Convergent Actions of IκB Kinase β and Protein Kinase Cδ Modulate mRNA Stability through Phosphorylation of 14-3-3β Complexed with Tristetraprolin (original) (raw)
Abstract
Regulation of gene expression at the level of mRNA stability is a major topic of research; however, knowledge about the regulatory mechanisms affecting the binding and function of AU-rich element (ARE)-binding proteins (AUBPs) in response to extracellular signals is minimal. The β1,4-galactosyltransferase 1 (β4GalT1) gene enabled us to study the mechanisms involved in binding of tristetraprolin (TTP) as the stability of its mRNA is regulated solely through one ARE bound by TTP in resting human umbilical vein endothelial cells. Here, we provide evidence that the complex formation of TTP with 14-3-3β is required to bind β4GalT1 mRNA and promote its decay. Furthermore, upon tumor necrosis factor alpha stimulation, the activation of both Iκβ kinase and protein kinase Cδ is involved in the phosphorylation of 14-3-3β on two serine residues, paralleled by release of binding of TTP and 14-3-3β from β4GalT1 mRNA, nuclear sequestration of TTP, and β4GalT1 mRNA stabilization. Thus, a key mechanism regulating mRNA binding and function of the destabilizing AUBP TTP involves the phosphorylation status of 14-3-3β.
Posttranscriptional regulation of gene expression offers a powerful mechanism to swiftly respond to extracellular stimuli and either up- or downregulate the expression of targeted proteins. An important posttranscriptional mechanism exerts its effects on mRNA stability. mRNA molecules contain specific _cis_-acting elements that are bound by _trans_-acting factors that affect the rate at which mRNA is degraded. The most common _cis_-acting elements in mammalian cells involved in rapid mRNA decay are the AU-rich elements (ARE). AREs contain one or more repeats of the pentanucleotide AUUUA sequence within the 3′ untranslated regions (3′UTR) of short-lived mRNA molecules. The regulation of gene expression through ARE-mediated modulation of mRNA plays a critical role during cell growth and differentation, in apoptosis, and in the immune response (2, 8, 34).
Various ARE-binding proteins (AUBPs) have been previously described. Both destabilizing and stabilizing factors have been identified which dynamically modulate mRNA turnover in an opposing fashion. Although the exact mechanisms by which the RNA/protein interactions affect the mRNA deadenylation and decay remain elusive, it has been reported that the destabilizing AUBPs directly or indirectly recruit the exosome, a multiprotein complex of 3′-to-5′ exoribonucleases, promoting mRNA decay (7, 15). Proteins belonging to the class of destabilizing AUBPs are AU-rich RNA-binding factor 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D; K homology splicing regulatory protein (KSRP); tristetraprolin (TTP); and the TTP-relative butyrate response factor-1 (BRF1) (3, 15, 32, 37). Proteins that have been shown to increase the stability of mRNA include Hu antigen R, nuclear factor 90, and nucleolin (4, 34, 36). These factors are thought to block mRNA decay by preventing binding of the destabilizing AUBPs and thus the recruitment of the exosome. Another layer of complexity is added by AUBPs like CUGBP2, which facilitate mRNA stabilization but at the same time inhibit translation (23).
Despite a large knowledge base about the AUBPs involved in regulating mRNA stability, how mRNA binds the factors and how the switch between destabilizing and stabilizing proteins is regulated remain ill defined. A variety of signaling pathways induced by inflammatory stimuli have been implicated in either stabilization or destabilization of mRNAs. Whereas the use of inhibitors and dominant negative mutants of signaling proteins has shown the involvement of TAK1, mitogen-activated protein (MAP) kinase kinase kinase 1, MAP kinase kinase 6, p38, p38 MAP kinase-activated protein kinase 2 (MK2; also known as MAPKAP2), c-jun N-terminal kinase, and extracellular signal-regulated kinase in modulating mRNA turnover, it is unclear how these mechanisms are linked (10, 11, 17, 22, 45). More specifically, MK2 has been shown to be involved in the phosphorylation of the destabilizing factor TTP, influencing the association of TTP with the chaperone protein 14-3-3, yet it remains unresolved how phosphorylation affects the mRNA binding and activity of TTP (9). In contrast, the phosphorylation of AUF1 has been shown to remodel local RNA structures and most likely regulates mRNA turnover by altering the recruitment of the degradation machinery; however, the signaling pathways and kinases responsible for AUF1 phosphorylation in vivo remain elusive (43, 44).
Recently, we reported that the stability of the β1,4-galactosyltransferase I (β4GalT1) mRNA is solely mediated through the second AU-rich element (AU2), which in resting primary human umbilical vein endothelial cells (HUVECs) is bound by a destabilizing factor. The proinflammatory cytokine tumor necrosis factor alpha (TNF-α) upregulates the expression of β4GalT1 via mRNA stabilization (13). To further understand how mRNA stability is modulated in response to extracellular stimuli, we investigated the signaling pathways and regulatory mechanisms involved in TNF-α-induced stabilization of β4GalT1 mRNA. It was found that in resting HUVECs, AU2 was bound by a destabilizing complex of TTP and 14-3-3β, resulting in rapid mRNA turnover. We observed that TNF-α induced two distinct signaling pathways, one mediated by inhibitor κB kinase β (IKKβ) and the other was mediated by protein kinase Cδ (PKCδ), which resulted in the dislodgment of the TTP/14-3-3β complex from AU2 and the nuclear translocation of TTP, paralleled by an increase in the β4GalT1 mRNA half-life. We provide a mechanism for these observations through the phosphorylation of 14-3-3β by IKKβ and PKCδ on serine residues Ser132 and Ser60, respectively, which interferes with its binding to TTP and hence the retention of TTP in the cytoplasm.
MATERIALS AND METHODS
HUVEC isolation, cell culture, and stimulation.
Endothelial cells were isolated from human umbilical cord veins by collagenase digestion as described previously (14). Cells were maintained in M199 medium supplemented with 10% human serum, 10% fetal calf serum, 1 mM l-glutamine, 5 U/ml heparin, 150 μg/ml basic fibroblast growth factor, 100 U/ml penicillin, and 100 U/ml streptomycin in 0.75% (wt/vol) gelatin-coated culture dishes.
The human cervical epithelial carcinoma cell line HeLa TetOff (HeLa TO) was purchased from BD Clontech (Palo Alto, CA) and maintained in high-glucose Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 0.1 mg/ml G418.
Prior to experiments, cells were serum starved for 2 h. TNF-α (CLB, Amsterdam, The Netherlands) was used at an end concentration of 100 U/ml. Preincubation with inhibitors occurred for 6 h (leptomycin B [LMB]) or 2 h (all other inhibitors). The inhibitors were used at the following end concentrations: 5 μM AG490 (JAK2 inhibitor; Alexis, Lausen, Switzerland), 5 μM Akt inhibitor (Alexis), 4 μM BAY11-7082 (IKK inhibitor; BioMol, Plymouth Meeting, PA), 0.2 μM FTI-277 (farnesyltransferase inhibitor; Calbiochem, Nottingham, United Kingdom), 20 μM SP600125 (c-jun N-terminal kinase inhibitor; BioMol), 20 μM LY294002 (phosphatidylinositol 3-kinase inhibitor; Cell Signaling Technology, Beverly, MA), 10 μM PD98059 (MEK1 inhibitor; Cell Signaling), 10 μM U0126 (MEK1/2 inhibitor; Cell Signaling), 20 nM Raf1 kinase inhibitor I (Calbiochem), 1 μM SB203580 (p38 inhibitor; Alexis), 50 μM rottlerin (PKCδ inhibitor; BioMol), and 10 ng/ml LMB (CRM1 inhibitor; LC Laboratories, Woburn, MA).
