The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations (original) (raw)
Abstract
Paleoanthropological evidence indicates that both the Levantine corridor and the Horn of Africa served, repeatedly, as migratory corridors between Africa and Eurasia. We have begun investigating the roles of these passageways in bidirectional migrations of anatomically modern humans, by analyzing 45 informative biallelic markers as well as 10 microsatellite loci on the nonrecombining region of the Y chromosome (NRY) in 121 and 147 extant males from Oman and northern Egypt, respectively. The present study uncovers three important points concerning these demic movements: (1) The E3b1-M78 and E3b3-M123 lineages, as well as the R1*-M173 lineages, mark gene flow between Egypt and the Levant during the Upper Paleolithic and Mesolithic. (2) In contrast, the Horn of Africa appears to be of minor importance in the human migratory movements between Africa and Eurasia represented by these chromosomes, an observation based on the frequency distributions of E3b*-M35 (no known downstream mutations) and M173. (3) The areal diffusion patterns of G-M201, J-12f2, the derivative M173 haplogroups, and M2 suggest more recent genetic associations between the Middle East and Africa, involving the Levantine corridor and/or Arab slave routes. Affinities to African groups were also evaluated by determining the NRY haplogroup composition in 434 samples from seven sub-Saharan African populations. Oman and Egypt’s NRY frequency distributions appear to be much more similar to those of the Middle East than to any sub-Saharan African population, suggesting a much larger Eurasian genetic component. Finally, the overall phylogeographic profile reveals several clinal patterns and genetic partitions that may indicate source, direction, and relative timing of different waves of dispersals and expansions involving these nine populations.
Introduction
Extant genetic diversity within Africa is particularly complex, not only because of its primary role in the evolution of our species but also because Africa was both a source and a recipient of transcontinental gene flow during different episodes of human history. In addition to the Strait of Gibraltar connecting Iberia and Maghreb (North Africa) (Hitti 1990; Newman 1995; Bosch et al. 2001), two other migration routes between Africa and Eurasia are generally considered, the Levantine corridor and the Horn of Africa (Cavalli-Sforza et al. 1994; Kivisild et al. 1999; Quintana-Murci et al. 1999; Stringer 2000; Underhill et al. 2001_b_; Bar-Yosef 2002; Nebel et al. 2002). These two passageways have their immediate termini at opposite ends of the Arabian Peninsula, the geographic nexus of Africa, Europe, and Asia. Although a principal-component analysis of African populations, based on 79 protein polymorphisms (Cavalli-Sforza et al. 1993, 1994) and mtDNA haplogroups (Salas et al. 2002), revealed pan-African substructure, only slight differences are suggested between the Nile River Valley in the north and regions near the Bab-el Mandeb channel located between the Horn of Africa and the Arabian Peninsula. Furthermore, Y chromosome data involving 63 samples from Cairo typed at 10 binary markers failed to detect any genetic barriers between Egypt and the Middle East (Manni et al. 2002). These studies suggest a regional genetic continuity among the Nile River Valley, the Middle East, and the Arabian Peninsula. In contrast, analysis of a larger set of Egyptian samples using the complex 49a,f polymorphic system revealed some geographic substructure in the Nile River Valley (Lucotte and Mercier 2003). However, direct correlation to PCR-compatible binary polymorphisms is often not straightforward, because of the limited resolution and the inherent potential for recurrent mutations in the 49a,f locus.
It is fortunate that the establishment of a well-resolved, unambiguous Y chromosome genealogy (Underhill et al. 2000) and its continuing refinement (Underhill et al. 2001_b_; Hammer 2002; Jobling and TylerSmith 2003) has provided new opportunities to evaluate temporal and spatial aspects concerning these movements in and out of Africa. The strong phylogeographic pattern observed with the haploid Y chromosome phylogeny indicates that this marker system is a more sensitive index of divergence relative to other loci. Consequently, to better understand population relationships in two pivotal contact zones between Eurasia and Africa, we investigated the nonrecombining Y chromosome (NRY) binary marker patterns of affinity and diversification in 147 North Egyptian and 121 Omani males. In addition, the NRY composition of Egypt and Oman are compared with those of various African collections. Specifically, 434 samples from seven sub-Saharan populations were examined to evaluate levels of affinity among geographically targeted regions. The phylogeographic profile that emerges reveals several areal patterns and genetic partitions that may, in some cases, indicate source, direction, and relative timing of different waves of dispersals and expansions involving these nine populations. Our findings are then viewed within the context of other Eurasian and African data. This integrative approach provides a thorough comparison of Middle Eastern and sub-Saharan NRY affinity levels in Egypt and Oman, two populations proximal to the Levantine and Horn of Africa migratory corridors, respectively.
Material and Methods
Populations Analyzed
A total of 702 male DNA samples was isolated from peripheral blood lymphocytes by standard protocol (Sambrook et al. 1989). Information concerning geography, ethnic origin, and linguistic affiliation of the populations examined is provided in table 1. Adherence to ethical guidelines was followed as stipulated by each of the universities involved.
Table 1.
