Structure of dimeric mitochondrial ATP synthase: Novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis (original) (raw)
Abstract
The F1F0–ATP synthase exists as a dimer in mitochondria, where it is essential for the biogenesis of the inner membrane cristae. How two ATP synthase complexes dimerize to promote cristae formation is unknown. Here we resolved the structure of the dimeric F1F0 ATP synthase complex isolated from bovine heart mitochondria by transmission electron microscopy. The structure of the ATP synthase dimer has an overall conic appearance that is consistent with the proposed role of the dimeric enzyme in mitochondrial cristae biogenesis. The ATP synthase dimer interface is formed by contacts on both the F0 and F1 domains. A cross-bridging protein density was resolved which connects the two F0 domains on the intermembrane space side of the membrane. On the matrix side of the complex, the two F1 moieties are connected by a protein bridge, which is attributable to the IF1 inhibitor protein.
Keywords: F1F0 dimer, electron microscopy, protein bridge
The F1F0 ATP synthase is a ubiquitous rotary motor enzyme that couples a transmembrane flow of protons through its F0 channel to ATP synthesis taking place on its F1 moiety (1). The mechanism of energy coupling between the F1 ATPase and the F0 proton channel involves a rotating central stalk as well as a peripheral stalk that is part of the stator. In the ATP synthase of Escherichia coli (EcF1F0), five different subunits of F1 (α3β3γδε) and three of F0 (_ab_2_c_10–12) form the core of the F1F0 motor structure. The mitochondrial enzyme, also called complex V, is more complicated in that it contains the accessory proteins IF1, ε, A6L, F6, d, e, f, and g in bovine heart. The enzyme of yeast mitochondria contains additional supernumerary subunits named i and k. With the exception of ε and IF1, which bind to F1, most of the additional subunits are associated with the F0 proton channel or the peripheral stalk. IF1 is a small-molecular-weight protein that controls the ATPase and ATP synthase activities of the enzyme (2). The structure of soluble F1–ATPase from bovine heart (MF1) reconstituted with IF1 corresponds to a F1–F1 dimer bridged by two IF1 molecules (3, 4). Low-resolution crystallographic studies of the whole F1F0 complex from yeast showed a ring of 10 rotary c-subunits, but the other F0 subunits were not resolved (5). Remarkably, electron microscopy (EM) studies of EcF1F0 (6) and the bovine heart complex V (MF1F0) (7) provided the unique view of the second stalk (8) and the position of some of its subunits. EM was also used to resolve the whole architecture of bovine MF1F0 in its typical monomeric form with the F1 and F0 parts connected by central and peripheral stalks (9). In addition, an ATP synthasome complex that contains monomeric F1F0 associated to adenine nucleotide translocator and Pi carriers has also been examined by EM (10). This complex is assumed to improve the efficiency of ATP synthesis by substrate–product channeling.
Recently it was shown by blue native gel electrophoresis (BN/PAGE) that the mitochondrial ATP synthase of yeast can exist as a dimer (11, 12) and that dimer formation depends on the accessory subunits e and g. Deletion of these subunits abolishes ATP synthase dimerization, and, remarkably, mitochondria of these yeast cells exhibit altered morphology, suggesting that biogenesis of mitochondrial cristae depends on formation of the F1F0 dimer (12, 13). Models in which F1F0 dimers oligomerize to form arc-like assemblies and multimeric ATP synthase chains that can induce membrane invaginations and eventually cristae have been proposed (12, 14, 15). The models are based on the existence of a dimeric F1F0 structure that remains, until now, unresolved. In this work, we purified dimeric F1F0–ATP synthase from bovine heart mitochondria and analyzed its structure by EM and single-particle image analysis. The images show that the dimer interface is formed predominantly by the two F0 domains in such a way that the long axes of the two ATP synthase monomers are arranged at an angle of ≈40°. The results indicate that the angle between the ATP synthase monomers determines the curvature observed in the cristae of the inner mitochondrial membrane upon further F1F0 polymerization.