Plasmids and site-directed mutagenesis.
pTet-β-globin-3′UTRβ4GT1and pTet-β-globin-mAU2 have been described before (13). These chimeric mRNAs are continuously transcribed in cell lines expressing the tetracycline-controlled transactivator, like HeLa TO. The expression plasmids pHM6/14-3-3β(wt), pHM6/14-3-3ɛ(wt), and pHM6/14-3-3η(wt) were kindly provided by N. Fujita (33). pcDNA3-His/TTP(wt) was a gift from W. Rigby (5). The S60A, S132A, S60/132A, and S132E mutants of 14-3-3β, the S59A mutants of 14-3-3ɛ and 14-3-3η, and the S186A mutant of TTP were generated using the QuickChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The expression plasmids pCMV5-HA/IKKα(S176/180A), pCMV5-HA/IKKα(S176/180E), pCMV5-flag/IKKβ(S177/181A), and pCMV5-flag/IKKβ(S177/181E) were gifts from R. Gaynor (28), while pcDNA3-flag/IKKγ(wt) and pcDNA3-flag/IKKγ(Δ246-365) were kindly provided by C. Scheidereit (40). Bacterial expression plasmids for the production of glutathione _S_-transferase (GST) fusion proteins were constructed by subcloning 14-3-3β(wt), 14-3-3β(S60A), 14-3-3β(S132A), and 14-3-3β(S60/132A) from the pHM6 expression plasmids into the EcoRI site of pGEX-4T/3 (Amersham Biosciences, Uppsala, Sweden). All constructs were verified by sequencing.
For the generation of stable knockdown HeLa TO cell lines for TTP and BRF1, target small interfering RNAs (siRNAs) were cloned into the BglII/HindIII sites of the pSuper.retro.puro vector, a kind gift of R. Agami (6). siRNA targets for TTP and BRF1 were 5′ TGTCTCCTGGTAACTGGAA 3′ and 5′ CCTTTGTGTTGGACTGCAA 3′, respectively.
Transfection.
HeLa TO cells were grown to 90% confluence before transfection. The transfection with clonfectin (BD Clontech) was performed as recommended by the manufacturer, using 2 μg pTetBBB plasmid, 2 μg reporter plasmid and 1 μg clonfectin per 2.5 × 105 cells. The cells were incubated with the mixture of DNA and clonfectin in serum-free medium for 3 h, after which the medium was changed for serum-containing medium again. Cells were used for experiments 24 h after transfection.
mRNA half-life determination.
HUVECs were stimulated with TNF-α as indicated, before the addition of 10 μg/ml actinomycin D (ActD; Sigma, St. Louis, MO) to block transcription. After stimulation of transfected HeLa TO cells as indicated, 1 μg/ml doxycycline (DOX; BD Clontech) was added to block tetracycline-controlled transactivator-dependent transcription of chimeric mRNA. At 0, 20, 40, 60, 90, and 120 min after the addition of either ActD or DOX, cells were lysed for mRNA isolation as described below. Quantification of mRNA expression by real-time PCR analysis after transcription stop, as described below, allows for the calculation of the mRNA half-life.
mRNA isolation, cDNA synthesis, and quantitative real-time PCR.
Cells were lysed and mRNA was isolated with the mRNA capture kit (Roche, Basel, Switzerland), following the manufacturer's guidelines. cDNA synthesis then proceeded in the streptavidin-coated tubes with the captured mRNA with the reverse transcriptase kit (Promega Corporation, Madison, WI) as recommended by the manufacturer.
For real-time PCR analysis, PCR amplification was performed with an ABI 7900HT sequence detection system (Applied Biosystems, Foster City, CA) with SYBR Green master mix (Applied Biosystems) as previously described (14). Specific primers for β4GalT1, TTP, AUF1, KSRP, 14-3-3β, 14-3-3ɛ, 14-3-3η, 14-3-3θ, 14-3-3ζ, 14-3-3γ, 14-3-3ζ, rabbit β-globin, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were designed using Primer Express 2.0 (Applied Biosystems). Specificity of PCR products was confirmed by restriction enzyme control reactions. For each sample, the normalized amount of target mRNA (Nt) was calculated from the obtained Ct values for both target and GAPDH mRNA with Nt = 2_Ct_(GAPDH)−Ct(target). The relative mRNA expression was obtained by setting Nt in unstimulated control samples at 1 within one experiment.
RNA interference.
Stable knockdown cell lines were generated by the transfection of HeLa TO cells with pSuper.retro.puro vectors containing either TTP, BRF1, or scrambled control siRNA targets. At 24 h after transfection, puromycin was added at a concentration of 0.3 μg/ml. Two weeks after transfection, single colonies of cells were picked and screened for silenced expression of TTP or BRF1 mRNA by real-time PCR analysis.
RNA immunoprecipitation (RIP) assay.
A total of 2.5 × 105 HUVECs or 1 × 106 transfected HeLa TO cells were fixed with 0.5% formaldehyde in phosphate-buffered saline, washed, resuspended in 250 μl RIP lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 μM EGTA, 1 mM Na3VO4, 0.05% sodium dodecyl sulfate [SDS], 2 U/μl RNAsin [Promega], 1% NP-40, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 0.4 mM phenylmethylsulfonyl fluoride), sheared through a 26-gauge needle, and incubated on ice for 1 h. Lysates were incubated with antibodies, and immunocomplexes were precipitated using protein A/G PLUS agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Immunocomplex-associated mRNAs contained on the beads and nonassociated mRNAs present in the supernatant were separated by centrifugation. Both fractions were subjected to heat treatment to reverse cross-links (2 h at 70°C), followed by incubation with proteinase K (1 h at 45°C). Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA). mRNA was isolated and subjected to real-time PCR as described above to establish the distribution of β4GalT1 mRNA between immunocomplex-associated and nonassociated fractions.
Polyclonal antibodies used were anti-TTP, anti-pan 14-3-3, anti-14-3-3β, anti-14-3-3ɛ, anti-14-3-3η, anti-14-3-3θ, anti-14-3-3γ, and anti-14-3-3σ (all from Santa Cruz Biotechnology) and anti-14-3-3ζ (IBL, Gunma, Japan).
Western blotting and ECL immunodetection.
Immunoprecipitations were performed using whole-cell extracts from either nontreated or formaldehyde-treated, unstimulated, or TNF-α-stimulated HUVECs with anti-TTP-specific antibodies directed against 14-3-3 isoforms as mentioned above or anti-phosphoserine/threonine antibodies (BD Transduction Laboratories, Palo Alto, CA). Immunocomplexes were subjected to SDS-polyacrylamide gel electrophoresis (PAGE); blotted onto polyvinylidene difluoride membranes; analyzed for TTP or 14-3-3 isoform expression with anti-TTP, anti-pan 14-3-3, or anti-14-3-3β antibodies, as indicated; and incubated with swine anti-rabbit immunoglobulin (Ig)-HRP (DAKO, Glostrup, Denmark), and assayed with the ECL detection system (Amersham Biosciences).
The overexpression of proteins in transfection experiments was checked by performing Western blot analysis on whole-cell lysates: 0.5 × 106 transfected HeLa TO cells were lysed in 200 μl WCL lysis buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1 μM EGTA, 1 mM Na3VO4, 1 mM NaF, 1% Triton X-100, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 0.4 mM phenylmethylsulfonyl fluoride) for 30 min on ice. A total of 35 μl was subjected to SDS-PAGE; blotted onto polyvinylidene difluoride membranes; analyzed for overexpression of either His-tagged proteins [TTP(wt) and TTP(S186A)], HA-tagged proteins [IKKα(S176/180A), IKKα(S176/180E), 14-3-3β(wt), 14-3-3β(S60A), 14-3-3β(S132A), and 14-3-3β(S132E)], or flag-tagged proteins [IKKγ(Δ246-365), IKKβ(S177/181A), and IKKβ(S177/181E)] with anti-His or anti-hemagglutinin (HA) (both Santa Cruz Biotechnology) or anti-flag (Sigma) antibodies, as indicated; incubated with the appropriate horseradish peroxidase-labeled secondary antibodies; and assayed with the ECL detection system.