Populations Analyzed
| Linguistic Affiliation | ||||
|---|---|---|---|---|
| Geographic Areaand Country | Population | Family | Sublevel | Reference |
| Central Africa: | ||||
| Central African Republic | Biaka Pygmies | Niger-Congo | Benué-Congo, Bantu | Underhill et al. 2000 |
| Democratic Republic of Congo | Mbuti Pygmies | Nilo-Saharan | Central Sudanic | Underhill et al. 2000 |
| Central West Africa: | ||||
| Cameroon (southern) | Bakaka | Niger-Congo | Benué-Congo, Bantu | Cruciani et al. 2002 |
| Cameroon (southern) | Bamileke | Niger-Congo | Benué-Congo, Bantoid | Cruciani et al. 2002 |
| Cameroon (southern) | Bamileke | Niger-Congo | Benué-Congo, Bantoid | Present study |
| Cameroon (southern) | Bantu | Niger-Congo | Benué-Congo, Bantu | Present study |
| Cameroon (southern) | Ewondo | Niger-Congo | Benué-Congo, Bantu | Cruciani et al. 2002 |
| Cameroon (northern) | Fali | Niger-Congo | Adamawa | Cruciani et al. 2002 |
| Cameroon (northern) | Mixed Nilo-Saharan | Nilo-Saharan | Central Sudanic/Saharan | Cruciani et al. 2002 |
| Cameroon (northern) | Tali | Niger-Congo | Adamawa | Cruciani et al. 2002 |
| East Africa: | ||||
| Tanzania | Wairak | Niger-Congo | Benué-Congo, Bantu | Present study |
| Kenya | Bantu | Niger-Congo | Benué-Congo, Bantu | Present study |
| Ethiopia | Ethiopian Jews | Afro-Asiatic | Cushitic | Cruciani et al. 2002 |
| Rwanda | Hutu | Niger-Congo | Benué-Congo, Bantu | Present study |
| Rwanda | Tutsi | Niger-Congo | Benué-Congo | Present study |
| North Africa: | ||||
| Morocco | Arabs | Afro-Asiatic | Semitic | Cruciani et al. 2002 |
| Morocco | Berbers | Afro-Asiatic | Berber | Cruciani et al. 2002 |
| Egypt | Arabs/Berbersa | Afro-Asiatic | Semitic | Present study |
| South Africa: | ||||
| South Africa | Khwe | Khoisan | Central | Cruciani et al. 2002 |
| South Africa | !Kung | Khoisan | Northern | Cruciani et al. 2002 |
| West Africa: | ||||
| Benin | Fon | Niger-Congo | Volta-Congo, Fon | Present study |
| Burkina Faso | Fulbe | Niger-Congo | West Atlantic | Cruciani et al. 2002 |
| Burkina Faso | Mossi | Niger-Congo | Voltaic | Cruciani et al. 2002 |
| Burkina Faso | Rimaibe | Niger-Congo | West Atlantic | Cruciani et al. 2002 |
| Near East/Asia: | ||||
| Oman | Arabs | Afro-Asiatic | Semitic | Present study |
Y-Haplogroup Analysis
Forty-five binary markers were genotyped by various standard methods, including denaturing high-performance liquid chromatography, RFLP, direct sequencing, and size detection of two insertional polymorphisms, the YAP (a Y-specific PAI or polymorphic Alu insertion [Rowold and Herrera 2000) and the 12f2 long insertion element (LINE). Lineages are referred to in the text by haplogroup and terminal mutation, according to the Y Chromosome Consortium (YCC) standardized nomenclature (Hammer 2002; Jobling and Tyler-Smith 2003). Details concerning primer sets and the allelic states of binary markers have been published elsewhere (Underhill et al. 2001_b_; Cruciani et al. 2002; Semino et al. 2002).
Statistical and Phylogenetic Analyses
Sixteen additional African populations from previous studies (table 1) were included in the statistical and phylogenetic analyses to generate a more complete picture of African Y-haplogroup variation and phylogeographic relationships. An analysis of molecular variance (AMOVA) (Excoffier et al. 1992), based on the resulting Y-haplogroup distributions, was executed using the Arlequin version 2.000 package. Two sets of AMOVAs were performed since the 25 populations were subdivided into statistical groups according to either geographic or linguistic criteria. An updated version of the maximum parsimony phylogeny illustrated in Cruciani et al. (2002) served as a reference to measure molecular distances (i.e., number of mutational steps) between haplogroups. A maximum-likelihood (ML) phylogeny constructed using the Phylip v3.6 program (Felsenstein 1989) and a correspondence analysis conducted with the NTSYSpc– 2.02i package by Rohlf (2002) are included as well.
Microsatellite Analysis
The Egyptian and Omani samples were also analyzed at 10 microsatellite loci as described by Cinnioğlu et al. (2004). In the expansion analysis, we have used a value of 25 years as intergeneration time to be congruent with previous studies (Cruciani et al. 2002; Cinnioğlu et al. (2004). We have also included results of an expansion analysis based on an intergeneration time of 30 years (see table 3) as recommended by Tremblay and Vézina (2000), for comparison purposes.
Table 3.
Y Chromosome Haplogroup Variance and Expansion Times Based on 10 STR Loci
| BATWING with 25-YearGeneration Timeb(ky) | BATWING with 30-YearGeneration Timec(ky) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Country andHaplogroup | N | Haplogroup Variance | Ta(ky) | Mean | Median | .025Quantile | .975Quantile | Mean | Median | .025Quantile | .975Quantile |
| Egypt: | |||||||||||
| E3b-M35 | 52 | .50 | 17.9 | 47.4 | 16.9 | .4 | 532.0 | 56.9 | 20.3 | .5 | 638.4 |
| E3b1-M78 | 26 | .42 | 15.0 | 12.6 | 7.8 | 1.4 | 105.7 | 15.1 | 9.4 | 1.7 | 126.8 |
| E3b2-M81 | 12 | .15 | 5.4 | 11.6 | 5.7 | .1 | 144.0 | 13.9 | 6.8 | .1 | 172.8 |
| E3b3-M123 | 10 | .41 | 14.6 | 25.3 | 10.8 | .2 | 352.2 | 30.4 | 12.9 | .3 | 422.6 |
| G-M201 | 13 | .36 | 12.9 | 13.7 | 6.2 | .1 | 281.2 | 16.5 | 7.4 | .1 | 337.4 |
| J-12f2 | 48 | .45 | 16.1 | 11.9 | 10.2 | 2.1 | 58.2 | 14.2 | 12.3 | 2.6 | 69.9 |
| J*-12f2d | 31 | .31 | 11.1 | 15.0 | 6.4 | .6 | 278.5 | 18.0 | 7.6 | .7 | 334.1 |
| J2-M172 | 17 | .46 | 16.4 | 27.5 | 17.9 | 1.7 | 188.4 | 33.0 | 21.5 | 2.1 | 226.1 |
| K2-M70 | 12 | .49 | 17.5 | 25.5 | 13.7 | .5 | 242.4 | 30.6 | 16.4 | .6 | 290.9 |
| Oman: | |||||||||||
| E3b-M35 | 17 | .14 | 5.0 | 6.4 | 2.4 | .0 | 112.1 | 7.6 | 2.9 | .0 | 134.5 |
| E3b3-M123 | 15 | .05 | 1.7 | 5.9 | 3.2 | .1 | 55.1 | 7.1 | 3.9 | .1 | 66.1 |
| J-12f2 | 58 | .40 | 14.2 | 23.6 | 7.7 | .2 | 428.4 | 28.4 | 9.2 | .2 | 514.1 |
| J*-12f2 | 46 | .27 | 9.6 | 3.4 | 2.3 | .6 | 29.2 | 4.0 | 2.8 | .7 | 35.0 |
| J2-M172 | 12 | .58 | 20.7 | 14.4 | 4.4 | .1 | 306.2 | 17.3 | 5.2 | .1 | 367.4 |
| K2-M70 | 10 | .18 | 6.4 | 9.0 | 1.6 | .0 | 318.2 | 10.8 | 1.9 | .0 | 381.9 |
| R1a1-M17 | 11 | .32 | 11.4 | 11.0 | 3.4 | .0 | 290.7 | 13.2 | 4.0 | .1 | 348.8 |
Microsatellite variances for all significant binary haplogroups were calculated using the vp equation of Kayser et al. (1997). Both the linear expansion and BATWING (Bayesian analysis of trees with internal node generation) (Wilson et al. 2000) procedures employ the stepwise mutation model (Ohta and Kimura 1973; Wehrhahn 1975; Di Rienzo et al. 1994; Kittles et al. 1998) and a mean STR mutation rate of 0.0007 per STR locus per generation (Kittles et al. 1998; Zhivotovsky et al. 2003).
