Disrupted in Schizophrenia 1 (DISC1): Association with Schizophrenia, Schizoaffective Disorder, and Bipolar Disorder (original) (raw)
Abstract
Schizophrenia, schizoaffective disorder, and bipolar disorder are common psychiatric disorders with high heritabilities and variable phenotypes. The Disrupted in Schizophrenia 1 (DISC1) gene, on chromosome 1q42, was originally discovered and linked to schizophrenia in a Scottish kindred carrying a balanced translocation that disrupts DISC1 and DISC2. More recently, DISC1 was linked to schizophrenia, broadly defined, in the general Finnish population, through the undertransmission to affected women of a common haplotype from the region of intron 1/exon 2. We present data from a case-control study of a North American white population, confirming the underrepresentation of a common haplotype of the intron 1/exon 2 region in individuals with schizoaffective disorder. Multiple haplotypes contained within four haplotype blocks extending between exon 1 and exon 9 are associated with schizophrenia, schizoaffective disorder, and bipolar disorder. We also find overrepresentation of the exon 9 missense allele Phe607 in schizoaffective disorder. These data support the idea that these apparently distinct disorders have at least a partially convergent etiology and that variation at the DISC1 locus predisposes individuals to a variety of psychiatric disorders.
Introduction
Schizophrenia (MIM 181500) and bipolar disorder are complex psychiatric disorders affecting ∼1.8% of the population worldwide (Sklar 2002). Family and twin studies have shown that there is a strong genetic component to these psychiatric diseases. Both disorders demonstrate complex patterns of inheritance and variable phenotypes. Linkage studies have identified multiple regions as potential susceptibility loci, with little or no overlap between the loci for each of the two disorders (Sklar 2002). However, chromosome 1q42 was initially identified as a possible site for a schizophrenia susceptibility locus in a large Scottish kindred exhibiting a wide spectrum of psychiatric disorders (St. Clair et al. 1990). In this family, a balanced (1;11)(q42.1;q14.3) translocation was shown to cosegregate with schizophrenia and other psychiatric disorders. Two novel genes, Disrupted in Schizophrenia 1 and 2 (DISC1 [MIM 605210] and DISC2 [MIM 605271]), were observed to be disrupted by the translocation (Millar et al. 2000, 2001; Blackwood et al. 2001). Analysis of other independent Scottish families with schizophrenia and bipolar disorder, using intragenic SNPs, failed to show significant association between disease and these two genes (Devon et al. 2001). However, linkage of a microsatellite marker located within the DISC1 gene to disease in families from the general Finnish population confirmed the 1q42 region as a potential susceptibility locus (Hovatta et al. 1999; Ekelund et al. 2001).
The DISC1 protein is a multifunctional protein capable of protein-protein interactions via distinct functional domains (Millar et al. 2003; Morris et al. 2003; Ozeki et al. 2003; Brandon et al. 2004). The inferred sequence of DISC1 predicts a secondary structure with a globular N-terminal region and a coil-coil C-terminus. In terms of function, two microtubule-binding proteins, MAP1A (MIM 600178) and MIPT3 (MIM 607380), interact with DISC1 through two separate domains in the N-terminal half of the DISC1 protein. NUDEL (MIM 607538) and NUDE, centrosome-associated proteins, interact with DISC1 through a C-terminal domain in DISC1 that is lost as a result of the truncation seen in the original Scottish family. In addition, the transcription factors ATF4 (MIM 604064) and ATF5 (MIM 606398) bind to the C-terminus of DISC1. Through these multiple interactions, DISC1 appears to be a component of the intracellular machinery that integrates multiple functions, including intracellular transport, neuronal migration and architecture, neuronal cell signaling, and gene expression. Alteration of the functioning of such a multifunctional protein would be expected to lead to pleiotropic molecular effects dependent on the location and type of sequence change and on the functional status of the different components of intracellular functions in which DISC1 plays a role. Such mechanisms are capable of explaining how pleiotropy of phenotypic expression leads to different clinical diseases. In addition, pleiotropy may result from alterations in functions of neuronal networks with roles in diverse behaviors—the final behavioral phenotype representing the integrated expression of the functioning of different brain circuits within a developmental trajectory and environmental context. These phenomena help account for both single-gene pleiotropy (different sequence variants of a gene may have different effects) and single-locus pleiotropy (the same sequence variant may have various effects). The Scottish family identified by Blackwood et al. (2001) demonstrated single-locus pleiotropy weighted toward recurrent major depression, but other sequence variants within DISC1 might be expected to result in different disease profiles.
In a family-based study using broad diagnostic criteria, Hennah et al. (2003) reported the undertransmission of a common haplotype to affected women in the general Finnish population. In the present study, we report data obtained from a case-control data set of white individuals from North America. Our data support the finding of undertransmission of a common haplotype at the 5′ end of the DISC1 gene, but we also find evidence of association of schizophrenia, schizoaffective disorder, and bipolar disorder with multiple haplotypes contained within four blocks extending between exon 1 and exon 9. In particular, we find a strong association between a missense variant in exon 9 and schizoaffective disorder. These data suggest a generalized role for DISC1 mutations in psychiatric disease in which the phenotypic outcome is determined in part by the nature of the alteration in the DISC1 gene and in part by the effects of other modifying loci and the environment.