Materials and Methods
Extraction of Dimeric F1F0–ATP Synthase. Mitochondria were isolated from bovine heart as described before (16). Isolated mitochondria were diluted to 10–15 mg/ml in solubilization buffer (750 mM 6-aminohexanoic acid/50 mM Bis-Tris, pH 7.0) and extracted with 2.5 mg/mg digitonin. Under these conditions, solubilization of dimeric F1F0 is favored (11). Afterward, the samples were centrifuged at 186,000 × g for 45 min at 4°C, and the soluble fraction was recovered. BN/PAGE was carried out as described in ref. 11, and subsequent 2D SDS/PAGE was carried out as described in ref. 16.
In-Gel ATPase Activity Staining. Mitochondrial extracts were subjected to BN/PAGE. Lanes of interest were excised and incubated overnight at room temperature with ATPase activity-developing buffer (10 mM ATP/30 mM CaCl2/50 mM Hepes, pH 8.0). Precipitated calcium phosphate was visualized as white bands on a dark background by densitometric scanning.
Enrichment of Dimeric F1F0 by Glycerol Gradient Centrifugation. Mitochondrial digitonin extracts were loaded on a discontinuous glycerol gradient (20–40% glycerol/20 mM Mes, pH 7.0/2 mM EDTA/2 mM ADP/5 mM digitonin) and centrifuged at 54,000 × g for 16 h at 4°C. Afterward, 0.5-ml fractions were collected from the gradient from top to bottom. Fractions containing monomeric and dimeric F1F0–ATP synthase were identified by BN/PAGE followed by SDS/PAGE.
EM. Samples of dimeric ATP synthase were diluted to 25 μg/ml and applied to carbon-coated copper grids. Grids were washed once with water and stained with 1% uranyl acetate. Grids were examined in a FEI Tecnai12 transmission electron microscope operating at 100 kV. Images were recorded on a 2,048 × 2,048 slow-scan charge-coupled device camera in low-dose mode with an underfocus of 500 nm and an electron optical magnification of 30,000, placing the first zero of the contrast transfer function at ≈0.05 Å–1. Images were analyzed with the imagic-5 package of programs (17) on a dual processor Octane workstation (Silicon Graphics, Mountain View, CA). A data set of 1,130 molecular images (140 × 140 pixels) was extracted interactively from a total of 111 charge-coupled device frames. Images were band-pass-filtered to eliminate unwanted spatial frequencies (<0.008 Å–1 and >0.13 Å–1) and normalized. Image analyses, including alignment by classification, multivariate statistical analysis, and multireference alignment were done as described in refs. 6 and 8.
Results and Discussion
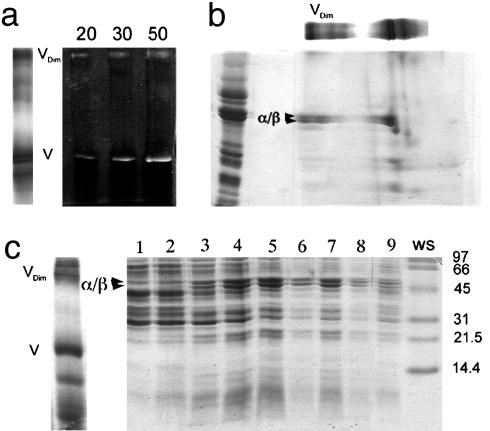
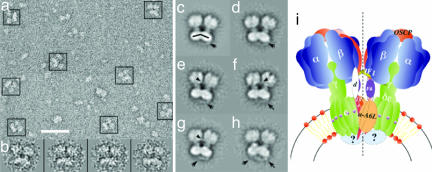
Dimeric ATP synthase was extracted from bovine heart mitochondria with digitonin and enriched by glycerol density gradient centrifugation. Fig. 1_a_ Left shows BN/PAGE of respiratory chain complexes and dimers and monomers of the ATP synthase. The presence of ATP synthase at the top and in the middle of the native gel was confirmed by ATPase activity staining (Fig. 1_a_ Right) and 2D SDS/PAGE (Fig. 1_b_). To obtain dimeric ATP synthase suitable for EM, the solubilized protein complexes were separated by centrifugation on glycerol gradients. The gradients were fractionated and analyzed by SDS/PAGE (Fig. 1_c_) where two ATP synthase peaks were observed (fractions 5 and 7). Analysis of the corresponding fractions by negative-stain EM revealed that the fraction near the bottom of the gradient (fraction 7) contained predominantly ATP synthase dimers (Fig. 2_a_). Representative images of the dimers are shown in Fig. 2_b_.