Immunofluorescence staining.
HUVECs were grown on gelatin-coated, glutaraldehyde-treated Lab-Tek II chamber slides (Nunc, Roskilde, Denmark), while transfected HeLa TO cells were grown on untreated chamber slides prior to stimulation as indicated. After fixation with 3% _para_-formaldehyde, cells were stained with anti-TTP or anti-His (Santa Cruz Biotechnology) antibodies, followed by incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit Ig (Jackson ImmunoResearch, West Grove, PA) and rabbit anti-mouse Ig (DAKO), respectively. Nuclei were stained with Hoechst (Molecular Probes, Eugene, OR). The cellular localizations of endogenous TTP and His-tagged TTP were determined under a Nikon eclipse E800 microscope.
Recombinant protein expression.
GST-14-3-3β proteins were expressed in Escherichia coli DH5α and purified using the B-PER GST spin purification kit (Pierce, Rockford, IL) according to the manufacturer's instructions.
IKKβ and PKCδ kinase assays.
Immunoprecipitations were performed using whole-cell extracts from either unstimulated or TNF-α-stimulated HUVECs with anti-IKKβ or anti-PKCδ antibodies (both from Santa Cruz Biotechnology). Immunocomplexes were resuspended in assay mixture (20 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol, 1 mM Na3VO4, 1 mM NaF, 15 μM ATP, and 5 μCi [γ-32P]ATP [3,000 Ci/mmol; Amersham Biosciences] for IKKβ; 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 400 μM CaCl2, 250 μM EGTA, 320 μg/ml phosphatidylserine, 32 μg/ml diacylglycerol, 100 μg/ml bovine serum albumin, 20 μM ATP, and 5 μCi [γ-32P]ATP for PKCδ). Either 1 μg GST-IκBα (Santa Cruz Biotechnology), GST-14-3-3β(wt), GST-14-3-3β(S60A), GST-14-3-3β(S132A), or GST-14-3-3β(S60/132A) was added as substrate, and the reactions were incubated at 30°C for 30 min. After SDS-PAGE, phosphorylated substrates were visualized using the Molecular Dynamics Storm system and ImageQuant software (Amersham Biosciences).
Statistical analysis.
Data are presented as means ± standard deviation derived from at least three independent experiments. Statistical analyses were performed on the data using Student's t test for paired observations. Statistical significance of the data was set at P < 0.05.
RESULTS
β4GalT1/AU2 is bound by TTP in resting HUVECs, whereas TNF-α signaling induces its release.
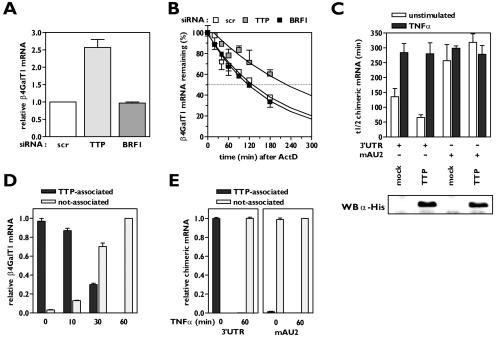
To identify the destabilizing factor binding to β4GalT1 mRNA in resting cells (13), we first examined what known destabilizing AUBPs are expressed in HUVECs. Quantitative real-time PCR analysis revealed that TTP and BRF1 mRNAs are present in HUVECs, whereas AUF1 and KSRP mRNAs are not (data not shown). For comparison, all four AUBP mRNAs were easily detectable in control samples from T lymphocytes (data not shown). To determine the role of TTP and BRF1 in the modulation of the β4GalT1 mRNA stability, stable knockdown cell lines for either of both factors in HeLa TO cells were established. Analysis at the level of mRNA expression by quantitative real-time PCR showed that the expression of TTP and BRF1 mRNAs was silenced by >90 and 95%, respectively (data not shown). Silencing of the expression of TTP resulted in a significant increase in the β4GalT1 mRNA expression level: it was 2.6 ± 0.23-fold (P < 0.01) higher than the expression levels in control cells expressing a scrambled siRNA (Fig. 1A). In accordance, the half-life of β4GalT1 mRNA was increased from 121 ± 16 min in control cells to 210 ± 25 min (P < 0.05) (Fig. 1B). In contrast, neither the expression level nor the half-life of β4GalT1 mRNA was changed in BRF1 knockdown cells compared with control cells (Fig. 1A and B). These results indicate that the TTP is involved in the modulation of β4GalT1 mRNA stability.
FIG. 1.
TTP binds to AU2 of β4GalT1 3′UTR, but is released upon TNF-α stimulation. (A) The expression of β4GalT1 mRNA is higher in HeLa TO cells that lack the expression of TTP but normal in HeLa TO cells lacking BRF1 expression. Stable HeLa TO knockdown cell lines for both TTP and BRF1 were generated as described in Materials and Methods; a scrambled (scr) control was included to monitor nonspecific effects. Quantitative real-time PCR analysis was performed on mRNA isolated from resting cells to determine the relative expression level of β4GalT1 mRNA. (B) The half-life of β4GalT1 mRNA is prolonged in TTP but not BRF1 knockdown HeLa TO cells. The half-life of β4GalT1 mRNA was determined by quantitative real-time PCR analysis of mRNA isolated from resting cells at different times after addition of ActD. (C) Overexpression of TTP accelerates the decay of chimeric mRNA containing the wild-type (wt) β4GalT1 3′UTR, but not the AU2 mutant. TTP was overexpressed in HeLa TO cells cotransfected to express chimeric mRNA containing either the wt β4GalT1 3′UTR or the AU2 mutant. At 24 h after transfection, the half-life of chimeric mRNAs was determined by quantitative real-time PCR analysis on mRNA isolated at different times after addition of doxycycline. The graphs (relative chimeric mRNA expression versus time after addition of DOX) for the determination of the half-lives are available in Fig. S1 in the supplemental material. The overexpression of His-tagged TTP was checked by Western blotting with an anti-His antibody as described in Materials and Methods. (D) TNF-α induces the release of binding of TTP from β4GalT1 mRNA. HUVECs, stimulated with 100 U/ml TNF-α as indicated, were treated with formaldehyde to preserve protein complexes bound to mRNA by reversible cross-links. Anti-TTP immunocomplexes were analyzed for the presence of β4GalT1 mRNA by quantitative real-time PCR. (E) TTP binds AU2. RNA/immunoprecipitation assays were performed on HeLa TO cells expressing chimeric mRNAs containing either the wt β4GalT1 3′UTR or the AU2 mutant as described in panel D.
Overexpression of TTP in HeLa TO cells cotransfected with a chimeric β-globin mRNA containing the β4GalT1 3′UTR (β-globin-3′UTRβ4GT1) resulted in a significant reduction of β-globin-3′UTRβ4GT1 mRNA stability: its half-life was determined at 66 ± 9 min (P < 0.01), compared with 135 ± 28 min for mock-transfected cells (Fig. 1C; see Fig. S1 in the supplemental material). Both in mock-transfected and in TTP-overexpressing cells, TNF-α stimulation increased the half-life of β-globin-3′UTRβ4GT1 mRNA to the same level (Fig. 1C; see Fig. S1 in the supplemental material). Overexpression of TTP in cells expressing a chimeric mRNA harboring a mutated AU2 within the β4GalT1 3′UTR (β-globin-mAU2) did not affect the stability of this chimeric mRNA, neither in unstimulated nor in TNF-α-stimulated cells (Fig. 1C; see Fig. S1 in the supplemental material).