Methods
Diagnosis and Subject Recruitment
Subjects were recruited from the clinical services of the Zucker Hillside Hospital, a division of the North Shore–Long Island Jewish Health System (NSLIJHS). After giving written informed consent to an NSLIJHS institutional review board–approved human-research protocol, each subject was assessed with the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID, version 2.0). All SCID interviews were conducted by trained raters whose reliability was confirmed by an expert diagnostician's observation of a minimum of three interviews. The subject’s report during the structured interview, current inpatient medical records, past records (when available), and collateral information from family members were then used to rate the SCID score. Data were then compiled by the interviewer into a detailed narrative case summary. These summaries included information in regard to the onset and course of axis I disorders, presence of any axis II pathology that may have contributed to symptom presentation, any axis III diagnoses, and a brief description of the subject’s psychosocial and occupational functioning during the course of their illness. The interviewer then presented the resulting case summary to a consensus diagnostic committee that comprises a minimum of three senior faculty with DSM-IV diagnostic expertise. In addition, the committee includes junior faculty, residents, fellows, graduate students, and other trained SCID raters. After the presentation of the case summary to the committee, each case was discussed to arrive at the consensus DSM-IV diagnosis. All participants were North American white individuals of European origin. The total sample of 586 individuals was composed of 217 healthy controls (90 male, 127 female; mean age 46.3 years), 196 subjects diagnosed with schizophrenia (141 male, 55 female; mean age 39.1 years), 82 subjects with bipolar disorder (43 male, 39 female; mean age 33.7 years), and 62 subjects with schizoaffective disorder (33 male, 29 female; mean age 39.8 years).
Genotyping
A total of 39 SNP loci were genotyped by 5′-exonuclease assay. Thirty-five primer-probe sets were obtained as Taqman Assays-on-Demand (Applied Biosystems). Four primer-probe sets corresponding to missense variants reported in the dbSNP database (build 119) were designed using Assays-by-Design software (ABI). Genomic DNA (5 ng) was amplified on a 9700 thermocycler (ABI). At the end point of amplification, genotypes were discriminated using SDS 2.0 software on an Applied Biosystems 7900 Analyzer. For amplification, initial incubation was at 95°C for 10 min, followed by 40 cycles at 92°C (15 s) and 60°C (1 min). All assays successfully discriminated three genotypes for each biallelic marker. Genotyping accuracy was assessed by regenotyping one in six samples, randomly selected, which produced an overall accuracy >99%; genotyping completeness was >0.99. Haplotype block structure was determined using the Haploview program (J. C. Barrett and M. Daly; see Haploview Web site). Blocks were defined in accordance with the criteria of Gabriel et al. (2002), but the criterion for pairwise linkage disequilibrium (LD) between markers was relaxed from an average D′ >95% to an average D′ >90% within a block. Haplotypes were assigned to individuals by use of PHASE 2.02 (Stephens et al. 2001; Stephens and Donnelly 2003). Individuals with incomplete genotyping data were excluded from haplotype analyses to prevent possible distortion of haplotype frequencies by the program's inference of absent genotypes.
Statistical Methods
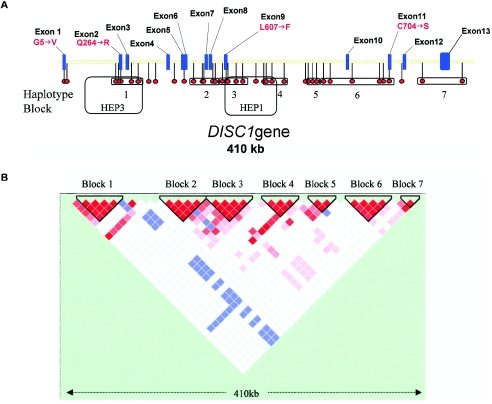
Nonparametric testing for significance values for allelic and genotypic association was performed using a standard χ2 test. Haplotype association was evaluated using the more stringent Fisher exact test, because haplotype frequencies sometimes equaled zero. (Note that it is the number of haplotypes expected that determines suitability for χ2 analysis, not the frequency observed). Bonferroni correction was not applied, because the seven DISC1 haplotype regions do not meet the criterion of independence. First, they are regions of the same gene, and, in fact, our data indicate that there may be more than one linkage signal corresponding to more than one functional locus at the gene. Second, pairwise LD values (see fig. 1B) indicate some evidence of extended LD (i.e., nonindependence) across haplotype-block boundaries.
Figure 1.
A, Schematic of the DISC1 gene, showing the 13 exons and the relative positions of the 39 SNP markers and haplotype blocks. HEP1 and HEP3 are the haplotype blocks identified by Hennah et al. (2003). B, Haploview-generated LD map of the DISC1 gene in North American white controls. Regions of high LD (D′ = 1 and _LOD_>2) are shown in bright red. Markers with lower LD (D′ < 1 and _LOD_>2) are shown in red through pink (color intensity decreases with decreasing D′ value). Regions with high LD but low informativeness (D′ = 1 and LOD<2) are shown in blue. Regions of low LD and low LOD scores (_D′_ < 2 and _LOD_<2) are shown in white. Seven haplotype blocks are identified in which average _D′_ > 0.90.