Fig. 1.
Gel analysis of dimeric ATP synthase. (a) Digitonin extracts of mitochondrial protein complexes were subjected to BN/PAGE and stained with Coomassie blue (Left) or developed for ATPase activity (Right, containing 20–50 μg of extracted protein). (b) An excised lane from the BN/PAGE was loaded as shown on top of a SDS/polyacrylamide gel together with an enriched F1F0 standard (left lane). Major F1 subunits (α and β) and dimeric F1F0 (Vdim) are indicated. (c) Monomeric (V) and dimeric (Vdim) F1F0 were identified by BN/PAGE in a mitochondrial digitonin extract (Left). The digitonin extract was subsequently loaded on a glycerol density gradient, and fractions obtained after centrifugation were analyzed by SDS/PAGE (Right). Dimeric F1F0 (fraction 7) was resolved from the monomer (fractions 4 and 5). The lane marked WS shows molecular mass standards. For details, see text.
Fig. 2.
EM and image analysis of ATP synthase dimer. Dimeric F1F0 enriched as described in the text and in Fig. 1 was used for EM analysis. (a) Area of a typical electron micrograph used for collecting single images. (Scale bar: 50 nm.) The ATP synthase dimers are indicated by boxes. (b) Representative images of the F1F0 dimer. (c) Total average of a data set of 1,130 ATP synthase dimers after the final alignment step. (d_–_h) Averages of the aligned data set after sorting by multivariate statistical analysis. Between 100 and 150 individual images were averaged to produce the images shown. (i) Current working model of the dimeric F1F0 ATP synthase complex. We propose that the association of the two F0 domains involves subunits e and g, which have been shown to be essential for dimer formation. The subunit arrangement is drawn according to cross-linking (16, 23) and EM data (7–10). F1 association might result from IF1 bridging in a similar way as seen in the crystal structure of the dimer of soluble F1–ATPase (3). The proposed twofold symmetry axis is indicated by the dotted line. For details, see text.
To determine the structure of the dimeric ATP synthase complex, a data set of 1,130 single images was collected and analyzed by computer-assisted single-particle image alignment and classification. Fig. 2_c_ shows the average of the total data set calculated after the final alignment step. Fig. 2 d_–_h shows the most characteristic averages of the data set after classification. Note that in Fig. 2 c_–_h essentially all of the F1F0 dimers are oriented on the carbon film to produce the side view projection in which the ATP synthase is seen perpendicular to the long axis of the complex. Closer inspection of the averages revealed that there are no image/mirror image pairs, suggesting that the two ATP synthase monomers associate to form a dimer with a twofold symmetry axis that passes through the dimer interface, parallel to the long axis of the complex.
The association of the two ATP synthase monomers occurs at the level of both F0 and F1 subunits (Fig. 2 c_–_h). Associated F0 portions show very close packing in the membrane interface of the dimer, whereas the two F1 parts are more distant. This arrangement produces an angle of ≈40° between the long axes of the two F1F0 monomers (see the black lines in the F0 domains, Fig. 2_c_). The two F1 parts of the dimer are connected by a bridge-like structure of low density (see arrowheads in Fig. 2 e_–_g). Among several subunits of F1, it is likely that this F1–F1 bridge corresponds to the inhibitor protein (IF1). Although IF1 is not essential for F1F0 dimerization, as shown by gene disruption experiments in yeast (18) and by physical removal of IF1 in bovine heart submitochondrial particles (19), the inhibitor protein does induce dimerization of the soluble F1 portion (3, 4, 20) by forming an IF1 bridge connecting the two F1 monomers (3, 4). Furthermore, when IF1 is overexpressed or reconstituted, the ratio of dimeric to monomeric F1F0–ATP synthase is significantly increased in digitonin extracts of rat liver mitochondria (J.J.G., unpublished data). Therefore, it is likely that the ATP synthase dimer can be formed in the absence of IF1 because the essential and closest contacts for F1F0 dimerization take place at the F0 level (refs. 11–13 and Fig. 2); however, the binding of IF1 could stabilize this ATP synthase dimer by forming the observed F1–F1 bridge.