To confirm the binding of TTP to β4GalT1 mRNA, we performed RIP assays after cross-linking of cellular RNA-protein and DNA-protein complexes with formaldehyde. All β4GalT1 mRNA in resting HUVECs was found in TTP-immunocomplexes (Fig. 1D). Similarly, all β-globin-3′UTRβ4GT1 mRNA in unstimulated transfected HeLa TO cells was found associated with TTP (Fig. 1E). Notably, no β-globin-mAU2 mRNA could be detected in TTP-immunocomplexes (Fig. 1E).
TNF-α stimulation gradually abrogated the binding of TTP to β4GalT1 mRNA: after 10 min of TNF-α treatment, nearly 87% of all cellular β4GalT1 mRNA was still bound by TTP, whereas after 30 min only approximately 30% was associated with TTP. After 1 h of TNF-α stimulation, no β4GalT1 mRNA could be found in association with TTP (Fig. 1D). This finding correlates with the gradual increase in the half-life of β4GalT1 mRNA that was observed after TNF-α stimulation (13). Thus, it can be concluded that the binding of TTP to AU2 of β4GalT1 mRNA is affected by TNF-α-induced signaling pathways, resulting in the release of the AUBP.
TNF-α induces β4GalT1 mRNA stabilization through two distinct signaling pathways mediated by IKKβ and PKCδ.
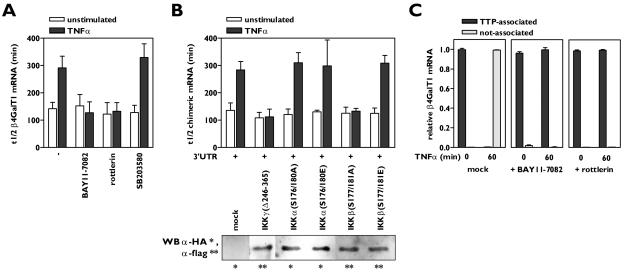
The TNF-α receptors (TNFRs) TNFR1 and TNFR2 trigger a variety of signaling pathways, leading to a large range of cellular effects (21). To identify the signaling pathways induced by TNF-α which are involved in the observed β4GalT1 mRNA stabilization, a series of inhibitors directed against proteins known to participate in TNF-α signal transduction routes were employed. The β4GalT1 mRNA had a half-life of 142 ± 23 min, which increased to 295 ± 48 min (P < 0.01) after 1 h of TNF-α stimulation (Fig. 2A; see Fig. S2A in the supplemental material) (13). Two of the inhibitors antagonized the TNF-α-induced stabilization of β4GalT1 mRNA without affecting the half-life in unstimulated HUVECs. The half-life of β4GalT1 mRNA in TNF-α-stimulated cells after treatment with BAY11-7082, an inhibitor of IκB phosphorylation, was determined at 127 ± 40 min (P < 0.01), while the half-life in TNF-α-stimulated cells after treatment with rottlerin, a specific inhibitor of the serine/threonine kinase PKCδ, was 133 ± 32 min (P < 0.01), similar to the half-life found in unstimulated cells (Fig. 2A; see Fig. S2A in the supplemental material). The TNF-α-induced suppression of β4GalT1 mRNA turnover was insensitive to the other inhibitors applied, including the p38/MK2 inhibitor SB203580 (see Material and Methods for all specific inhibitors used) (Fig. 2A; see Fig. S2A in the supplemental material).
FIG. 2.
Activation of both IKKβ and PKCδ by TNF-α is required for β4GalT1 mRNA stabilization. (A) Inhibitors of IKK (BAY11-7082) and PKCδ (rottlerin) antagonize TNF-α-induced β4GalT1 mRNA stabilization, while the p38 inhibitor SB203580 has no effect. HUVECs were treated with various inhibitors as indicated, before the half-life of β4GalT1 mRNA was determined as described in the legend to Fig. 1B. The graphs (relative β4GalT1 mRNA expression versus time after the addition of ActD) for the determination of the half-lives are available in Fig. S2A in the supplemental material. (B) IKK activity is required but not sufficient to reduce the β4GalT1 mRNA turnover. Dominant-interfering [IKKγ(Δ246-365), IKKα(S176/180A), and IKKβ(S177/181A)], and kinase-active [IKKα(S176/180E), IKKβ(S177/181E)] mutants of IKK components were overexpressed in HeLa TO cells expressing chimeric mRNA containing the wt β4GalT1 3′UTR. Its half-life was determined as in the results shown in Fig. 1C. The graphs (relative chimeric mRNA expression versus time after addition of DOX) for the determination of the half-lives are available in Fig. S2B in the supplemental material. The overexpression of flag- or HA-tagged IKK mutants was checked by Western blotting with anti-flag and anti-HA antibodies as described in Materials and Methods. (C) Inhibitors of either IKK or PKCδ (BAY11-7082 and rottlerin, respectively) block the TNF-α-induced release of binding of TTP from β4GalT1 mRNA. RNA/immunoprecipitation assays were performed on HUVECs in either the absence or presence of inhibitors as described in the legend to Fig. 1D.
BAY11-7082 is an inhibitor of IκB phosphorylation, a process that is mediated by the multiprotein IKK complex. The core IKK holoenzyme is composed of three distinct subunits, the two catalytically active kinases IKKα and IKKβ, and the noncatalytic IKKγ. IKKγ is obligatory for oligomerization and hence transautophosphorylation-mediated activation of the IKK complex (20, 40). Dominant-interfering and kinase-active IKK mutants were employed to examine which of the IKK components are required for TNF-α-induced β4GalT1 mRNA stabilization. The overexpression of the IKK mutants after transfection of HeLa TO cells was checked by Western blotting and was similar for all mutants in all experiments (data not shown). The overexpression of an IKKγ mutant lacking the domain responsible for oligomerization of the IKK complex, IKKγ(Δ246-365), in HeLa TO cells completely blocked the increase in the half-life of β-globin-3′UTRβ4GT1 mRNA induced by TNF-α, indicating that IKK activity is required for TNF-α to increase the stability of β4GalT1 mRNA (Fig. 2B; see Fig. S2B in the supplemental material). Similarly, the overexpression of a kinase-inactive IKKβ mutant, IKKβ(S177/181A), efficiently impeded the TNF-α-induced stabilization of β-globin-3′UTRβ4GT1 mRNA (Fig. 2B; see Fig. S2B in the supplemental material). In contrast, the overexpression of a kinase-inactive mutant of IKKα, IKKα(S176/180A), did not affect β-globin-3′UTRβ4GT1 mRNA stabilization after TNF-α stimulation (Fig. 2B; see Fig. S2B in the supplemental material). Strikingly, cells overexpressing a constitutively active IKKβ mutant, IKKβ(S177/181E), still required TNF-α stimulation to induce stabilization of the chimeric mRNA (Fig. 2B; see Fig. S2B in the supplemental material), implying that the activation of IKKβ is necessary but not sufficient for TNF-α to exert its modulatory effects of mRNA stability.
These data together suggested that the TNF-α-mediated activation of both IKKβ and PKCδ was required for the release of TTP from mRNA which precedes β4GalT1 mRNA stabilization; therefore, we next performed RNA immunoprecipitation assays using anti-TTP antibodies on both resting and TNF-α-stimulated cells in the absence or presence of the BAY11-7082 or rottlerin inhibitors. In the presence of the IKK inhibitor BAY11-7082, which would still allow the activation of PKCδ, the TNF-α-induced signaling pathways were not sufficient to dissociate TTP from β4GalT1 mRNA, as all cellular β4GalT1 mRNA was still found associated with TTP after TNF-α stimulation (Fig. 2C). Similarly, all cellular β4GalT1 mRNA remained bound by TTP after TNF-α stimulation in the presence of the PKCδ-inhibitor rottlerin, which would still allow the activation of the IKK complex (Fig. 2C). These results indicate that the activation of either IKKβ or PKCδ by TNF-α is insufficient to induce the release of TTP from β4GalT1 mRNA and induce its stabilization, whereas the TNF-α-induced activation of both kinases at the same time does result in TTP release from and stabilization of β4GalT1 mRNA.
