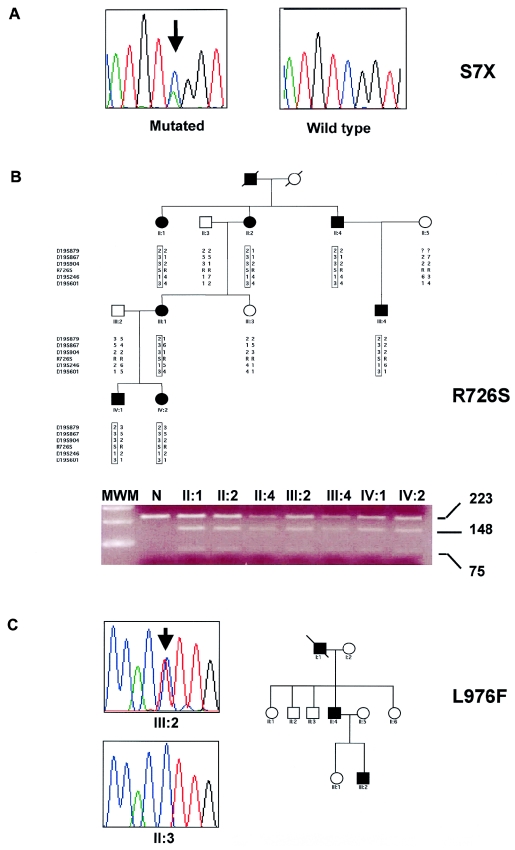
The mutational screening of the MYH14 gene allowed us to identify one nonsense and three missense mutations (table 1). The first mutation is a C→A nucleotide change at position 20, which replaces a serine residue at position 7 with a stop codon (S7X) (fig. 3_A_). This mutation was detected in the German 4-generation kindred Tü-N that mapped to the DFNA4 locus and defined a 2.7-Mb candidate region containing the MYH14 gene (M.P., unpublished data). In this pedigree, the affected individuals show a progressive sensorineural hearing impairment that starts in the 1st or 2nd decade of life and leads to severe-to-profound hearing loss in the 4th decade of life.
Table 1.
Mutations Identified in the MYH14 Gene
| Mutationa | Exon | Effecton Protein | Protein Domain |
|---|---|---|---|
| C20A | 1 | S7X | Motor domain |
| G1126T | 9 | G376C | Motor domain |
| C2176A | 16 | R726S | Tail |
| C2926T | 22 | L976F | Tail |
Figure 3.
MYH14 nonsense and missense mutations. A, Sequence data of the nonsense S7X mutation. B, Pedigree of the Belgian family with the R726S mutation and results showing its presence after _Alw_NI restriction-enzyme digestion. Haplotypes were constructed with markers around the MYH14 gene. Shared haplotypes are indicated with rectangles. MWM = molecular weight marker; N = normal. C, Pedigree of the Italian family with the L976F mutation and results showing sequence data in the proband (III:2) and in a healthy member (II:3).
A G→T nucleotide change was detected at position 1126 of the cDNA, leading to a glycine→cysteine amino acid substitution at position 376 (G376C) in the motor domain (data not shown). It was detected in a 9-year-old Italian patient affected by a moderate bilateral sensorineural hearing loss without vestibular involvement. There was no family history of hearing loss, and neither parent had the G376C mutation. Repeated analysis in the proband confirmed the presence of this mutated allele, which affects a residue highly conserved across species (fig. 4), was absent in control chromosomes and is, therefore, described as a de novo alteration. Paternity has been confirmed using a panel of highly informative microsatellite markers.
Figure 4.
ClustalW (European Bioinformatics Institute) alignment of the amino acid sequences of smooth muscle and nonmuscle myosins of class II; sequence alignments for MYH14 missense mutations. The residues mutated in NMHC IIC and their conservation across species are underlined. Hs = Homo sapiens; Rn = Rattus norvegicus; Mm = Mus musculus.
Another C→A change affecting nucleotide 2176 was detected in a large Belgian kindred. This change leads to substitution of a highly conserved arginine residue with serine at position 726 (R726S) (fig. 4). The presence of the mutation can be easily detected by PCR and subsequent cleavage with the restriction endonuclease _Alw_NI. We constructed haplotypes for a 3-cM region surrounding the MYH14 gene, including the mutation and a total of five dinucleotide markers (fig, 3_B_).
We found that all affected family members shared the same haplotype from markers D19S879 (centromeric) to D19S601 (telomeric), indicating that this family showed linkage with the DFNA4 region and that the mutation cosegregated with hearing impairment. Patients show a mild-to-moderate progressive hearing loss without vestibular involvement.