A salient feature of the images in Fig. 2 is that a novel bridging protein density was observed in the intermembrane space side of the complex; this is right underneath the interacting F0 parts (see arrows in Fig. 2 c_–_f). This protein density, which is not seen in the monomeric mitochondrial ATP synthase (7, 9), forms a hanging bridge-like structure connecting the two F0 domains of the dimer. The identity of the bridging protein(s) is not clear at this point, but in all likelihood it is formed by some of the dimer-specific accessory proteins of mitochondrial ATP synthase. The hanging bridge-like structure probably contains part of subunit e, which is known to be essential for dimer formation (18, 21). Subunit e contains a transmembrane GxxxG dimerization motif and a coiled-coil forming region in the C-terminal domain, which is predicted to protrude into the intermembrane space (21–23). Another subunit that was predicted to be bound on the intermembrane space of the yeast enzyme is subunit k (11). However, it remains to be established if a homologous protein exists in the ATP synthase complex from bovine heart mitochondria. It is also noted that MF1F0 also forms an ATP synthasome complex containing complex V and adenine nucleotide translocator and Pi carriers (10); therefore, it is possible that other proteins contribute in the formation of the F0–F0 bridge.
It should be pointed out that the second stalk of the F1F0 complex has been previously resolved by EM of the bacterial (6) and mitochondrial (11, 12) enzymes. Our data do not show the peripheral stalk; it is possible that this is occluded in the dimer interface. Fig. 2_i_ shows our current working model of the F1F0–ATP synthase dimer. We have depicted the dimer-forming subunits (e and g) in the F0–F0 interface close to subunits 6 and 8 on the base of the second stalk according to cross-linking studies (23). The F0 bridging protein seems to reinforce the F0–F0 interface, thereby stabilizing the dimeric arrangement of the ATP synthase.
The most striking feature of the ATP synthase dimer is the conic geometry with which the two ATP synthase monomers are arranged. This conic shape of the dimer is consistent with the current models of cristae biogenesis, in that the inner mitochondrial membrane adopts a curvature comparable to the 40° angle formed by the tilting of the F1F0 monomers. As put forth by Allen (14), polymerization of conic F1F0 dimers could induce budding of the inner mitochondrial membrane, which would ultimately result in the observed tubular structure of the membrane cristae. Other authors have made similar proposals on the basis of genetic disruption of the dimer forming ATP synthase subunits e, g, and k (12, 13, 15, 21). The F1F0 polymer should also add stability to the monomeric F1F0 and resistance to the rotational drag of the central rotor to carry out ATP synthesis more efficiently.
In conclusion, the EM images of the mitochondrial F1F0–ATP synthase dimer here presented reveal the geometric arrangement of the two monomers of the complex. The data show that the two ATP synthase monomers are joined by protein bridges on both the intermembrane space and matrix sides of the complex. The tilting of the two monomers indicates that the formation of F1F0 polymers could be a central issue in the formation of mitochondrial cristae, which are membrane structures designed to increase the overall surface available for oxidative phosphorylation. Further studies will be required to define the identity of the bridging subunits of the ATP synthase dimer and to confirm the role of this dimer in cristae biogenesis.
Acknowledgments
The valuable suggestions of Prof. Armando Gómez-Puyou (National University of México, Mexico City) on the revised version of this work are gratefully acknowledged. This work was supported by the University of California Institute for Mexico and the United States–Consejo Nacional de Ciencia y Tecnología (CONACyT) Collaborative Grant CN-02-44 (to J.J.G. and S.W.), CONACyT (México) Grants J34744-N and V43814-M (to J.J.G.), and National Institutes of Health Grant GM68600 (to S.W.). F.M.-S. was partially supported by Programa de Apoyo para Estudios de Posgrado Grant 202362 from the National University of México.
Author contributions: F.M.-S., S.W., and J.J.G. performed research; F.M.-S., S.W., and J.J.G. analyzed data; F.M.-S., S.W., and J.J.G. wrote the paper; S.W. and J.J.G. designed research; and S.W. and J.J.G. contributed new reagents/analytic tools.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: BN/PAGE, blue native gel electrophoresis.