Nuclear sequestration of TTP induces β4GalT1 mRNA stabilization.
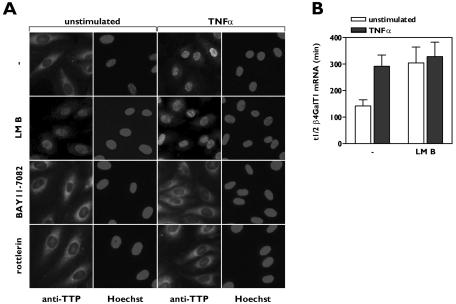
Nucleocytoplasmic shuttling has been demonstrated for several AUBPs, like TTP, AUF1, and Hu antigen R, and their cytoplasmic accumulation in response to extracellular signals is suggested to be linked to mRNA stability modulation (26, 31, 41). The subcellular localization of endogenous TTP in HUVECs was examined by immunocytochemistry. Serum starvation as applied in all experiments did not affect the localization of endogenous TTP (data not shown). TTP was predominantly localized in the cytoplasm in resting HUVECs. However, after TNF-α treatment of the cells, the localization of TTP was restricted to the nucleus (Fig. 3A).
FIG. 3.
Nuclear sequestration of TTP induces β4GalT1 mRNA stabilization. (A) TNF-α-induced nuclear translocation of TTP is inhibited by inhibitors of IKK and PKCδ. HUVECs were grown in chamber slides, treated with inhibitors, and stimulated for 1 h with TNF-α as indicated. Subcellular localizaton of TTP was determined by immunofluorescence microscopy after staining with anti-TTP antibodies. Hoechst staining indicates the position of nuclei. (B) Inhibition of TTP nuclear export by LMB stabilizes β4GalT1 mRNA. HUVECs were treated with LMB, before the half-life of β4GalT1 mRNA was determined as described in the legend to Fig. 1B. The graphs (relative β4GalT1 mRNA expression versus time after addition of ActD) for the determination of the half-lives are available in Fig. S3 in the supplemental material.
TTP is subject to constant nucleocytoplasmic shuttling due to the presence of both a nuclear localization signal (NLS) and a nuclear export signal. Nuclear export of TTP is dependent on the nuclear export receptor CRM1, while nuclear import of TTP is an energy-driven process involving the import receptors importin α/β (24, 26). Inhibition of the nuclear export system by LMB, which specifically binds to CRM1, resulted in the nuclear accumulation of TTP (Fig. 3A). In parallel, we observed that LMB significantly increased the half-life of β4GalT1 mRNA to 304 ± 70 min (P < 0.03) in unstimulated HUVECs, mimicking the effect of TNF-α stimulation with respect to β4GalT1 mRNA stabilization (Fig. 3B; see Fig. S3 in the supplemental material).
Inhibition of either TNF-α-induced IKKβ or PKCδ activity with BAY11-7082 or rottlerin, respectively, blocked the nuclear translocation of TTP in HUVECs as induced by TNF-α in the absence of inhibitors (Fig. 3A). These results led to the working hypothesis that the TNF-α-induced IKKβ/PKCδ-mediated event leading to the release of the binding of TTP to mRNA is one and the same as the event resulting in the nuclear translocation of TTP.
TTP is complexed with 14-3-3β while bound to β4GalT1 mRNA.
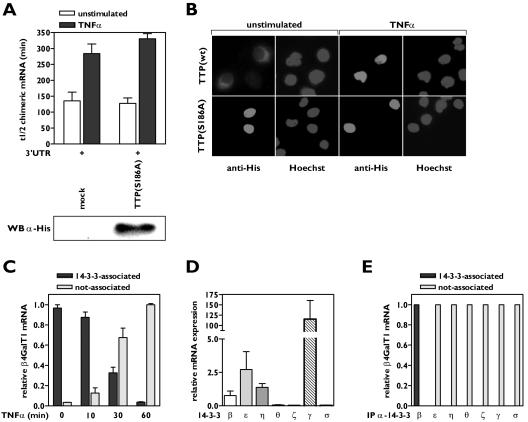
The cytoplasmic localization of TTP is influenced by binding of the chaperone protein 14-3-3 to a phosphoserine residue at position 186 of TTP, which supposedly masks the NLS of TTP (19, 24). Overexpression of a His-tagged TTP mutant (S186A), which is unable to interact with 14-3-3, in HeLa TO cells was unable to accelerate the decay of β-globin-3′UTRβ4GT1 mRNA (Fig. 4A; see Fig. S4 in the supplemental material), unlike its wild-type counterpart (Fig. 1C), although the expression levels of both proteins were comparable (data not shown). Immunocytochemistry showed that TTP(S186A) was sequestered in the nucleus (Fig. 4B), in accordance with the reported requirement for the association between TTP and 14-3-3 to retain TTP in the cytoplasm, enabling TTP to bind to target mRNAs there.
FIG. 4.
Complex formation of TTP with 14-3-3β is required for and intact on mRNA binding. (A) A TTP mutant lacking the ability to complex with 14-3-3 has no effect on β4GalT1 mRNA stability. His-TTP(S186A) was overexpressed in HeLa TO cells expressing chimeric mRNA containing the wt β4GalT1 3′UTR. Its half-life was determined as described in the legend to Fig. 1C. The graphs (relative chimeric mRNA expression versus time after the addition of DOX) for the determination of the half-lives are available in Fig. S4 in the supplemental material. The overexpression of His-tagged TTP(S186A) was checked by Western blotting with an anti-His antibody as described in Materials and Methods. (B) TTP(S186A) is localized in the nucleus. HeLa TO cells expressing His-TTP(S186A) were grown in chamber slides. Subcellular localization of mutant TTP was determined by immunofluorescence microscopy after staining with anti-His antibodies. Hoechst staining indicates the position of nuclei. (C) TNF-α induces the dissociation of 14-3-3 from β4GalT1 mRNA. HUVECs, stimulated with TNF-α as indicated, were treated with formaldehyde to preserve protein complexes bound to mRNA by reversible cross-links. Anti-pan 14-3-3 immunocomplexes were analyzed for the presence of β4GalT1 mRNA by quantitative real-time PCR. (D) HUVECs express a subset of 14-3-3 isoforms. Quantitative real-time PCR analysis of 14-3-3 isoforms was performed on mRNA isolated from resting HUVECs to determine the relative expression levels. (E) 14-3-3β is the only isoform associated with β4GalT1 mRNA. RNA/immunoprecipitation assays were performed on HUVECs as described in the legend to Fig. 1D, using specific antibodies directed against the different isoforms instead of the anti-pan 14-3-3 antibodies.
Next, it was investigated whether TTP and 14-3-3 remain associated when bound to β4GalT1 mRNA in HUVECs. RNA/protein complexes were immunoprecipitated using anti-14-3-3 antibodies and analyzed for the presence of β4GalT1 mRNA. All β4GalT1 mRNA in resting HUVECs was detected in association with 14-3-3 (Fig. 4C). The association between 14-3-3 and β4GalT1 mRNA was gradually lost (Fig. 4C), with kinetics similar to the dissociation of TTP from β4GalT1 mRNA due to TNF-α treatment (Fig. 1D).