Results
Phylogeography
Thirty-eight binary markers were found to be polymorphic, and these define 31 paternal lineages. The apportionments of these lineages by population are displayed in a hierarchical phylogeny (fig. 1) that shows the correspondence between the previous haplotype categories of Underhill et al. (2000) and the YCC nomenclature (Hammer 2002; Jobling and Tyler-Smith 2003). Figure 1 also lists the Y-specific SNP (Y-SNP)/YAP/12f2 frequency profiles of the nine populations sampled in the present study. Figure 2 depicts geographic distribution of the observed YCC groups and major subclades. The most obvious aspect of the biogeographic pattern is a north versus south—as well as an east versus west—partitioning of NRY diversification. The Afro-Asiatic populations of Egypt and Oman exhibit, by far, the highest number of both groups and haplogroups (a mean of 9.5 groups and 20 haplogroups), followed by the East African collections of Tanzania, Kenya, Rwandan Hutu, and Rwandan Tutsi (a mean of 3 groups and 8 haplogroups). There is a considerable decline in diversity from east to west along a Central African corridor to Benin. The West African populations are represented by an average of only two groups and three haplogroups.
Figure 1.
Maximum-parsimony hierarchy and frequency table. A total of 44 binary markers and 36 haplogroups are represented; 31 of these haplogroups are detected in 702 African and Omani males. Markers not typed are shown in italics. Om = Oman, Eg = Egypt, SC = southern Cameroon, Kn = Kenya, Tn = Tanzania, Bn = Benin, Rw = Rwanda.
Figure 2.
Geographic frequency distribution of binary markers in eight African populations and one Omani population
Nearly all of the Y chromosomes in the sub-Saharan collections belong to groups A, B, and E. Furthermore, the vast majority of these individuals (92.2%) are members of group E, the only group observed in all nine populations.
J is the most common group of the Omani collection (frequency 47.9%), followed in succession by E (23.1%), R (10.7%), and K2 (8.3%). In Egypt, the order of the polymorphic groups is slightly different: E (39.5%), J (32.0%), G (8.8%), K2 (8.2%), and R (7.5%). Noteworthy is the asymmetrical representation of groups A, B, and E in the sub-Saharan versus the Afro-Asiatic samples (98.8% vs. 32.1%, respectively).
A3b2-M13, the lone representative of group A in our collection, occurs in Kenya (13.8%), Tanzania (7.0%), Egypt (2.7%), and Oman (0.8%). In contrast, group B is seen in all sub-Saharan collections except Benin and Bamileke. In the Tutsi, the frequency of B2b-M112 is considerably higher than that of B2a-M150 (13.8% vs. 1.1%, respectively), whereas the frequencies are similar in the Hutu (1.4% and 2.9%, respectively) and Tanzania (4.7% and 2.3%, respectively). Only B2a-M150 chromosomes were detected in southern Cameroon and Kenya (7.1% and 3.4%, respectively).
For group E, we observe a geographic gradient from west to east as well as a partitioning from south to north. The sample collections from the western sub-Saharan populations (Benin and Bamileke) are represented exclusively by group E, whereas the frequencies of these chromosomes are somewhat lower in the east (94.2%, 85.1%, 81.4%, and 82.8% for the Hutu, the Tutsi, Tanzania, and Kenya, respectively) and drop sharply in the northern-most populations of Egypt (39.5%) and Oman (23.1%). There exists a west-to-east as well as a south-to-north clinal distribution with respect to E3a-M2. Bamileke and Benin display the highest frequencies (100% and 95.0%, respectively), Kenya and Tanzania show intermediate values, and Oman (7.4%) and Egypt (2.8%) exhibit relatively low percentages of this subclade. In sub-Saharan Africa, the east-to-west clinal distribution of E3b-M35 is inverse to that displayed by E3a-M2. The percentage of these M35 haplogroups is >35% in Tanzania and Egypt, whereas it is less than half of that value in Oman and Kenya. The level of this mutation is very low in the Tutsi and the Hutu samples (<3% in both) and drops to zero in the more western populations of Cameroon and Benin. Nearly all of the E3b-M35 chromosomes in the Egypt (92%) and Oman (100%) collections harbor downstream mutations (E3b1-M78, E3b2-M81, and E3b3-M123), which are absent in the sub-Saharan populations.
In the majority of the nine populations sampled, the M75 subclade is either absent or nearly so (frequencies of <5%). The Rwandan Hutu and Kenya are the exceptions, with M75 found in 8.6% and 17.2% of their respective samples. The E1-M33 polymorphism, distinguishing the remaining subclade of E, is detected in two Egyptian males (1.4%).
Group C appears to be relatively rare in the populations sampled in this study. Only four Omani individuals (3.3% of the Omani) harbor the RPS4Y mutation. There appears to be a sharp north-south partitioning of F-related lineages (i.e., groups F, G, H, I, and J). Although these are completely absent in the sub-Saharan collections, they constitute the majority of the Omani (51.2%) samples as well as a relatively high proportion of Egyptian males (41.5%). Of these lineages, the J-12f2 clade is by far the most prevalent (32% frequency in Egypt and 47.9% in Oman). K2-M70 chromosomes are detected in Tanzania (3.8%), Egypt (8.2%), and Oman (8.3%). M173 chromosomes (group R) are observed in the Bantu of southern Cameroon (14.3%), Oman (10.7%), Egypt (6.8%), and the Hutu (1.4%). Whereas the R1*-M173 undifferentiated lineage is present in all four populations, the two downstream mutations, M17 and M269, are confined to Egypt and Oman.