Results
We genotyped 39 SNPs (table 1) that span the 410-kb DISC1 gene from exon 1 to exon 13. Each marker was analyzed by χ2 analysis for allelic and genotypic association with schizophrenia, schizoaffective disorder, and bipolar disorder (table 1). Five markers—rs2812393, rs1322784, rs1322783, rs2255340, and _rs2738864_—show significant allelic association with schizophrenia (_P_=.048, .013, .047, .006, and .005, respectively). A single marker, rs6675281, shows significant allelic association with schizoaffective disorder (P_=.0000023). Five markers—_rs1322784, rs2255340, rs2738864, and hCV1650723, all of which also show allelic association, and _hCV9628138_—show genotypic association with schizophrenia (P_=.008, .016, .012, .003, and .037, respectively. Four markers—_rs6675281 (which also shows allelic association), hCV1650713, rs999710, and _hCV1650723_—show genotypic association with schizoaffective disorder.
Table 1.
Allele-Based and Genotype-Based Association with Schizophrenia, Schizoaffective Disorder, and Bipolar Disorder, Showing the Celera SNP and dbSNP Identifiers for Each Genotyping Marker[Note]
| Association P Valuea | Allele Frequency | ||||||
|---|---|---|---|---|---|---|---|
| Celera SNP | dbSNP | Allelic | Genotypic | Coding Change | Controls | Cases | Risk Ratio |
| hCV27474272 | rs3738400 | … | … | Gly5Met | … | … | … |
| hCV12001977 | rs1895225 | … | … | … | … | … | … |
| hCV12001946 | rs1572899 | … | .048c | … | .298 | .397 | 1.185 |
| hCV12001945 | rs1538975 | … | … | … | … | … | … |
| hCV25641899 | rs3738401 | … | … | Gln264Arg | … | … | … |
| hCV12001940 | rs1954175 | … | … | … | … | … | … |
| hCV9626784 | rs1340982 | … | … | … | … | … | … |
| hCV16113533 | rs2812379 | … | … | … | … | … | … |
| hCV16114160 | rs2793094 | … | … | … | … | … | … |
| hCV12001932 | rs1538977 | … | … | … | … | … | … |
| hCV16114126 | rs2793101 | … | … | … | … | … | … |
| hCV16113570 | rs2812393 | .048b | … | … | .336 | .404 | 1.203 |
| hCV12001930 | rs1322784 | .013b | .008b | … | .680 | .759 | 1.117 |
| hCV12001929 | rs1322783 | .047b | … | … | .849 | .899 | 1.059 |
| hCV1650649 | rs2255340 | .006b | .016b | … | .702 | .787 | 1.121 |
| hCV1650650 | rs2738864 | .005b | .012b | … | .700 | .787 | 1.125 |
| hCV1650657 | rs1407598 | … | .046d | … | .198 | .227 | 1.150 |
| hCV1650667 | rs6675281 | .0000023c | .000056c | Leu607Phe | .132 | .319 | 2.417 |
| hCV1650669 | rs1407598 | … | .039d | … | .195 | .221 | 1.133 |
| hCV9627536 | rs1535529 | … | .035d | … | .200 | .221 | 1.106 |
| hCV1650688 | rs1000731 | … | … | … | … | … | … |
| hCV1650709 | rs734551 | … | … | … | … | … | … |
| hCV1650713 | … | … | .024c | … | .337 | .327 | 1.031 |
| hCV9628231 | rs999710 | … | .003c | … | .691 | .612 | 1.129 |
| hCV1650723 | … | .01b | .003b,c | … | .309 | .410 | 1.326 |
| hCV9628176 | rs821723 | … | … | … | … | … | … |
| hCV1650750 | rs2038636 | … | … | … | … | … | … |
| hCV1650754 | rs821717 | … | … | … | … | … | … |
| hCV1433206 | rs821577 | … | … | … | … | … | … |
| hCV9628138 | … | … | .037b | … | .477 | .527 | 1.106 |
| hCV9628093 | rs1417866 | … | … | … | … | … | … |
| hCV9628061 | rs821592 | … | … | … | … | … | … |
| hCV1433188 | rs821597 | … | … | … | … | … | … |
| hCV1433174 | rs821653 | … | … | … | … | … | … |
| hCV9627519 | rs843979 | … | … | … | … | … | … |
| hCV1433135 | rs821616 | … | … | Cys704Ser | … | … | … |
| hCV1433111 | rs3524 | … | .037d | … | .498 | .617 | 1.240 |
| hCV26095 | rs2806465 | … | … | … | … | … | … |
| hCV9626960 | rs1411776 | … | … | … | … | … | … |
The DISC1 haplotype block structure (individual blocks being termed haploblocks) was determined separately for control and affected groups. Conservatively, seven blocks were identified across DISC1 in controls (fig. 1B). This block structure is altered within block 3 in individuals with schizophrenia. Marker rs6675281 (Leu607Phe) is in strong LD with neighboring markers in controls but not in individuals with schizophrenia or bipolar disorder (data not shown).