In mammalian cells, seven highly conserved isoforms of 14-3-3 have been identified (1). Quantitative real-time PCR analysis showed that 14-3-3β, 14-3-3ɛ, and 14-3-3η are expressed at intermediate levels in HUVECs; 14-3-3γ is highly expressed; and 14-3-3ζ, 14-3-3θ, and 14-3-3σ are undetected (Fig. 4D). The HeLa TO cells used to study chimeric mRNA stability only expressed 14-3-3β and the epithelial specific isoform 14-3-3σ (data not shown). To examine whether 14-3-3 isoforms show preferences in binding towards TTP, RNA immunoprecipitation assays were performed with antibodies specific for all seven isoforms. Only 14-3-3β was found associated with β4GalT1 mRNA in resting HUVECs (Fig. 4E), and this association was reversed after 1 h of TNF-α stimulation (Fig. 4C). Western blotting experiments confirmed that the lack of association of other 14-3-3 isoforms with β4GalT1 mRNA was not due to failure of the antibodies to recognize the proteins after formaldehyde-induced cross-linking (data not shown). All together, these data suggest that a complex of TTP with 14-3-3β is formed to retain TTP in the cytoplasm, where it binds to β4GalT1 mRNA to induce rapid mRNA decay.
IKKβ phosphorylates serine residue Ser132 and PKCδ phosphorylates serine residue Ser60 of 14-3-3β.
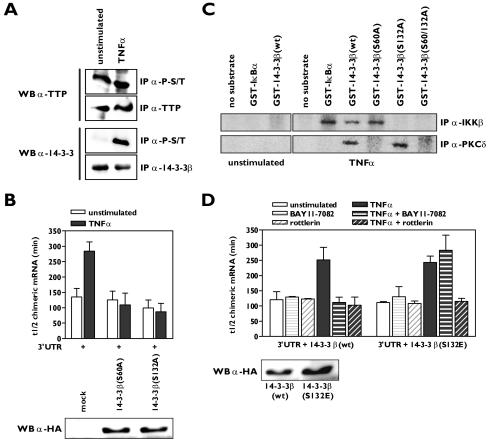
To understand mechanistically how the TNF-α-induced signaling pathways mediated by the serine/threonine kinases IKKβ and PKCδ affect the association of 14-3-3β with TTP and consequently binding of the complex to mRNA, we set out to find target phosphorylation sites on either TTP or 14-3-3β. Western blotting experiments clearly showed that TTP was phosphorylated on serine/threonine residues in resting HUVECs, while serine/threonine phosphorylation of 14-3-3β could not be detected. Upon TNF-α stimulation, phosphorylated 14-3-3β was easily detectable (Fig. 5A), indicating that most likely 14-3-3β is the substrate for both IKKβ and PKCδ. Since it has been reported that dimerization of 14-3-3 is required for ligand binding and that phosphorylation of a serine residue within the dimer interface of some of the 14-3-3 isoforms disrupts the 14-3-3 dimer (35, 46), we mutated Ser60 to alanine to generate a mutant of 14-3-3β that is resistant to dimer disruption. Overexpression of 14-3-3β(S60A) in HeLa TO cells completely inhibited the TNF-α-induced stabilization of β-globin-3′UTRβ4GT1 mRNA (Fig. 5B; see Fig. S5A in the supplemental material). This effect was restricted to the 14-3-3β mutant as mutants of two other isoforms, 14-3-3ɛ and 14-3-3η, harboring identical Ser-to-Ala substitutions within their dimerization domain were unable to block the effect of TNF-α on β-globin-3′UTRβ4GT1 mRNA stability (data not shown).
FIG. 5.
Phosphorylation of 14-3-3β by IKKβ and PKCδ promotes β4GalT1 mRNA stabilization. (A) TNF-α stimulation induces serine/threonine phosphorylation of 14-3-3β, while TTP is already heavily phosphorylated on serine/threonine residues in resting HUVECs. Phospho-serine/threonine (P-S/T)-containing proteins were immunoprecipitated from lysates of unstimulated HUVECs or HUVECs stimulated for 15 min with TNF-α. Western blotting was performed using anti-TTP and anti-14-3-3β antibodies. Total levels of TTP and 14-3-3β were also checked by control immunoprecipitations. (B) 14-3-3β mutants carrying S60A or S132A substitutions interfere with TNF-α-induced β4GalT1 mRNA stabilization. 14-3-3β(S60A) or 14-3-3β(S132A) was overexpressed in HeLa TO cells expressing chimeric mRNA containing the wt β4GalT1 3′UTR. Its half-life was determined as described in the legend to Fig. 1C. The graphs (relative chimeric mRNA expression versus time after the addition of DOX) for the determination of the half-lives are available in Fig. S5A in the supplemental material. The overexpression of HA-tagged 14-3-3β(S60A) and 14-3-3β(S132A) was checked by Western blotting with an anti-HA antibody as described in Materials and Methods. (C) Ser60 and Ser132 of 14-3-3β are phosphorylated by PKCδ and IKKβ in vitro. PKCδ and IKKβ were immunoprecipitated from lysates of u nstimulated HUVECs or HUVECs stimulated for 15 min with TNF-α. Bacterially expressed GST-14-3-3β fusion proteins were used as substrates in kinase assays. (D) Acquired resistance to rottlerin in 14-3-3β(S132E)-expressing cells with regard to TNF-α-induced β4GalT1 mRNA stabilization. 14-3-3β(S132E) was overexpressed in HeLa TO cells expressing chimeric mRNA containing the wt β4GalT1 3′UTR. The cells were treated with BAY11-7082 or rottlerin as indicated, before the half-life of the chimeric mRNA was determined as described in the legend to Fig. 1C. The graphs (relative chimeric mRNA expression versus time after the addition of DOX) for the determination of the half-lives are available in Fig. S5B in the supplemental material. The overexpression of HA-tagged 14-3-3β(wt) and 14-3-3β(S132E) was checked by Western blotting with an anti-HA antibody as described in Materials and Methods.
Phosphorylation of Ser130 of rat 14-3-3β (the equivalent of Ser132 in human 14-3-3β), a serine residue unique to the 14-3-3β isoform, has been shown to negatively regulate ligand binding (42). Likewise, as 14-3-3β(S60A), the overexpression of 14-3-3β(S132A) counteracted the TNF-α-induced stabilization of β-globin-3′UTRβ4GT1 mRNA (Fig. 5B; see Fig. S5A in the supplemental material).
To determine if IKKβ and PKCδ could phosphorylate these serine residues of 14-3-3β in vitro, we produced 14-3-3β mutants fused to GST and used these proteins as substrates in IKKβ and PKCδ immunocomplex kinase assays. We found that both kinases phosphorylated wild-type 14-3-3β, whereas the double mutant 14-3-3β(S60/132A) remained unphosphorylated (Fig. 5C). IKKβ phosphorylated only the 14-3-3β(S60A) mutant, while PKCδ phosphorylated only 14-3-3β(S132A) (Fig. 5C). To verify these findings, we overexpressed either wild-type 14-3-3β or 14-3-3β(S132E), a mutant mimicking phosphorylation of Ser132, in HeLa TO cells and assessed the sensitivity of the TNF-α-mediated stabilization of β-globin-3′UTRβ4GT1 mRNA to the IKK and PKCδ inhibitors. Both BAY11-7082 and rottlerin blocked the increase in the half-life of β-globin-3′UTRβ4GT1 mRNA after TNF-α stimulation in the cells expressing wild-type 14-3-3β (Fig. 5D; see Fig. S5B in the supplemental material), in agreement with their effects on the stabilization of β4GalT1 mRNA in HUVECs (Fig. 2A). However, cells expressing 14-3-3β(S132E) were insensitive to treatment with BAY11-7082 with regard to the TNF-α-induced β-globin-3′UTRβ4GT1 mRNA stabilization, indicating that the kinase responsible for phosphorylation of Ser60 was still functional in these cells (Fig. 5D; see Fig. S5B in the supplemental material). In contrast, TNF-α treatment no longer resulted in a reduced turnover of β-globin-3′UTRβ4GT1 mRNA in the 14-3-3β(S132E)-expressing cells in the presence of rottlerin, indicating that rottlerin inhibited the kinase responsible for phosphorylation of Ser60 (Fig. 5D; see Fig. S5B in the supplemental material). Taken together, these results strongly suggest that the phosphorylation of 14-3-3β on Ser60 and Ser 132 by PKCδ and IKKβ, respectively, in response to TNF-α stimulation is responsible for the dissociation of 14-3-3β from TTP.