AMOVA
The results of the AMOVA analysis are listed in table 2. The overall _F_st (among all 25 collections) is estimated to be 0.333 (P<.0001), indicating a significant degree of Y-haplogroup interpopulation diversity throughout the sample area. Upon assignment of the populations within six broad geographic regions (North Africa, East Africa, Central Africa, Central West Africa, West Africa, and South Africa), we obtained an _F_ct of 0.192 (_P_=.003) and an _F_sc of 0.174 (P<.0001), which suggests significant geographical structuring involving both inter- and intraregional heterogeneity, respectively. We observed similar results when the populations were grouped by language (Afro-Asiatic, Nilo-Saharan, Atlantic Congo, Volta-Congo, and Khoisan). In this case, the _F_ct is 0.204 (_P_=.002), and the _F_sc equals 0.170 (P<.0001), indicating significant inter- and intralinguistic structure, respectively.
Table 2.
Results of AMOVA
| Sample | No. ofPopulations | No. ofGroups | _F_st (P Value) | _F_ct (P Value) | _F_sc (P Value) |
|---|---|---|---|---|---|
| Geographic region: | |||||
| Overall | 25 | 6 | .333 (.000) | .192 (.003) | .174 (.000) |
| North Africa | 4 | 1 | .209 (.000) | ||
| East Africa | 3 | 1 | .107 (.002) | ||
| West Africa | 4 | 1 | .198 (.000) | ||
| Central-West Africa | 8 | 1 | .175 (.000) | ||
| Northern Cameroon | 3 | 1 | .086 (.018) | ||
| Southern Cameroon | 5 | 1 | .081 (.003) | ||
| Central Africa | 4 | 1 | .080 (.000) | ||
| South Africa | 2 | 1 | .105 (.007) | ||
| Sub-Saharan Africa | 21 | 1 | .186 (.000) | ||
| Language group: | |||||
| Overall | 25 | 6 | .339 (.000) | .204 (.002) | .170 (.000) |
| Afro-Asiatic | 5 | 1 | .214 (.000) | ||
| Nilo-Saharan | 2 | 1 | .052 (.185) | ||
| West Atlantic | 2 | 1 | .119 (.029) | ||
| Volta-Congo | 3 | 1 | .165 (.000) | ||
| Benué-Congo | 10 | 1 | .106 (.000) | ||
| Khoisan | 2 | 1 | .105 (.007) |
ML Phylogeny
An ML analysis (fig. 3) performed on the NRY frequencies in table 1 reveals patterns of geographic associations and population affinities. Strong geographic structure is apparent, since, for the most part, the three main clusters (West African/Central West African, Central African, and North African/East African/South African) represent distinct regions of the African continent, with Oman clustering with the North African groups.
Figure 3.
ML radial phylogeny based on Y-haplogroup frequencies of 25 populations (24 African and 1 Omani). The nodal values represent the number of bootstrap replicates out of 1,000 that share the corresponding bifurcations. “PS*” denotes the present study.
Correspondence Analysis (CA)
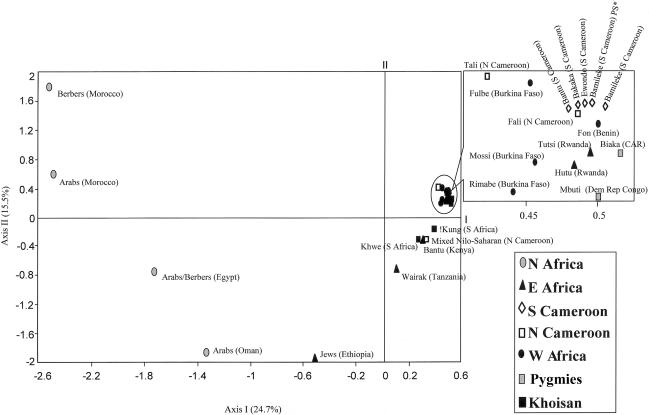
Figure 4 is a CA involving 25 populations and 31 haplogroups. Dimensions 1, 2, and 3 account for 25%, 16%, and 12% of the total variation, respectively. Most notable is an Afro-Asiatic/sub-Saharan partitioning along axis 1. The negative plot positions of North Africa, Oman, and Ethiopia most likely reflect the common occurrence of several E3b-M35 derivatives as well as the G and J components. The clustering of the remaining 20 populations to the right of the origin may be influenced by the presence of one or more of the B-M60, E, and R1*-M173 lineages. The North African and Omani groups are well differentiated along both axis 2 and axis 3 (not shown) but are close with respect to axis 1. The two western Afro-Asiatic populations (Moroccan Arabs and Berbers) are located in the positive region of axis 2 and in the negative region of axis 3, whereas the more eastern Afro-Asiatic populations (Egypt and Oman) occupy opposite positions along these dimensions. The graphical separation of these two factions may be due to differences with respect to both E3b3-M123 and J frequencies.
Figure 4.
CA based on the Y-SNP haplotype frequency data of 25 populations (24 African and 1 Omani). Percent of total inertia is shown for each axis. “PS*” denotes the present study.
Y-Microsatellite Diversity
The microsatellite results for Egyptian and Omani samples are given in table 3. The variance, continuous expansion, and median BATWING values of the Egyptian M35 lineages are considerably larger than those of Oman. This is also true for K2-M70. However, for either the collective J-12f2 or J*-12f2, the disparity is not so large. The expansion times of collective E3b-M35 lineages of the Egyptian sample are substantially older than those of the J-12f2, whereas, in Oman, the order is reversed. On the average, the median BATWING values based on the 30-year generation time are 20% greater than those calculated using a generation interval of 25 years.