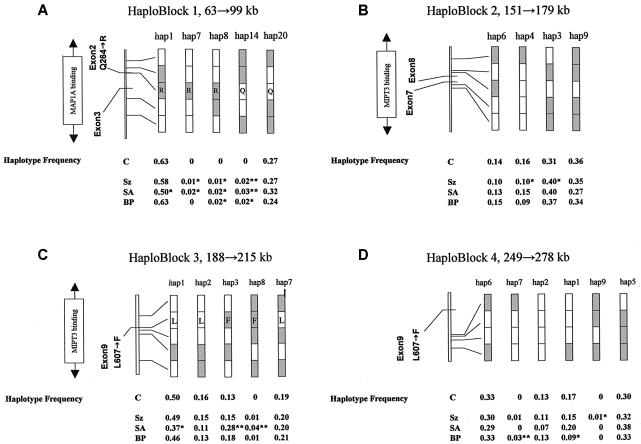
Figure 2A_–2_D shows common haplotypes for haploblocks 1–4, together with the haplotype frequency for the control group and for each case group (haplotypes, frequencies, and case/control numbers shown in table 2). Of the seven blocks we identified, four carried haplotypes that were underrepresented either in cases or in controls. Block 1 (fig. 2A) revealed a common haplotype (hap1) that was underrepresented only in patients with schizoaffective disorder (_P_=.013). Similarly, a common haplotype (hap1) in block 3 (fig. 2C) was also underrepresented in patients with schizoaffective disorder (_P_=.015). Block 1 and block 3 overlap the haplotype blocks HEP3 and HEP1, respectively, reported elsewhere (Hennah et al. 2003). HEP3 was reported to be undertransmitted to affected females in the broadest liability class, whereas HEP1, which carries the T allele of SNP rs6675281 (Phe607), was overtransmitted (Hennah et al. 2003) (fig. 1A and 1B). In block 1 (fig. 2A), two haplotypes (hap8 and hap14) showed association to schizophrenia (_P_=.021 and .0095, respectively) and schizoaffective (_P_=.047 and .001) and bipolar (_P_=.0192 and .0192) disorders. In block 1, hap7 was absent from the bipolar group but showed association with schizophrenia (_P_=.045) and schizoaffective disorder (_P_=.047). In individuals with schizophrenia, two common haplotypes in block 2 (fig. 2B) were distorted in their distribution between cases and controls. In haploblock 2, hap3 showed significant association with disease (_P_=.025), but hap4 (21111) was significantly underrepresented in schizophrenic patients (_P_=.021). In haploblock 3 (fig. 2C), two haplotypes (hap3 and hap8) show association with schizoaffective disorder (_P_=.00045 and .000054, respectively). In haploblock 4 (fig. 2D), two haplotypes show association with disease—hap8 with bipolar disorder (_P_=.0048) and hap9 with schizophrenia (_P_=.03). It should be noted that a common haplotype (hap1) in haploblock 4 was also underrepresented in patients with bipolar disorder (_P_=.027) (fig. 2D).
Figure 2.
Schematic of common haplotypes and of haplotypes showing significant association with disease, in haploblocks 1, 2, 3, and 4 (major alleles [_unshaded_] and minor alleles [_shaded_]). Haplotype frequencies for controls and for patients with schizophrenia (Sz), schizoaffective disorder (SA), and bipolar disorder (BP) are shown below the diagrams. Amino acid changes are shown within the given haplotypes for haploblock 1 (Gln264Arg) and haploblock 3 (Leu607Phe). *P<.05. **P<.01.
Table 2.
Common DISC1 Haplotypes in Cases and Controls[Note]
| No. of Individuals | |||||||
|---|---|---|---|---|---|---|---|
| Block andHaplotypeNumber | Haplotype | Frequency | Controls | Schizophrenia | SchizoaffectiveDisorder | BipolarDisorder | Total |
| Block 1: | |||||||
| 1 | 12211 | .60 | 261 | 205 | 57 | 96 | 619 |
| 2 | 12212 | .02 | 7 | 3 | 4 | 5 | 19 |
| 4 | 12222 | .03 | 15 | 8 | 2 | 2 | 27 |
| 7 | 11211 | .01 | 0 | 4 | 2 | 0 | 6 |
| 8 | 11221 | .01 | 0 | 5 | 2 | 3 | 10 |
| 11 | 22211 | .01 | 4 | 6 | 3 | 2 | 15 |
| 14 | 22112 | .01 | 0 | 6 | 3 | 3 | 12 |
| 18 | 21111 | .01 | 4 | 6 | 0 | 1 | 11 |
| 19 | 21121 | .01 | 2 | 3 | 1 | 0 | 6 |
| 20 | 21122 | .27 | 112 | 95 | 36 | 37 | 280 |
| All | … | … | 412 | 354 | 114 | 152 | 1,032 |
| Block 2: | |||||||
| 3 | 12122 | .37 | 100 | 144 | 46 | 59 | 349 |
| 4 | 21111 | .12 | 51 | 36 | 17 | 15 | 119 |
| 5 | 21122 | .03 | 7 | 12 | 3 | 4 | 26 |
| 6 | 21211 | .12 | 46 | 35 | 15 | 23 | 119 |
| 8 | 22111 | .01 | 0 | 4 | 0 | 1 | 5 |
| 9 | 22122 | .34 | 114 | 128 | 31 | 54 | 327 |
| All | … | … | 320 | 362 | 114 | 158 | 954 |
| Block 3: | |||||||
| 1 | 12121 | .48 | 210 | 176 | 43 | 72 | 501 |
| 2 | 12122 | .15 | 68 | 54 | 13 | 20 | 155 |
| 3 | 11121 | .16 | 55 | 53 | 32 | 28 | 168 |
| 7 | 22212 | .20 | 81 | 73 | 23 | 33 | 210 |
| 8 | 21212 | .01 | 0 | 2 | 5 | 2 | 9 |
| All | … | … | 418 | 362 | 116 | 156 | 1,052 |
| Block 4: | |||||||
| 1 | 1112 | .16 | 67 | 44 | 20 | 13 | 144 |
| 2 | 1111 | .12 | 51 | 31 | 7 | 18 | 107 |
| 4 | 1212 | .06 | 25 | 18 | 2 | 12 | 57 |
| 5 | 1222 | .32 | 117 | 89 | 37 | 47 | 290 |
| 6 | 2112 | .32 | 130 | 86 | 28 | 47 | 291 |
| 7 | 2111 | .01 | 0 | 2 | 0 | 4 | 6 |
| 10 | 2222 | .02 | 3 | 8 | 3 | 1 | 15 |
With the exception of the allelic (_P_=.000027) and genotypic (_P_=.0066) associations of marker rs6675281 and of hap3 and hap8 in haploblock3 (_P_=.047 and .0057, respectively) with schizoaffective disorder, none of the observed associations remains significant if the Bonferroni correction—highly conservative, given the LD in the DISC1 region—is made. These observations remain suggestive of association of DISC1 variation with schizophrenia, schizoaffective disorder, and bipolar disorder.