Finally, the sequence from the affected member (III-2) of a small Italian family (fig. 3_C_) displayed the heterozygous nucleotide transition C→T at position 2926 in exon 22. This leads to the amino acid substitution L976F affecting the NMHC IIC tail. Although leucine and phenylalanine are both nonpolar residues, this particular position appears to be evolutionarily conserved in the species available in the databases (fig. 4). The patient inherited the mutation from his hearing-impaired father (II-4). Sequence analysis confirmed the absence of this mutation in the healthy members of the pedigree. Affected individuals present with a mild-to-moderate bilateral sensorineural hearing impairment without vestibular symptoms.
The presence of these mutated alleles was tested on 200 normal chromosomes of individuals from the same geographical area as that of patients. None of the alleles was detected in any normal chromosome. We also detected two polymorphic variants, I266V and N1559S, 12 synonymous substitutions, and 7 intronic nucleotide substitutions (table 2). Since these latter changes do not create putative cryptic splice sites and/or did not segregate with deafness (data not shown), they have been excluded as causative mutations for deafness in our cohort of patients.
Table 2.
Nucleotide Substitutions Identified in the Coding Region of the MYH14 Gene[Note]
| NucleotideVariant | Exon | Effecton Protein |
|---|---|---|
| T474C | 2 | Silent |
| A796G | 6 | I266V |
| C1128T | 9 | Silent |
| C1731T | 13 | Silent |
| C1863T | 14 | Silent |
| C2082T | 15 | Silent |
| G1506A | 21 | Silent |
| A2895G | 21 | Silent |
| G4722A | 32 | Silent |
| A4676G | 32 | N1559S |
| A5307G | 36 | Silent |
| G5406T | 37 | Silent |
| T5745C | 39 | Silent |
| A5889T | 40 | Silent |
The MYH14 gene was an excellent candidate for hearing impairment because of its chromosomal location within the DFNA4 critical region and its high expression in the cochlea. One nonsense and three missense heterozygous mutations have been identified in 4 patients belonging to a cohort of 300 affected individuals from different European countries. Apart from patients carrying the nonsense allele who presented a profound hearing loss from the 4th decade of life, patients with the missense mutations were affected by sensorineural bilateral hearing loss of variable degree, usually ranging from moderate to severe. This finding might indicate a genotype-phenotype correlation, in which a more-severe clinical course is most likely to be associated with reduced levels of NMHC IIC.
It is interesting that one of the three missense mutation is a de novo allele altering an evolutionarily conserved residue. This mutated allele was never detected in healthy control individuals, and paternity was confirmed at molecular levels. The patient is the only affected member of his family, as negative records (i.e., audiological and clinical data) have been obtained for all members. These findings strongly support the pathogenic role of G376C. Two other missense mutations were detected in families affected by autosomal dominant hearing loss. Similar to most of the MYH9 mutations (Seri et al. 2003), the majority of mutations identified in the MYH14 gene comprises amino acid substitutions, suggesting that they might exert a dominant negative effect. For the three missense mutations found, there is a remarkable conservation of the residues' positions across species, which probably highlights their functional importance in the peptide (fig. 4). However, a functional analysis of all missense mutations will definitively prove their molecular mechanisms on pathogenesis.
Myosins of class II are hexameric enzymes composed of two heavy chains and two pairs of light chains. Dimerization of the heavy chains yields a polar structure. The N-termini form two globular heads with actin- and ATP-binding domains required for motor activity, whereas the α-helical C-termini form a single rodlike tail, which allows the molecules to polymerize into bipolar filaments in both muscle and nonmuscle cells (Sellers 2000). Besides the well-characterized role of myosin in contraction and force production in muscles, little is known about the specific functional role of nonmuscle myosins, although they are likely to participate in motility, cytokinesis, phagocytosis, maintenance of cell shape, and organelle/particle trafficking.
The NMHC IIC myosin has only recently been characterized, and information on its biological role is very limited. The expression pattern defined in human and mouse tissues showed that the gene is ubiquitously expressed at higher levels in adults than during development (Buxton et al. 2003; Leal et al. 2003; Golomb et al. 2004). In the mouse cochlea, with the exception of the Reissner membrane, the protein is localized in all cells of the scala media wall, with a relatively higher level in the organ of Corti and the stria vascularis. It is remarkable that the NMHC IIC distribution is different from the expression of the MYH9 (Lalwani et al. 2000), which immunolocalizes specifically in outer cells of the organ of Corti, spiral ligament, and Reissner membrane. Although the role of both nonmuscle myosins in the cochlear cells remains unclear, taken together, the expression data indicate that different physiopathogenetic mechanisms are probably implicated in hearing impairment owing to mutations in the MYH9 and MYH14 genes.
Finally, the identification of four different mutations suggests that the MYH14 gene distinctly contributes to autosomal dominant cases of hearing impairment. As a matter of fact, only a few genes among those so far described present with mutations in more than one single family (see Hereditary Hearing Loss Homepage). Additional population-based and functional studies are required to determine the precise contribution of MYH14 to deafness.