Discussion
Y-Chromosome Distribution in Sub-Saharan Africa
The sub-Saharan NRY haplogroup distribution displays distinctive regional patterns. Of particular interest is an east-west partitioning of sub-Saharan haplogroup diversity. To the west, Benin, Bamileke, and southern Cameroon are represented predominantly by chromosomes carrying the E3a-M2 mutation, a signature of the recent expansion of Bantu populations (Passarino et al. 1998; Scozzari et al. 1999; Underhill et al. 2001_b_; Cruciani et al. 2002). Although the E3a-M2 subclade is prevalent in our East African groups (Tutsi, Hutu, Kenya, and Tanzania) as well, these collections contain several additional Y-chromosomal types and, thus, demonstrate a much higher level of NRY diversity. Therefore, unlike its hegemony in the west, E3a-M2’s contribution to the genetic landscape of East Africa was not great enough to completely erase pre-existing Y haplogroups and may have been diluted further by subsequent migratory movements from the north involving other Y chromosomes. The patterns we have observed are, for the most part, consistent with and may provide additional support for published hypotheses concerning African population dynamics (Underhill et al. 2001_b_; Cruciani et al. 2002).
When taken in context with previous studies, the current NRY data seem to reflect the linguistic boundaries demarcating southern Kenya as the northern limit of the Bantu speakers as they progressed eastward through the Central African corridor and southward along the Swahili coast. Kenya displays an E3a-M2 frequency of 52%, whereas the more northern populations, such as Ethiopia (Underhill et al. 2000; Semino et al. 2002), the Ethiopian Jews (Cruciani et al. 2002), and Sudan (Underhill et al. 2000), are characterized by frequencies close to or at zero. The genetic composition of the people inhabiting the area east of the East African Ridge is especially complex and may reflect the superposition of multiple demic events over the past 5,000 years. This complexity is depicted by the results of the CA (fig. 4). The two East African Bantu populations, Kenya and Tanzania, are positioned between Ethiopia and other sub-Saharan African collections, perhaps representing an NRY composition reflecting geographical proximity and linguistic affiliation, respectively. The Bantu contribution to both the Kenyan and Tanzanian NRY composition is obvious (52% and 42% of E3a-M2, respectively).
The joint occurrence of E3a-M2 and E3b-M35 chromosomes in the East African (Tanzania and Kenya) and Central African (Hutu and Tutsi) populations (fig. 1) represents a convergence of independent demographic events. The E3b-M35–related lineages may be a legacy left by earlier inhabitants. As indicated above, E3a-M2 was most likely introduced into the region later by Bantu speakers from the west.
Genetic contributions from the Neolithic and Bantu expansions are observed in East Africa and are demarcated by a reciprocal north-south partitioning of their respective markers. As stated above, Kenya is the northern limit of E3a-M2, whereas J-12f2, described as a marker of the Neolithic expansion (Semino et al. 2000), extends southward only as far as Ethiopia. The geographic distribution of these chromosomes is consistent with the arrangement of the Ethiopian Jews, and the Kenyan and Tanzanian populations in the CA (fig. 4) along the first dimension. These groups lie between the sub-Saharan and North African populations in the plot. The intermediate status of these three East African groups is mirrored in the ML phylogeny (fig. 3) as well, since, in the North African/East African/South African cluster, the Ethiopian Jews, the Tanzanians, and the Kenyans are wedged between the North African (including Oman) and either the Central African (as is the case with the Ethiopian Jews and Tanzanians) or South African groups (see the position of the Kenyans in the ML tree).
The _F_ct estimates based on geographic and linguistic groups are very similar (0.192 and 0.204, respectively; table 2). From these results, we argue that language affiliation and geographic location contribute more or less equally to the overall African Y-haplogroup substructure. However, in some cases one criterion prevails over the other. For example, although the 10 Bantoid-speaking populations occupy a wide geographic range, they show an _F_st value close to—and, in some cases, lower than—that of other, more geographically restricted language groups with a much smaller number of populations. Perhaps this relative NRY-haplogroup homogeneity, consistent with the values obtained from autosomal markers (Cavalli-Sforza et al. 1994), reflects the fact that the Bantu language is a cultural hallmark of a people who, via a pervasive and protracted demographic expansion across a large tract of sub-Saharan Africa (Newman 1995), has shaped a substantial portion of its current genetic landscape with the M2 lineage. Nevertheless, the Bantoid _F_st (0.106; table 2) indicates that differences among the 10 groups are still significant (P<.0005), and this variation most likely arises from the differing genetic contributions from previous inhabitants and/or subsequent migrations of non-Bantu speakers into these regions. These NRY differences are apparent in the topology of the ML radial phylogeny since the Bantu is the only linguistic group found in all three geographically based clades.
In contrast with our findings, results of AMOVA for mtDNA data in African populations (Salas et al. 2002) suggest a lower importance of language versus geography in defining differences among the main groups in the female genetic pool. This may reflect a general pattern of cultural assimilation of the indigenous females during the Bantu expansion. This scenario would explain the greater linguistic differentiation reflected in the Y chromosome.
Middle Eastern/African Affinities of Oman and Egypt
Egypt and Oman demonstrate a much higher NRY diversity than do the sub-Saharan groups. A total of 12 groups and 25 haplogroups each are detected in these two populations, versus 5 groups and 14 haplogroups in the seven sub-Saharan sample collections. The substantial intrapopulation variation of Egyptian and Omani assemblages reflects the multiple demographic events that have occurred in these regions (i.e., in northeastern Africa and the Near East), in part because of their geographical positions near strategic crossroads between Africa and Eurasia.
The NRY composition of the Egyptian and Omani collections exhibits a greater Middle Eastern versus sub-Saharan affinity. The cumulative frequency of typical sub-Saharan lineages (A, B, E1, E2, E3a, and E3b*) is 9% in Egypt and 10% in Oman, whereas the haplogroups of Eurasian origin (Groups C, D, and F–Q) account for 59% and 77%, respectively. These profiles display levels of diversity similar to those of the nine Turkish populations reported by Cinnioğlu et al. (2004) (an average of 9 groups and 19 haplogroups per population) and also include polymorphic frequencies of many of the lineages observed in Turkey (i.e., E3b1-M78, Eb3-M123, G-M201, the collective J-12f2, J2-M172, and R1a1-M17). Many of these haplogroups are common throughout the Middle East and Europe as well, and several are thought to have arisen somewhere within this range (Underhill et al. 2001_b_).