Discussion
The translocation in the original Scottish family demonstrated that disruption of the DISC1 gene, although sufficient to predispose an individual to psychiatric disorders, was, in itself, insufficient to predict any particular disorder. The majority of affected individuals within the Scottish kindred were affected with recurrent major depression, and some individuals carrying the translocation had no diagnosed psychiatric disorder. The Finnish study, which detected undertransmission of HEP3 (a haplotype encompassing the whole of block 1 defined in our study), used a broad phenotype for the haplotype analysis that included schizophrenia, schizoaffective disorder, and schizophrenia spectrum diagnosis, as well as bipolar disorder and major depression. In the present study, we analyzed the disease classes separately and have shown that, for haploblock 1 (HEP3 from Hennah et al. [2003]), the signal for underrepresentation/undertransmission of the most common haplotype is solely derived from the individuals with schizoaffective disorder, in our population (fig. 2). In both the Finnish study (Hennah et al. 2003) and our study, the undertransmitted haplotype carries the A allele of marker rs3738401, suggesting that the haplotype involved may be the same in both cases.
The associations we have identified are unlikely to have arisen as a result of any admixture within our test group, in part because they are congruent with results from the Finnish population and do not reflect DSC1 allele-frequency differences between populations (U.S. whites and African-Americans) we have observed. This problem will also be definitively resolved by genotyping a large genomic control panel across the case and control samples in this data set.
In the Finnish population, a later reanalysis of data supporting the association of HEP1 (corresponding to haploblock 3 defined in our study) with disease suggested that the initial association resulted from a linkage artifact (Hennah et al. 2003). However, in our population, we have found a strong association of two psychiatric disorders—bipolar disorder and schizoaffective disorder—with the T-allele (Phe607) SNP rs6675281 and the haplotypes carrying it (fig. 2C), including the most common haplotype (hap3; frequency >10%), which shows significant association with schizoaffective disorder. This data was obtained using only individuals for whom all necessary genotyping information was available, to avoid the problems that arise due to the inference of missing data. The most common haplotype carrying this allele shows association only with schizoaffective disorder, and this may explain the failure of the original HEP1 observation to pass the validation tests performed (Hennah et al. 2003).
Both hap3 and hap8 of block 3 carry Phe607. The effect of this amino acid substitution on protein function is currently unknown. The human DISC1 polypeptide has an overall amino acid identity to the mouse and rat sequences of 55.2% and 52.2%, respectively (Ma et al. 2002). Leu607 is the ancestral allele and is conserved across the mouse (Mus musculus), rat (Rattus norvegicus), pufferfish (Fugu rubripes), and zebrafish (Danio rerio). Such sequence conservation across an evolutionary time frame of >100 million years suggests functional significance of the Leu607Pro polymorphism found in the human. Furthermore, Leu607 lies within the regions identified as necessary for binding NUDEL, MIPT3, ATF4, and ATF5. Leu607 also lies at the end of a putative leucine zipper domain, although the function of this putative domain is unknown, and the sequences encoding it are absent from an alternatively spliced murine mRNA. Of the three other missense variants listed in the public database (Gly5Met, Gln264Arg, and Cys704Ser), only Gly5 is conserved among humans, rodents, and pufferfish (Ma et al. 2002). We identified the Met5 allele in an Asian/Pacific islander population at a frequency of <3% (data not shown). Rats and mice carry a serine residue at the position corresponding to human residue 704 and carry a threonine at the position corresponding to human residue 264 (Ma et al. 2002; Taylor et al. 2003).