DISCUSSION
The important role for the regulation of gene expression at the level of mRNA stability has been recognized for many years. A large knowledge base about the proteins affecting mRNA turnover exists, yet the regulatory mechanisms involved remain to be elucidated. The β4GalT1 gene provided us with a unique opportunity to study the regulatory mechanism affecting TTP binding to AU-rich elements and function, since the stability of its mRNA is regulated through a single ARE bound by TTP in resting HUVECs. Our findings provide evidence for a mechanism regarding the IKKβ- and PKCδ-dependent phosphorylation of 14-3-3β in an mRNA-bound complex with TTP, a destabilizing AUBP, which induces the release of the complex from AREs and consequently stabilization of their target mRNAs. The mechanism described might influence the mRNA stability of all mRNAs containing TTP-binding elements.
Our results indicate that cytoplasmic retention of TTP is effected through association with the chaperone protein 14-3-3β and, more importantly, is required for TTP to bind to its target mRNAs and induce their exosome-mediated decay. We now show for the first time that the association between 14-3-3β and TTP remains intact upon binding to mRNA. This observation is of importance, as it adds a potential target for the modulation of mRNA stability by extracellular signals. TNF-α stimulation of HUVECs induces the nuclear translocation of TTP, in parallel with stabilization of β4GalT1 mRNA, whose stability is solely regulated through a TTP-dependent mechanism. Based on our studies with 14-3-3β mutants, we propose a mechanism in which 14-3-3β becomes phosphorylated upon TNF-α stimulation, resulting in both the disruption of the 14-3-3 dimer and the association between 14-3-3β and TTP. The binding of 14-3-3β to TTP retains TTP within the cytoplasm, supposedly by masking its NLS (19, 24). The dissociation of 14-3-3β from TTP will once again expose the NLS, forcing the release of TTP from the mRNAs to which it is bound, followed by its nuclear translocation. Presumably, stabilizing AUBPs will bind the abandoned mRNAs, but the identity of the factors involved remains to be established.
Two distinct signaling pathways triggered by TNF-α are involved in the phosphorylation of 14-3-3β, and both are required to interfere with the interaction between 14-3-3β and TTP. One pathway induces the activation of the IKK complex, while the other involves the activation of PKCδ, both known downstream modulators of TNFR ligation (21). Surprisingly, we found no regulatory role for MK2 in the association between 14-3-3 and TTP, although MK2 has been implicated in phosphorylation of both proteins (9, 27).
The activation of the IKK complex leads to the phosphorylation of 14-3-3β on Ser132 by IKKβ, while PKCδ activation results in phosphorylation of Ser60. Ser60 resides within the dimer interface of 14-3-3β. It has previously been reported that dimerization of 14-3-3 isoforms is required for phosphorylation-dependent binding of its ligands (35). Binding of 14-3-3β to TTP is controlled through a phosphoserine at position 186. Mutation of this serine residue abolishes binding of TTP to 14-3-3β, leading to sequestration of TTP in the nucleus, as shown by immunocytochemistry. Previous reports had already identified sphingosine-dependent protein kinase 1, which comprises the 40-kDa kinase domain of PKCδ, as a 14-3-3-phosphorylating kinase (16). Phosphorylation of Ser60 disrupts the dimer formation of 14-3-3β, interfering with the binding of TTP. However, our findings indicate that phosphorylation of Ser60 alone is insufficient to antagonize the destabilization of mRNA as induced by the bound 14-3-3β/TTP complex: phosphorylation of Ser 132 is also necessary.
Ser132 is identified as a target phosphorylation site for IKKβ in response to TNF-α stimulation. This is the first time that 14-3-3β has been shown to be a substrate for IKKβ. The sequence in which Ser132 (boldface) is present, RX2SX3S, matches the putative recognition sequence found in the few known IKKβ substrates (12, 18). Ser132 is located closely to the binding pocket of 14-3-3β, which is formed by Lys51, Arg58, Arg129, and Tyr130 (29). We hypothesize that phosphorylation of Ser132 affects but does not block the binding of TTP in the binding pocket of 14-3-3β, either by steric hindrance or through induced conformational changes. We propose a model in which dimerization of 14-3-3 is crucial for ligand binding by making the binding groove accessible for phosphorylated ligands; however, it is not necessary to maintain ligand binding. On the other hand, nonoptimal binding of ligand in the binding pocket due to phosphorylation of Ser132 is stabilized by the conformation adopted by dimerization and ligand binding. It has been demonstrated for 14-3-3ζ that ligand binding in the binding pocket results in conformational changes within the C-terminal tail (25), hypothetically stabilizing the ligand binding or locking the ligand in place.
The expression of many other genes besides β4GalT1 will be affected by the proposed mechanism. AREs have been detected in many cytokine, growth factor, and chemokine mRNAs. Furthermore, many glycosyltransferase mRNAs besides β4GalT1 contain AREs, although it remains to be established whether these are functional (J. J. García-Vallejo and S. I. Gringhuis, unpublished data). The importance of regulating mRNA binding by TTP and understanding the underlying mechanism is especially showcased by the TNF-α gene: mice lacking TTP develop an autoimmune syndrome as a consequence of excessive TNF-α production, due to the increased stability of TNF-α mRNA (39). Of course, many labile mRNAs are regulated through binding of more than one AUBP, and the additive modulatory mechanisms affecting the binding of those AUBPs will have to be considered for each gene individually.
During the preparation of the manuscript, Stoecklin and colleagues described that complex formation between 14-3-3 and TTP prevented the decay of ARE-containing mRNAs (38). Although at first glance these results seem to be in contrast with our results, they might actually reflect another layer of complexity to the mechanisms regulating mRNA stability through TTP. Preliminary results show that not TTP but 14-3-3β is responsible for the recruitment of the exosome to the β4GalT1 transcripts. Stoecklin et al. did not discriminate between the seven 14-3-3 isoforms, but it is tempting to speculate that the observed differences lie in a differential expression pattern of 14-3-3 isoforms within different cell types. The highly homologous isoforms show only variability within their C-terminal stretches, which might reflect their ability to recruit the exosome. In accordance with this hypothesis is the finding that components of the exosome can be isolated from HeLa cell lysates via 14-3-3 affinity chromatography (30). Further investigations are ongoing to analyze in molecular detail the modulation of mRNA stability through different TTP/14-3-3 complexes, which will greatly enhance our knowledge of the regulatory mechanisms behind mRNA stability modulation.