The Levantine Corridor versus the Horn of Africa
Both the Horn of Africa and the Levantine corridor have been proposed as main passageways for migrations of anatomically modern humans out of Africa (Cavalli-Sforza et al. 1994; Lahr and Foley 1994). The distributions of haplogroups C and D suggest early dispersals (50–45 thousand years [ky] ago) of ancestors of these lineages from the Horn of Africa to southern Asia (Underhill et al. 2001_b_), most likely in conjunction with mtDNA haplogroup M (Quintana-Murci et al. 1999; Kivisild et al. 1999). The presence of haplogroup C chromosomes found in Oman (3%) is consistent with this hypothesis, although more-recent back flow from the east cannot be ruled out. The use of the southern route, however, may have been restricted to intervals of low sea levels and mild monsoonal conditions (Underhill et al. 2001_b_). Posterior demic movements, such as the one represented by the dissemination of the ancestral M89 chromosomes and the 10873T mtDNA lineage to Eurasia, most likely occurred via the Levantine corridor, ∼45 ky ago (Underhill et al. 2001_b_; Quintana-Murci et al. 1999).
A more recent dispersal out of Africa, represented by the E3b-M35 chromosomes, expanded northward during the Mesolithic (Underhill et al. 2001_b_). The East African origin of this lineage is supported by the much larger variance of the E3b-M35 males in Egypt versus Oman (0.5 versus 0.14; table 3). Consistent with the NRY data is the mtDNA expansion estimate of 10–20 ky ago for the East African M1 clade. Local expansions of this clade and subsequent demic movements may have resulted in the irregular presence of the M1 haplogroup in the Mediterranean area (Quintana-Murci et al. 1999).
M35 chromosomes are seen in the Oman, North African, and East African populations, as well as in the South African Khoisans (Underhill et al. 2000; Cruciani et al. 2002; present study). There are three distinctive sublineages (E3b1-M78, E3b2-M123, and E3b3-M81) that display nonrandom distributions (fig. 1). E3b1-M78 predominates in Egypt and Ethiopia, E3b3-M123 in Oman, and E3b2-M81 in northwestern Africa. Importantly, these three sublineages are restricted to regions north of the equator. In contrast, the E3b*-M35 lineages appear to be confined almost exclusively to the sub-Saharan populations, except for a very low incidence in Egypt (2.7%) and a somewhat larger frequency in Ethiopia (7%, as reported by Underhill et al. [2000]). The highest levels of E3b*-M35 are in Tanzania (37.2%), Kenya (13.8%), and the Khoisans (11% in !Kung and 31% in Khwe).
The present-day Egyptian E3b-M35 distribution most likely results from a juxtaposition of various demic episodes. Since the E3b*-M35 lineages appear to be confined mostly to the sub-Saharan populations, it is conceivable that the initial migrations toward North Africa from the south primarily involved derivative E3b-M35 lineages. These include E3b1-M78, a haplogroup especially common in Ethiopia (23%), and, perhaps, E3b2-M123 (2%), which is present as well (Underhill et al. 2000; Cruciani et al. 2002; Semino et al. 2002). The data suggest that two later expansions may have followed: one eastward along the Levantine corridor into the Near East and the other toward northwestern Africa. The extant North African and Middle Eastern distribution (Underhill et al. 2001_b_; Cruciani et al. 2002; present study) of these lineages suggests that both routes are associated with the dissemination of E3b1-M78. However, the E3b3-M123 chromosomes may have spread predominantly toward the east, whereas E3b2-M81, which is present in relatively high levels in Morocco (33% and 69% in Moroccan Arabs and Moroccan Berbers, respectively [Cruciani et al. 2002]), dispersed mainly to the west. This proposal is in accordance with a population expansion involving E3b2-M81 believed to have occurred in northwestern Africa ∼2 ky ago (Cruciani et al. 2002). The considerably older linear expansion estimate of the Egyptian E3b2-M81 (5.4 ky ago) is also compatible with this scenario. The Turkish collection displays a near absence of E3b2-M81 (1 in 523 males) but displays polymorphic levels of the other two M35 derivatives (5.5% for E3b3-M123 and 5% for E3b1-M78 [Cinnioğlu et al. 2004]). This M35 profile, combined with the substantially older BATWING expansion times of the corresponding Egyptian lineages (E3b1-M78: 7.8 ky in Egypt vs. 4.8 ky in Turkey [Cinnioğlu et al. 2004]; E3b3-M123: 10.8 ky in Egypt vs. 3.7 ky in Turkey [Cinnioğlu et al. 2004]), is highly consistent with a northbound migration through the Levantine corridor reflected in M35 males as far north as Turkey. The complexity of the E3b-M35 fraction in Egypt may have been enhanced by several episodes of backflow, beginning with the introduction of agriculture into Africa, and, later, various historical events, such as the Greek, Roman, and Arab occupations (Lucotte and Mercier 2003). These migrations, along with the much older existence of the Egyptian M35 lineage, may have contributed to the substantially higher variances observed among the Egyptian M35 chromosomes versus those of Oman (0.5 vs. 0.14, respectively).
The distribution pattern of M35 derivatives suggests that the diffusion of M35 into Oman did not, to any great extent, involve the Horn of Africa route. This hypothesis is based on the observation that Tanzania and Kenya, the two collections in the present study that are closest to the Bab-el Mandeb Channel, lack the most abundant Omani M35 lineage (i.e., E3b3-M123 at 12%) but carry respectable levels of E3b*-M35 (37.2% in Tanzania and 14% in Kenya; see fig. 1), which is not detected in Omani males. Furthermore, the M35 profile of Ethiopia (Underhill et al. 2000) is quite different from that of Oman. The Ethiopian collection exhibits a polymorphic level of undifferentiated M35 (7%), as well as a much lower frequency of E3b3-M123 (2%) versus that of E3b1-M78 (23%), a lineage that is nearly absent in Oman (1.7%).
It is possible that the arrival of the E3b1-M78 and E3b3-M123 chromosomes into Oman may have coincided with that of M201 and 12f2 (which collectively represent 50% of the Omani sample), since all four mutations are recognized as signatures of the Neolithic expansion from the Middle East (Underhill et al. 2001_b_; King and Underhill 2002). Although the relatively younger expansion times of the collective E3b-M35 lineages (including both the M78 and M123 derivatives) in comparison with that displayed by the various J lineages (table 3) may hint at a somewhat later appearance of M35 in Oman, the older J-12f2 expansion times may also be a result of multiple sources for these chromosomes. The presence of the G and J lineages in Egypt (40%; fig. 1) probably represents a southern branch of the Neolithic agricultural diffusion, which may have returned some E3b-M35 chromosomes as well. The J-12f2 BATWING expansion estimates in Egypt and Oman (10.2 and 7.7 ky, respectively) fit well with this hypothesis. The BATWING expansion times of both the Turkish G and J lineages are substantially older than those of the Egyptian (20.3 and 13.9 ky in Turkey [Cinnioğlu et al. 2004] vs. 6.2 and 10.2 ky in Egypt, for G-M201 and J-12f2, respectively). This finding is compatible with an expected earlier introduction of Neolithic lineages into Turkey and an entry into Africa by way of the Levantine corridor. In addition, the low level of presumptive 12f2 chromosomes in East Africa (Ethiopia and Sudan [Underhill et al. 2000] and Tanzania and Kenya [present study]), except for a relatively low frequency in the more recently admixed Ethiopian Jews (5%) (Cruciani et al. 2002), argues against an introduction of this mutation into Africa via the Horn of Africa passage during the Neolithic migration.