Schizophrenia and bipolar disorder are generally considered to be separate entities, but patients who exhibit multiple symptoms of both disorders are often given the hybrid diagnosis of schizoaffective disorder (Blacker and Tsuang 1992). This has helped give rise to the argument that schizophrenia and bipolar disorder are variant expressions of a diathesis, in part because of the similar disease frequencies, ages at onset, and absence of sex bias of the two disorders. Supporting this theory are studies that have linked the G72/G30 locus at 13q33 to both bipolar disorder (Hattori et al. 2003; Chen et al. 2004) and schizophrenia (Chumakov et al. 2002; Schumacher et al. 2004) and that have linked bipolar disorder to 22q11-13 and to 8p22, 10p14, and 18p11.2, regions previously linked to schizophrenia (Gill et al. 1996; Blouin et al. 1998; Faraone et al. 1998; Schwab et al. 1998, 2000; Straub et al. 1998; Detera-Wadleigh et al. 1999; Berrettini 2000, 2003; Foroud et al. 2000; Ophoff et al. 2002). In addition, possible common risk loci for the two disorders have been identified on chromosomes 4p16 and 3q27 (Bailer et al. 2002; Als et al. 2004). The argument of shared neurobiology is also supported by the effectiveness of antipsychotic drugs in the treatment of psychotic symptoms of both disorders (Adler and Strakowski 2003; Sundram et al. 2003). Countering this argument is the genetic epidemiological data (Badner and Gershon 2002; Sklar 2002; Levinson et al. 2003; Lewis et al. 2003; Segurado et al. 2003), which show little or no evidence for cosegregation of schizophrenia and bipolar disorder in families, although families both of patients with schizophrenia and of patients with bipolar disorder are at increased risk for schizoaffective disorder and unipolar depression (Baron 2001). Also, the two disorders show marked differences in their responses to lithium, anticonvulsants, and antidepressants. Imaging studies and postmortem analyses have shown altered neurostructure, with reduced brain weight and increased ventricular cerebrospinal fluid suggestive of cortical atrophy, seen only in schizophrenia and not in bipolar or the affective disorders. Indeed, it has been demonstrated in the parahippocampal region of individuals with schizophrenia that there is cytoarchitectural disorganization of the entorhinal region, with too few neurons in the superficial layers and too many neurons in the deeper layers (Jakob and Beckmann 1986; Arnold et al. 1991), and that there is a similar laminar shift in the cingulated cortex, suggesting a generalized failure of neurons to migrate and settle in the appropriate cortical locations (Benes et al. 1991; Akbarian et al. 1993). Our data suggest that the presence of particular haplotypes at the 5′ end of the DISC1 gene may predispose individuals to any of these three psychiatric disorders. Three haplotypes in haploblock 1 showed association with all three disorders (hap7 was not represented in the group with bipolar disorder and therefore showed no association).
As discussed above, there are candidate functional variants of DISC1. However, the identity of the functional DISC1 variant(s) that may be predisposing to psychiatric disease is still unknown. The effects of disrupting DISC1 in the original Scottish family appear to arise as a result of haploinsufficiency rather than loss of function, since none of the members of the family were reported to be homozygous for the translocation, although a possible dominant negative mechanism cannot be ruled out. Therefore, in addition to mutations that affect protein function, any mutation or polymorphism that affects the rate at which the DISC1 gene is transcribed might also be expected to predispose an individual to disease. It is possible that the predisposing haplotypes identified in blocks 1 and 2 are also associated with lower levels of transcription as a result of changes in putative intronic enhancer elements. An alternative is that the associating haplotypes may carry currently unknown missense or nonsense mutations in the coding sequence. These possibilities can be resolved by pulse-chase experiments to determine transcriptional activity and by resequencing. The strongest association of a DISC1 haplotype with disease was in haploblock 3, in which haplotypes carrying the allele encoding Phe607—in place of the evolutionarily conserved Leu607 within the third leucine zipper domain—show strong association with schizoaffective disorder. Leu607Phe could affect DISC1 interactions with multiple proteins, because this residue is a constituent of multiple-binding domains: NUDEL, MAP1A, ATF4, and ATF5. However, the functional significance of Leu607 is currently unknown. The DISC2 gene overlaps DISC1 and is thought to encode a nontranslated mRNA. DISC2 is also disrupted in the translocation observed in the Scottish family. The function of this gene is unknown, but marker _rs6675281_—which shows robust association with schizoaffective disorder—also lies within DISC2, and it can be argued that marker rs6675281 could be in strong LD with an important DISC2 polymorphism. The haplotype block HEP1 (Hennah et al. 2003) showed suggestive association with schizophrenia and also encompassed portions of both DISC1 and DISC2. The presence of a coding polymorphism in DISC1 makes this gene the leading candidate, but a functional consequence of the Leu607Phe transition needs to be demonstrated or an as-yet unknown missense or splicing variant needs to be identified by resequencing for this possibility to be confirmed.
The converging data from multiple studies suggest that all three disorders—schizophrenia, schizoaffective disorder, and bipolar disorder—have an overlapping root in altered DISC1 function or expression, but it is possible that different variants within the DISC1 gene determine variation in phenotype through corresponding variation in interactions with DISC1’s binding partners. It should also be noted that, with the exception of block 1, there is only an association overlap between schizoaffective disorder and bipolar disorder. This raises the possibility that different mechanisms involving DISC1 can cause schizophrenia and to bipolar and schizoaffective disorders.