Acknowledgments
We thank Robert S. Adelstein, for antibodies against NMHC IIC; Valeria Marigo, for helpful discussions; and the RNA In Situ Hybridization Core at Telethon Institute of Genetics and Medicine, for technical assistance. This work was supported by Telethon (P.G. and A.S.); the Istituto Banco di Napoli (P.G.); Fondo per gli Investimenti della Ricerca di Base grant (P.G.); Cofin 2002 (P.G.); Italian Ministry of Health (A.S.), The Flemish Fund for Scientific Research (G.V.C.); the Interuniversity Attraction Poles programme P5/19 of the Federal Office for Scientific, Technical, and Cultural Affairs, Belgium; and the Kröner-Fresenius Stiftung and the Kuhn Stiftung (M.P.). P.N. was supported by National Genome Research Network grant 01GR0104. X.E. was supported by National Institutes of Health grant 5R01 DC001402-09, La Marato de TV3 grant 993610, and Fondo de Investigación Sanitario–Instituto de Salud Carols III grants CO3/07 and GO/203.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw/ (for ClustalW)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human MYH14 [accession number NM_024729])
- Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh/
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for type 1B Usher syndrome and DFNA17)
References
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA (1995) The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11:369–375 [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE (2001) A millennial myosin census. Mol Biol Cell 12:780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton DB, Golomb E, Adelstein RS (2003) Induction of nonmuscle myosin heavy chain II-C by butyrate in RAW 264.7 mouse macrophages. J Biol Chem 278:15449–15455 10.1074/jbc.M210145200 [DOI] [PubMed] [Google Scholar]
- Chen AH, Ni L, Fukushima K, Marietta J, O’Neill M, Coucke P, Willems P, Smith RJ (1995) Linkage of a gene for dominant non-syndromic deafness to chromosome 19. Hum Mol Genet 4:1073–1076 [DOI] [PubMed] [Google Scholar]
- Chirivi RG, Garofalo A, Crimmin MJ, Bawden LJ, Stoppacciaro A, Brown PD, Giavazzi R (1994) Inhibition of the metastatic spread and growth of B16-BL6 murine melanoma by a synthetic matrix metalloproteinase inhibitor. Int J Cancer 58:460–464 [DOI] [PubMed] [Google Scholar]
- Donaudy F, Ferrara A, Esposito L, Hertzano R, Ben-David O, Bell RE, Melchionda S, Zelante L, Avraham KB, Gasparini P (2003) Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet 72:1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS (2004) Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem 279:2800–2808 10.1074/jbc.M309981200 [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN (2000) Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet 67:1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal A, Endele S, Stengel C, Huehne K, Loetterle J, Barrantes R, Winterpacht A, Rautenstrauss B (2003) A novel myosin heavy chain gene in human chromosome 19q13.3. Gene 312:165–171 10.1016/S0378-1119(03)00613-9 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD (1997) Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 17:268–269 [DOI] [PubMed] [Google Scholar]
- Melchionda S, Ahituv N, Bisceglia L, Sobe T, Glaser F, Rabionet R, Arbones ML, Notarangelo A, Di Iorio E, Carella M, Zelante L, Estivill X, Avraham KB, Gasparini P (2001) MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 69:635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirghomizadeh F, Bardtke B, Devoto M, Pfister M, Oeken J, Konig E, Vitale E, Riccio A, De Rienzo A, Zenner HP, Blin N (2002) Second family with hearing impairment linked to 19q13 and refined DFNA4 localisation. Eur J Hum Genet 10:95–99 10.1038/sj.ejhg.5200769 [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA (1998) Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 280:1444–1447 10.1126/science.280.5368.1444 [DOI] [PubMed] [Google Scholar]
- Sellers JR (2000) Myosins: a diverse superfamily. Biochim Biophys Acta 1496:3–22 10.1016/S0167-4889(00)00005-7 [DOI] [PubMed] [Google Scholar]
- Seri M, Pecci A, Di Bari F, Cusano M, Savino M, Panza E, Nigro A, Noris P, Gangarossa S, Rocca B, Gresele P, Bizzarro N, Malatesta P, Koivisto PA, Longo I, Musso R, Pecoraro C, Iolascon A, Magrini U, Soriano JR, Renieri A, Ghiggeri GM, Ravazzolo R, Balduini CL, Savoia A (2003) MYH9-related disease: May-Hegglin anomaly, Sebastian syndrome, Fechtner syndrome, and Epstein syndrome are not distinct entities but represent a variable expression of a single illness. Medicine 82:203–215 10.1097/00005792-200305000-00006 [DOI] [PubMed] [Google Scholar]
- Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L (1991) Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res 69:530–539 [DOI] [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB (2002) From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA 99:7518–7523 10.1073/pnas.102091699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280:1447–1451 10.1126/science.280.5368.1447 [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193 [DOI] [PubMed] [Google Scholar]