More-recent gene flow between Africa and the Arabian Peninsula is indicated by the presence of M2 lineages in Oman. However, these episodes most likely involve the seafaring routes used in the East African slave trade in the last millennium or so (Anderson 1995). Since substantial frequencies of the E3b*-M35 chromosomes persist in East Africa to the present day, centuries after the Bantu-mediated introduction of E3a-M2, it is expected that the East Africans contributed the E3b*-M35 types along with any E3a-M2 haplogroups during these more recent demic movements. Hence, the occurrence of E3a-M2 lineages in Omani males is not likely to be due to gene flow from East Africa since only derivative M35 chromosomes are detected in the Omani sample. On the other hand, the presence of M2 combined with the absence of E3b*-M35 is a profile common in central sub-Saharan Africa. A scenario consistent with these findings involves the collection of slaves, from a location perhaps as far westward as Central Africa, to be dispatched to the Arabian Peninsula from various points along the Swahili coast.
A study addressing the issue of gene flow from sub-Saharan Africa into Near Eastern Arab populations shows a higher level of female migration compared with male gene flow through this area (Richards et al. 2003). In that investigation, the mtDNAs of sub-Saharan origin occur at a frequency of ∼35% in the Yemen Hadramawt and 10%–15% in other Arab populations, whereas the NRY data indicate that the level of recent male gene flow was substantially lower. The most likely explanation for this pattern is sex bias in the Arab slave trade from Africa. In addition, it may reflect differences in reproductive fitness between the sexes, resulting from male sterilization and/or the incorporation of women into Arab societies as concubines. However, our sample from Oman displays the highest frequency of E3a-M2 in Arab Near Eastern populations (7%), comparable only to the 4% in Yemen Hadramawt and significantly higher than in the rest of this area (0%–1%) (Richards et al. 2003), and may indicate different levels of male slave assimilation in different Arab populations.
Integration of our results with previous data expands the known R1*-M173 distribution and yields additional support for a previous hypothesis suggesting that the presence of this lineage in Africa signals a backflow from Asia (Cruciani et al. 2002) that has also been associated with the existence of the Eurasian mtDNA haplogroups U6 and H in the Fulbe population of Nigeria (Salas et al. 2002). The Oman collection in the present study is the only population outside of Africa in which R1*-M173 has been found. Up until now, these undifferentiated lineages have been detected only in Egypt and in some Central and West African populations (Cruciani et al. 2002; present study). It is plausible that the African and Omani R1*-M173 chromosomes may be relics of an ancient back migration from Asia to Africa, which may have been a southern branch of the Upper Paleolithic westward expansion of this clade after its emergence in northern Asia ∼30 ky ago (Underhill et al. 2001_b_). The antiquity of the M173 backflow is implied by the total lack in sub-Saharan Africa of downstream mutations associated with the post–Last Glacial Maximum (LGM) reinhabitation of Eurasia (R1a1-M17 and R1b-M269) (Semino et al. 2000) or, later, with the Neolithic expansion (J-12f2 and G-M201) (Hammer et al. 2000; Semino et al. 2000; King and Underhill 2002; Cinnioğlu et al. 2004).
Egypt is the only African population that is known to harbor all three M173 subtypes (R1b-M269, R1*-M173, and R1a1-M17). This unique status is most likely due to Egypt’s strategic location and its long history of interaction with Eurasia. Oman, like Egypt, also exhibits all three M173 haplogroups. The relatively high frequency of R1a1-M17 (9%) may result from the post-LGM expansion associated with this mutation. The expansion estimates of this haplogroup (11.4–3.4 ky; see table 3) support this hypothesis. The above data strongly suggest that the Levantine corridor was, by far, more important than the Horn of Africa passage in the original African dispersal of undifferentiated M173 chromosomes as well as the more recent introduction into Africa of its derivatives, since the M173 mutation is nearly absent in East, Central, and South African collections, except for a 1% frequency in both the Ethiopian (Underhill et al. 2000) and the Hutu assemblages.
K2-M70 is believed to have originated in Asia after the emergence of the K-M9 polymorphism (45–30 ky) (Underhill et al. 2001_a_). As deduced from the collective data (Underhill et al. 2000; Cruciani et al. 2002; Semino et al. 2002; present study), K2-M70 individuals, at some later point, proceeded south to Africa. These chromosomes are seen in relatively high frequencies in Egypt, Oman, Tanzania, Ethiopia, and Morocco and are especially prominent in the Fulbe (18% [Scozzari et al. 1997, 1999]), the highest concentration of this haplogroup found so far. The current patchy distribution of K2-M70 in Africa may be a remnant of a more widespread occupation. Subsequent demic events introducing chromosomes carrying the E3b-M35, E3a-M2, G-M201, and J-12f2 haplogroups may have overwhelmed the K2-M70 representatives in some areas. Like the R1*-M173 males, the M70 individuals could represent the relics of an early back migration to Africa from Asia, since these chromosomes are not associated with the G-M201, J-12f2, and R1-M173 derivatives, lineages that represent more-recent Eurasian genetic contributions (Semino et al. 2000; Underhill et al. 2001_b_). The K2-M70 expansion estimates in Egypt (17.5–13.7 ky; see table 3) are consistent with an early African diaspora. From the present-day African distribution of K2-M70, it is difficult to determine which of the two Africa/Asia migratory passages, if any, prevailed in its southward journey. However, the BATWING expansion estimates of both the Egyptian and Turkish K2-M70 lineages (13.7 ky and 9.0 ky, respectively) are much older than that of Oman (1.6 ky), which suggests that the Levantine corridor may have been used more extensively in the African dissemination of this lineage as well.