Brain imaging (Weinberger and Lipska 1995) and neuroanatomical data (Jakob and Beckmann 1986; Arnold et al. 1991; Benes et al. 1991) suggest that schizophrenia arises from an early neuronal deficit leading to aberrant folding of the cortical regions. Studies of DISC1 protein expression in rodents have indicated that its expression is developmentally regulated, with the highest expression during late embryogenesis (Ozeki et al. 2003). Loss of DISC1 expression in early brain development or loss of the ability of DISC1 to bind NUDEL, whose gene is known to be associated with diseases of cortical development (reviewed by Ross and Walsh [2001]), allows us to propose a neurodevelopmental model for the role of DISC1 in the development of schizophrenia. In this model, NUDEL-mediated microtubule organization is altered because of reduced DISC1 binding. The defect in microtubule function leads to deficits in neuronal migration, axonal extension, and neurite outgrowth. Reduced neurite outgrowth has been shown in cultured PC12 cells expressing mutant forms of DISC1 (Ozeki et al. 2003). The affective disorders and bipolar disorder, which exhibit none of the structural alterations seen in the brains of patients with schizophrenia, may arise solely as a result of disruption of DISC1 protein-protein interactions in mature neurons. For these intriguing possibilities to be resolved, it will be necessary to identify the exact nature of DISC1 alterations in affected individuals and to model those changes both in tissue culture, in which changes in protein-protein interaction can be evaluated, and in knockout mice, in which the developmental implications of particular DISC1 alleles can be determined. Given the heterogeneity of phenotypes in all three disease classes, it is also possible that alterations in DISC1 function or expression account for only a fraction of cases observed and that other mechanisms can give rise to similar phenotypes.
Acknowledgments
This project was supported by grants from the National Institutes of Mental Health (K23MH001760), the National Alliance for Research on Schizophrenia and Depression, and the Stanley Medical Research Institute (to A.K.M.) and by the National Institute on Alchohol Abuse and Alcoholism Intramural Research Program. The authors also wish to acknowledge the efforts of all the patients and staff at the Zucker Hillside Hospital who participated in this study.
Electronic-Database Information
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Haploview (by J. C. Barrett and M. Daly), http://www-genome.wi.mit.edu/personal/jcbarret/haploview/downloads/hapinstall.exe
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia, DISC1, DISC2, MAP1A, MIPT3, NUDEL, ATF4, and ATF5)
References
- Adler CM, Strakowski SM (2003) Boundaries of schizophrenia. Psychiatr Clin North Am 26:1–23 [DOI] [PubMed] [Google Scholar]
- Akbarian S, Vinuela A, Kim JJ, Potkin SG, Bunney WE Jr, Jones EG (1993) Distorted distribution of nicotinamideadenine dinucleotide phosphate-diaphorase neurons in temporal lobe of schizophrenics implies anomalous cortical development. Arch Gen Psychiatry 50:178–187 [DOI] [PubMed] [Google Scholar]
- Als TD, Dahl HA, Flint TJ, Wang AG, Vang M, Mors O, Kruse TA, Ewald H (2004) Possible evidence for a common risk locus for bipolar affective disorder and schizophrenia on chromosome 4p16 in patients from the Faroe Islands. Mol Psychiatry 9:93–98 [DOI] [PubMed] [Google Scholar]
- Arnold SE, Hyman BT, Van Hoesen GW, Damasio AR (1991) Some cytoarchitectural abnormalities of the entorhinal cortex in schizophrenia. Arch Gen Psychiatry 48:625–632 [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411 [DOI] [PubMed] [Google Scholar]
- Bailer U, Leisch F, Meszaros K, Lenzinger E, Willinger U, Strobl R, Heiden A, Gebhardt C, Doge E, Fuchs K, Sieghart W, Kasper S, Hornik K, Aschauer HN (2002) Genome scan for susceptibility loci for schizophrenia and bipolar disorder. Biol Psychiatry 52:40–52 [DOI] [PubMed] [Google Scholar]
- Baron M (2001) Genetics of schizophrenia and the new millennium: progress and pitfalls. Am J Hum Genet 68:299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, McSparren J, Bird ED, SanGiovanni JP, Vincent SL (1991) Deficits in small interneurons in prefrontal and cingulate cortices of schizophrenic and schizoaffective patients. Arch Gen Psychiatry 48:996–1001 [DOI] [PubMed] [Google Scholar]
- Berrettini WH (2000) Are schizophrenic and bipolar disorders related? a review of family and molecular studies. Biol Psychiatry 48:531–538 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Evidence for shared susceptibility in bipolar disorder and schizophrenia. Am J Med Genet 123C:59–64 [DOI] [PubMed] [Google Scholar]
- Blacker D, Tsuang MT (1992) Contested boundaries of bipolar disorder and the limits of categorical diagnosis in psychiatry. Am J Psychiatry 149:1473–1483 [DOI] [PubMed] [Google Scholar]
- Blackwood DHR, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ (2001) Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet 69:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–3 [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ (2004) Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci 25:42–55 [DOI] [PubMed] [Google Scholar]
- Chen Y-S, Akula N, Detera-Wadleigh SD, Schulze TG, Thomas J, Potash JB, DePaulo JR, McInnis MG, Cox NJ, McMahon FJ (2004) Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry 9:87–92 [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, et al (2002) Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 99:13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI Jr, Gershon ES (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 96:5604–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devon RS, Anderson S, Teague PW, Burgess P, Kipari TM, Semple CA, Millar JK, Muir WJ, Murray V, Pelosi AJ, Blackwood DH, Porteous DJ (2001) Identification of polymorphisms within Disrupted in Schizophrenia 1 and Disrupted in Schizophrenia 2, and an investigation of their association with schizophrenia and bipolar affective disorder. Psychiatr Genet 11:71–78 [DOI] [PubMed] [Google Scholar]
- Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, Suhonen J, Ellonen P, Chan G, Sinsheimer JS, Sobel E, Juvonen H, Arajarvi R, Partonen T, Suvisaari J, Lonnqvist J, Meyer J, Peltonen L (2001) Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet 10:1611–1617 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, Markel P, Lee H, Harkavy Friedman J, Kaufmann C, Cloninger CR, Tsuang MT (1998) Genome scan of European-American schizophrenia pedigrees: results of the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 81:290–295 [PubMed] [Google Scholar]
- Foroud T, Castelluccio PF, Koller DL, Edenberg HJ, Miller M, Bowman E, Rau NL, Smiley C, Rice JP, Goate A, Armstrong C, Bierut LJ, Reich T, Detera-Wadleigh SD, Goldin LR, Badner JA, Guroff JJ, Gershon ES, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Nurnberger JI Jr (2000) Suggestive evidence of a locus on chromosome 10p using the NIMH genetics initiative bipolar affective disorder pedigrees. Am J Med Genet 96:18–23 [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, McGuffin P, et al (1996) A combined analysis of D22S278 marker alleles in affected sib-pairs: support for a susceptibility locus for schizophrenia at chromosome 22q12: Schizophrenia Collaborative Linkage Group (Chromosome 22). Am J Med Genet 67:40–45 [DOI] [PubMed] [Google Scholar]
- Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES (2003) Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet 72:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Varilo T, Kestilä M, Paunio T, Arajärvi R, Haukka J, Parker A, Martin R, Levitzky S, Partonen T, Meyer J, Lönnqvist J, Peltonen L, Ekelund J (2003) Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet 12:3151–3159 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin M-L, Väisänen L, Mannila H, Lönnqvist J, Peltonen L (1999) A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 65:1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob H, Beckmann H (1986) Prenatal developmental disturbances in the limbic allocortex in schizophrenics. J Neural Transm 65:303–326 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Levinson MD, Segurado R, Lewis CM (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: methods and power analysis. Am J Hum Genet 73:17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Liu Y, Ky B, Shughrue PJ, Austin CP, Morris JA (2002) Cloning and characterization of Disc1, the mouse ortholog of DISC1 (Disrupted-in-Schizophrenia 1). Genomics 80:662–672 [DOI] [PubMed] [Google Scholar]
- Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, Devon RS, Arveiler B, Muir WJ, Blackwood DH, Porteous DJ (2001) Genomic structure and localisation within a linkage hotspot of Disrupted in Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry 6:173–178 [DOI] [PubMed] [Google Scholar]
- Millar JK, Christie S, Porteous DJ (2003) Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun 311:1019–1025 [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9:1415–1423 [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP (2003) DISC1 (Disrupted-in-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet 12:1591–1608 [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Escamilla MA, Service SK, Spesny M, Meshi DB, Poon W, Molina J, Fournier E, Gallegos A, Mathews C, Neylan T, Batki SL, Roche E, Ramirez M, Silva S, De Mille MC, Dong P, Leon PE, Reus VI, Sandkuijl LA, Freimer NB (2002) Genomewide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. Am J Hum Genet 71:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A (2003) Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to Nude-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA 100:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ME, Walsh CA (2001) Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci 24:1041–1070 [DOI] [PubMed] [Google Scholar]
- Schumacher J, Abon Jamra R, Freudenburg J, Becker T, Ohlraun S, Otte ACJ, Tullius M, Kovalenko S, Van Den Bogaert A, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S (2004) Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar disorder. Mol Psychiatry 9:203–207 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, Trixler M, Falkai P, Rietschel M, Maier W, Wildenauer DB (2000) A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10p and 6. Mol Psychiatry 5:638–649 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Lerer B, Albus M, Borrmann M, Hönig S, Strauß M, Segman R, Lichtermann D, Knapp M, Trixler M, Maier W, Wildenauer DB (1998) Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am J Hum Genet 63:1139–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI Jr, Craddock N, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 73:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P (2002) Linkage analysis in psychiatric disorders: the emerging picture. Annu Rev Genomics Hum Genet 3:371–413 [DOI] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosdeen C, Evans HJ (1990) Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336:13–16 [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Martin RB, Ma Y, Myakishev MV, Harris-Kerr C, Webb BT, O’Neill FA, Walsh D, Kendler KS (1998) A schizophrenia locus may be located in region 10p15-p11. Am J Med Genet 81:296–301 [DOI] [PubMed] [Google Scholar]
- Sundram S, Joyce PR, Kennedy MA (2003) Schizophrenia and bipolar affective disorder: perspectives for the development of therapeutics. Curr Mol Med 3:393–407 [DOI] [PubMed] [Google Scholar]
- Taylor MS, Devon RS, Millar JK, Porteous DJ (2003) Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics 81:67–77 [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Lipska BK (1995) Cortical maldevelopment, anti-psychotic drugs, and schizophrenia: a search for common ground. Schizophr Res 16:87–110 [DOI] [PubMed] [Google Scholar]