Supplementary Material
[Supplemental material]
Footnotes
REFERENCES
- 1.Aitken, A., H. Baxter, T. Dubois, S. Clokie, S. Mackie, K. Mitchell, A. Peden, and E. Zemlickova. 2002. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 30**:**351-360. [DOI] [PubMed] [Google Scholar]
- 2.Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 195**:**356-372. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear, P. J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30**:**945-952. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58**:**266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, S. A., J. E. Connolly, R. J. Diegel, R. A. Fava, and W. F. Rigby. 2002. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 46**:**1362-1370. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296**:**550-553. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107**:**451-464. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20**:**465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chrestensen, C. A., M. J. Schroeder, J. Shabanowitz, D. F. Hunt, J. W. Pelo, M. T. Worthington, and T. W. Sturgill. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279**:**10176-10184. [DOI] [PubMed] [Google Scholar]
- 10.Esnault, S., and J. S. Malter. 2002. Extracellular signal-regulated kinase mediates granulocyte-macrophage colony-stimulating factor messenger RNA stabilization in tumor necrosis factor-alpha plus fibronectin-activated peripheral blood eosinophils. Blood 99**:**4048-4052. [DOI] [PubMed] [Google Scholar]
- 11.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23**:**425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Z., D. Hwang, F. Bataille, M. Lefevre, D. York, M. J. Quon, and J. Ye. 2002. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J. Biol. Chem. 277**:**48115-48121. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Vallejo, J. J., W. van Dijk, I. van Die, and S. I. Gringhuis. 2005. Tumor necrosis factor-α up-regulates the expression of β1,4-galactosyltransferase I in primary human endothelial cells by mRNA stabilization. J. Biol. Chem. 280**:**12676-12682. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Vallejo, J. J., B. van het Hof, J. Robben, J. A. Van Wijk, I. Van Die, D. H. Joziasse, and W. van Dijk. 2004. Approach for defining endogenous reference genes in gene expression experiments. Anal. Biochem. 329**:**293-299. [DOI] [PubMed] [Google Scholar]
- 15.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14**:**571-583. [DOI] [PubMed] [Google Scholar]
- 16.Hamaguchi, A., E. Suzuki, K. Murayama, T. Fujimura, T. Hikita, K. Iwabuchi, K. Handa, D. A. Withers, S. C. Masters, H. Fu, and S. Hakomori. 2003. Sphingosine-dependent protein kinase-1, directed to 14-3-3, is identified as the kinase domain of protein kinase C delta. J. Biol. Chem. 278**:**41557-41565. [DOI] [PubMed] [Google Scholar]
- 17.Holtmann, H., J. Enninga, S. Kalble, A. Thiefes, A. Dorrie, M. Broemer, R. Winzen, A. Wilhelm, J. Ninomiya-Tsuji, K. Matsumoto, K. Resch, and M. Kracht. 2001. The MAPK kinase kinase TAK1 plays a central role in coupling the interleukin-1 receptor to both transcriptional and RNA-targeted mechanisms of gene regulation. J. Biol. Chem. 276**:**3508-3516. [DOI] [PubMed] [Google Scholar]
- 18.Hu, M. C., D. F. Lee, W. Xia, L. S. Golfman, F. Ou-Yang, J. Y. Yang, Y. Zou, S. Bao, N. Hanada, H. Saso, R. Kobayashi, and M. C. Hung. 2004. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117**:**225-237. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, B. A., J. R. Stehn, M. B. Yaffe, and T. K. Blackwell. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 277**:**18029-18036. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18**:**621-663. [DOI] [PubMed] [Google Scholar]
- 21.MacEwan, D. J. 2002. TNF receptor subtype signalling: differences and cellular consequences. Cell Signal. 14**:**477-492. [DOI] [PubMed] [Google Scholar]
- 22.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17**:**6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay, D., C. W. Houchen, S. Kennedy, B. K. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11**:**113-126. [DOI] [PubMed] [Google Scholar]
- 24.Murata, T., Y. Yoshino, N. Morita, and N. Kaneda. 2002. Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11. Biochem. Biophys. Res. Commun. 293**:**1242-1247. [DOI] [PubMed] [Google Scholar]
- 25.Obsilova, V., P. Herman, J. Vecer, M. Sulc, J. Teisinger, and T. Obsil. 2004. 14-3-3zeta C-terminal stretch changes its conformation upon ligand binding and phosphorylation at Thr232. J. Biol. Chem. 279**:**4531-4540. [DOI] [PubMed] [Google Scholar]
- 26.Phillips, R. S., S. B. Ramos, and P. J. Blackshear. 2002. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J. Biol. Chem. 277**:**11606-11613. [DOI] [PubMed] [Google Scholar]
- 27.Powell, D. W., M. J. Rane, B. A. Joughin, R. Kalmukova, J. H. Hong, B. Tidor, W. L. Dean, W. M. Pierce, J. B. Klein, M. B. Yaffe, and K. R. McLeish. 2003. Proteomic identification of 14-3-3zeta as a mitogen-activated protein kinase-activated protein kinase 2 substrate: role in dimer formation and ligand binding. Mol. Cell. Biol. 23**:**5376-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prajapati, S., and R. B. Gaynor. 2002. Regulation of IκB kinase (IKK)γ /NEMO function by IKKβ-mediated phosphorylation. J. Biol. Chem. 277**:**24331-24339. [DOI] [PubMed] [Google Scholar]
- 29.Rittinger, K., J. Budman, J. Xu, S. Volinia, L. C. Cantley, S. J. Smerdon, S. J. Gamblin, and M. B. Yaffe. 1999. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4**:**153-166. [DOI] [PubMed] [Google Scholar]
- 30.Rubio, M. P., K. M. Geraghty, B. H. Wong, N. T. Wood, D. G. Campbell, N. Morrice, and C. Mackintosh. 2004. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 379**:**395-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar, B., J. Y. Lu, and R. J. Schneider. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 278**:**20700-20707. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23**:**6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato, S., N. Fujita, and T. Tsuruo. 2002. Regulation of kinase activity of 3-phosphoinositide-dependent protein kinase-1 by binding to 14-3-3. J. Biol. Chem. 277**:**39360-39367. [DOI] [PubMed] [Google Scholar]
- 34.Sengupta, T. K., S. Bandyopadhyay, D. J. Fernandes, and E. K. Spicer. 2004. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 279**:**10855-10863. [DOI] [PubMed] [Google Scholar]
- 35.Shen, Y. H., J. Godlewski, A. Bronisz, J. Zhu, M. J. Comb, J. Avruch, and G. Tzivion. 2003. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell 14**:**4721-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim, J., H. Lim, J. R. Yates, and M. Karin. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10**:**1331-1344. [DOI] [PubMed] [Google Scholar]
- 37.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21**:**4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23**:**1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, G. A., E. Carballo, D. M. Lee, W. S. Lai, M. J. Thompson, D. D. Patel, D. I. Schenkman, G. S. Gilkeson, H. E. Broxmeyer, B. F. Haynes, and P. J. Blackshear. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4**:**445-454. [DOI] [PubMed] [Google Scholar]
- 40.Tegethoff, S., J. Behlke, and C. Scheidereit. 2003. Tetrameric oligomerization of IκB kinase γ (IKKγ) is obligatory for IKK complex activity and NF-κB activation. Mol. Cell. Biol. 23**:**2029-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran, H., F. Maurer, and Y. Nagamine. 2003. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol. 23**:**7177-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der Hoeven, P. C., J. C. Van Der Wal, P. Ruurs, M. C. van Dijk, and J. Van Blitterswijk. 2000. 14-3-3 isotypes facilitate coupling of protein kinase C-zeta to Raf-1: negative regulation by 14-3-3 phosphorylation. Biochem. J. 345**:**297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, G. M., J. Lu, K. Sutphen, Y. Suarez, S. Sinha, B. Brewer, E. C. Villanueva-Feliciano, R. M. Ysla, S. Charles, and G. Brewer. 2003. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 278**:**33039-33048. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, G. M., J. Lu, K. Sutphen, Y. Sun, Y. Huynh, and G. Brewer. 2003. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 278**:**33029-33038. [DOI] [PubMed] [Google Scholar]
- 45.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18**:**4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodcock, J. M., J. Murphy, F. C. Stomski, M. C. Berndt, and A. F. Lopez. 2003. The dimeric versus monomeric status of 14-3-3zeta is controlled by phosphorylation of Ser58 at the dimer interface. J. Biol. Chem. 278**:**36323-36327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]