References
- 1.Boyer, P. D. (2000) Biochim. Biophys. Acta 1458**,** 252–262. [DOI] [PubMed] [Google Scholar]
- 2.Pullman, M. E. & Monroy, G. C. (1963) J. Biol. Chem. 238**,** 3762–3769. [PubMed] [Google Scholar]
- 3.Cabezón, E., Montgomery, M. G., Leslie, A. G. & Walker, J. E. (2003) Nat. Struct. Biol. 10, 744–750. [DOI] [PubMed] [Google Scholar]
- 4.Cabezon, E., Arechaga, I., Jonathan, P., Butler, G. & Walker, J. E. (2000) J. Biol. Chem. 275**,** 28353–28355. [DOI] [PubMed] [Google Scholar]
- 5.Stock, D., Leslie, A. G. & Walker, J. E. (1999) Science 286**,** 1687–1688. [DOI] [PubMed] [Google Scholar]
- 6.Wilkens, S., Zhou, J., Nakayama, R., Dunn, S. D. & Capaldi, R. A. (2000) J. Mol. Biol. 295**,** 387–391. [DOI] [PubMed] [Google Scholar]
- 7.Karrasch, S. & Walker, J. E. (1999) J. Mol. Biol. 290**,** 379–384. [DOI] [PubMed] [Google Scholar]
- 8.Wilkens, S. & Capaldi, R. A. (1998) Nature 393**,** 29. [DOI] [PubMed] [Google Scholar]
- 9.Rubinstein, J. L., Walker, J. E. & Henderson, R. (2003) EMBO J. 22**,** 6182–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C., Ko, Y., Delannoy, M., Ludtke, S. J., Chiu, W. & Pedersen, P. L. (2004) J. Biol. Chem. 279**,** 31761–31768. [DOI] [PubMed] [Google Scholar]
- 11.Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R. A. & Schägger, H. (1998) EMBO J. 17**,** 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paumard, P., Vaillier, J., Coulary, B., Schaeffer, J., Soubannier, V., Mueller, D. M., Brethes, D., di Rago, J. P. & Velours, J. (2002) EMBO J. 21**,** 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arselin, G., Vaillier, J., Salin, B., Schaeffer, J., Giraud, M. F., Dautant, A., Brethes, D. & Velours, J. (2004) J. Biol. Chem. 279**,** 40392–40399. [DOI] [PubMed] [Google Scholar]
- 14.Allen, R. D. (1994) Protoplasma 189**,** 1–8. [Google Scholar]
- 15.Gavin, P. D., Prescott, M., Luff, S. E. & Devenish, R. J. (2004) J. Cell Sci. 117**,** 2333–2343. [DOI] [PubMed] [Google Scholar]
- 16.Minauro-Sanmiguel, F., Bravo, C. & García, J. J. (2002) J. Bioenerg. Biomembr. 34**,** 433–443. [DOI] [PubMed] [Google Scholar]
- 17.Van Heel, M., Harauz, G., Orlova, E. V, Schmidt, R. & Schatz, M. (1996) J. Struct. Biol. 116**,** 17–24. [DOI] [PubMed] [Google Scholar]
- 18.Dienhart, M., Pfeiffer, K., Schagger, H. & Stuart, R. M. (2002) J. Biol. Chem. 277**,** 39289–93925. [DOI] [PubMed] [Google Scholar]
- 19.Tomasetig, L., Di Pancrazio, F., Harris, D. A., Mavelli, I. & Lippe, G. (2002) Biochim. Biophys. Acta 1556**,** 133–141. [DOI] [PubMed] [Google Scholar]
- 20.Domínguez-Ramírez, L., Mendoza-Hernandez, G., Cárabez-Trejo, A., Gómez-Puyou, A. & Tuena de Gómez-Puyou, M. (2001) FEBS Lett. 507, 191–194. [DOI] [PubMed] [Google Scholar]
- 21.Arselin, G., Giraud, M. F., Dautant, A., Vaillier, J., Brethes, D., Coulary-Salin, B., Schaffer, J. & Velours, J. (2003) Eur. J. Biochem. 270**,** 1875–1884. [DOI] [PubMed] [Google Scholar]
- 22.Everard-Gigot, V., Dunn, C. D., Dolan, B. M., Brunner, S., Jensen, R. E. & Stuart, R. A. (2005) Eukaryot. Cell. 4**,** 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belogrudov, G. I., Tomich, J. M. & Hatefi, Y. (1996) J. Biol. Chem. 271**,** 20340–20345. [DOI] [PubMed] [Google Scholar]