Conclusion
The diverse NRY haplotypes observed in Egypt and Oman are, to a large extent, distinctive from those of sub-Saharan collections and establish a substantial base for comparisons with other regional populations. NRY markers typical of the current sub-Saharan Africa (E3a*-M2 and derivatives) are represented by low frequencies in Egypt and Oman and, thus, may be a recent acquisition, at least in part, from the slave trade. In contrast, markers signaling the Neolithic expansion from the Middle East (12f2, M201, and M35 derivatives) constitute the predominant component in these two Afro-Asiatic populations. The situation is further complicated by the fact that, unlike 12f2 and the M201, which are Eurasian in origin, the undifferentiated M35 lineage can be traced back to the Mesolithic in East Africa. In Egypt, known M35 derivatives are present at polymorphic levels and there is a near absence of undifferentiated M35. It is reasonable to believe that the Levantine corridor may have played an important role in the dispersal from Africa reflected by these chromosomes (involving both forward and backward flow). The lack of E3b*-M35, a common East African haplogroup, in Oman, and the asymmetrical presence of the two Omani M35 derivatives (E3b3-M123 has a greater frequency than E3b1-M78), as well as the differential distribution of M173 and 12f2 lineages in the integrated collection, reinforce the idea that the migratory movements between Eurasia and Africa involving these chromosomes occurred mainly across the Levantine corridor and that genetic flow through the Horn of Africa during these demic episodes was very limited. Nevertheless, previous studies support the importance of the Horn of Africa as a passageway in earlier human migrations.
Acknowledgments
This work was partially supported by National Institutes of Health grants GM 08205 (to R.J.H.) and GM 61347 (to D.J.R.). In addition, Xunta de Galicia grant XUGA20011/B98 provided funds in the form of a predoctoral fellowship to M.R.. We would also like to express our deepest appreciation to Kristina Prince of Stanford University for her significant contributions.
References
- Anderson SE (1995) The black holocaust for beginners. African World Press, Trenton, NJ [Google Scholar]
- Bar-Yosef O (2002) The Upper Paleolithic revolution. Annu Rev Anthropol 31:363–393 10.1146/annurev.anthro.31.040402.085416 [DOI] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1993) Demic expansions and human evolution. Science 259:639–646 [DOI] [PubMed] [Google Scholar]
- ——— (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Cinnioğlu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri Gl, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA (2004) Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 114:127–148 [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari, R., Underhill, PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- Hammer M (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 10.1073/pnas.100115997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti P (1990) The Arabs: a short history. Gateway Editions, Washington, DC [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, et al (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133 10.1007/s004140050051 [DOI] [PubMed] [Google Scholar]
- King R, Underhill P (2002) Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages. Antiquity 76:707–714 [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Bamshad M, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins W, Dixon M, Papiha S, Mastana S, Mir M, Ferak V, Villems R (1999) Deep common ancestry of Indian and Western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 10.1016/S0960-9822(00)80057-3 [DOI] [PubMed] [Google Scholar]
- Lahr M, Foley R (1994) Multiple dispersals and modern human origins. Evol Anthropol 3:48–60 [Google Scholar]
- Lucotte G, Mercier G (2003) Y-chromosome haplotypes in Egypt. Am J Phys Anthropol 121:63–66 10.1002/ajpa.10190 [DOI] [PubMed] [Google Scholar]
- Manni F, Leonardi P, Barakat A, Rouba H, Heyer E, Klintschar M, McElreavey K, Quintana-Murci L (2002) Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa. Hum Biol 74:645–658 [DOI] [PubMed] [Google Scholar]
- Nebel A, Landau-Tasseron E, Filon D, Oppenheim A, Faerman M (2002) Genetic evidence for the expansion of Arabian tribes into the Southern Levant and North Africa. Am J Hum Genet 70:1594–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J (1995) The peopling of Africa: a geographic interpretation. Yale University Press, New Haven [Google Scholar]
- Ohta T, Kimura M (1973) The model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a genetic population. Genet Res 22:201–204 [DOI] [PubMed] [Google Scholar]
- Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS (1998) Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 62:420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H, Passarino G, McElreavey K, Santachira-Benerecetti (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 10.1038/70550 [DOI] [PubMed] [Google Scholar]
- Richards M, Rengo C, Cruciani F, Gratrix F, Wilson J, Scozzari R, Macaulay V, Torroni A (2003) Extensive female-mediated gene flow from sub-Saharan Africa into Near Eastern Arab populations. Am J Hum Genet 72:1058–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F (2002) NTSYSpc. Exeter Publishing, Setauket, NY [Google Scholar]
- Rowold DJ, Herrera RJ (2000) Alu elements and the human genome. Genetica 108:57–72 10.1023/A:1004099605261 [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu MV, Sobrino B, Sanchez-Diz P, Macaulay V, Carracedo A (2002) The making of the African mtDNA landscape. Am J Hum Genet 71:1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scozzari R, Cruciani F, Malaspina P, Santolamazza P, Ciminelli BM, Torroni A, Modiano D, et al (1997) Differential structuring of human populations for homologous X and Y microsatellite loci. Am J Hum Genet 61:719–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Malaspina P, Torroni A, Sellitto D, Arredi B, Destro-Bisol G, De Stefano G, Rickards O, Martinez-Labarga C, Modiano D, Biondi G, Moral P, Olckers A, Wallace DC, Novelletto A (1999) Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 65:829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA (2002) Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 70:265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C (2000) Palaeoanthropology: coasting out of Africa. Nature 405:24–5, 27 10.1038/35011166 [DOI] [PubMed] [Google Scholar]
- Tremblay M, Vézina H (2000) New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet 66:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Marzuki S, Oefner PJ, Cavalli-Sforza LL, Chambers GK (2001_a_) Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Hum Mutat 17:271–280 10.1002/humu.23 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001_b_) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 10.1046/j.1469-1809.2001.6510043.x [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Wehrhahn C (1975) The evolution of selectively similar electrophoretically detectable alleles in finite populations. Genetics 80:375–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I, Weale M, Balding D (2000) BATWING: Bayesian analysis of trees with internal node generation. Department of Mathematical Sciences, University of Aberdeen, Aberdeen [Google Scholar]
- Zhivotovsky L, Rosenberg N, Feldman M (2003) Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet 72:1171–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]