Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases (original) (raw)
Abstract
Phytate (inositol hexakisphosphate, IP6) is a regulator of intracellular signaling, a highly abundant animal antinutrient, and a phosphate store in plant seeds. Here, we report a requirement for inositol polyphosphate kinases, AtIPK1 and _AtIPK2_β, for the later steps of phytate synthesis in Arabidopsis thaliana. Coincident disruption of these kinases nearly ablates seed phytate without accumulation of phytate precursors, increases seed-free phosphate by 10-fold, and has normal seed yield. Additionally, we find a requirement for inositol tetrakisphosphate (IP4)/inositol pentakisphosphate (IP5) 2-kinase activity in phosphate sensing and root hair elongation. Our results define a commercially viable strategy for the genetic engineering of phytate-free grain and provide insights into the role of inositol polyphosphate kinases in phosphate signaling biology.
Keywords: genetically engineered crops, low phytate, signal transduction, phosphate sensing
As a relatively inexpensive source of energy-dense and nutrient-rich food, cereal grains contribute >50% of world-wide energy intake, thus representing the most important food supply to the world population (1). The high nutritional value of seeds comes from the deposition of starch, lipids, proteins, and essential minerals during seed development. In this process, there is a corresponding accumulation of phytate, or inositol hexakisphosphate (IP6), which typically comprises >1% of the dry weight and is responsible for approximately two-thirds of total seed phosphorus (2). Although the role of phytate reserves in plant seed is poorly understood, its abundance in grain feed is known to cause nutritional and environmental problems. Monogastric animals are unable to digest phytate, which poses problems in the release of phosphate, inositol, and essential minerals. In developing countries, high-phytate grain-based diets are feared to exacerbate iron and zinc malnutrition. In developed countries, where livestock is fed primarily grain-based feed, the excreted phytate contributes to environmental phosphorus pollution by washing into surface waters, where it accelerates eutrophication (3). Collectively, these problems have provided a strong impetus to develop seed that is reduced in phytate and its inositol polyphosphate (IP) precursors. Several low phytic acid (lpa) mutants have been generated by random mutagenesis in various crops (2, 4-6). Mapping of some of these lpa mutants to early steps in inositol metabolism may explain the pleiotropic phenotypes and may indicate that targeting later steps in phytate synthesis may be prudent.
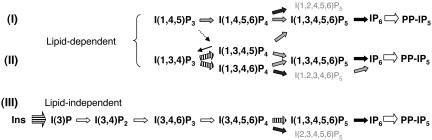
The molecular basis for phytate synthesis in plants is controversial and poorly defined. Three IP6 biosynthetic pathways have been proposed to exist in plants (Fig. 1). Pathways (I) and (II) are inositol lipid-dependent, whereas pathway (III) does not require lipid synthesis. Pathway (I) initiates through phospholipase C and subsequent sequential phosphorylation of inositol 1,4,5-trisphosphate [I(1,4,5)P3] by IP kinases (IPKs), IPK2 (7-12) and IPK1 (13-15). Pathway (II) is also a phospholipase-dependent pathway whereby I(1,4,5)P3 is phosphorylated to inositol 1,3,4,5-tetrakisphosphate [I(1,3,4,5)P4] by either Ipk2 or I(1,4,5)P3 3-kinase, dephosphorylated to inositol 1,3,4-trisphosphate [I(1,3,4)P3], and phosphorylated by a I(1,3,4)P3 5/6-kinase, Ipk2, and Ipk1, respectively, through the indicated intermediates (14, 16-18). The existence of pathway (II) in plants is supported by the cloning of a functional I(1,3,4)P3 5/6 kinase (19), the inositol 1,3,4,6-tetrakisphosphate [I(1,3,4,6)P4] 5-kinase activity of Arabidopsis Ipk2 (11), and the maize lpa2 mutation, which is defective in the I(1,3,4)P3 5/6-kinase that causes a 30% reduction of IP6 (20). However, the failure to identify a I(1,4,5)P3 3-kinase activity or gene ortholog in plants has challenged the existence of pathway (II). Pathway (III) has been proposed in Dictyostelium discoideum and Duckweed (21, 22), is lipid/lipase-independent, and occurs through the sequential phosphorylation of I(3)P or free _myo-_inositol. Recent work has identified a kinase activity with possible relevance to Pathway (III) (23). Here, we define a molecular basis for the later steps of plant phytate biosynthesis and discover insights into a role for plant IPKs in phosphate sensing and signaling.
Fig. 1.
Biosynthetic pathways of phytate in plants. (A) Three distinct pathways have been proposed for the synthesis of IP6. Pathways I and II originate from the hydrolysis of PI(4,5)P2 by phospholipase C and are therefore lipid-dependent. Pathway III is a lipid-independent pathway that involves the sequential phosphorylation of I(3)P or inositol (21, 38). The majority of the participating kinases in this pathway are unknown at the molecular level. Gray arrows designate AtIpk2 activities; black arrows designate AtIpk1 activities; striped arrows designate I(1,3,4)P3 5/6-kinase/I(3,4,5,6)P4 1-kinase activities; the thin dashed arrow designates the I(1,4,5)P3 3-kinase that has not been identified in plants but exists in other higher eukaryotes; the thin solid arrow represents IP 5-phosphatase; IP6 kinase is indicated for which no gene has been formally identified but which are likely to exist given their conservation; the horizontal strip indicates _myo-_inositol kinase; and white arrows indicate unknown or tentatively assigned kinases.
Methods
Cloning of AtIPK1. cDNA clone M40H3 from the Arabidopsis Biological Resource Center was identified as AtIPK1 through blast sequence searches. The ORF was amplified by PCR by using 25 pmol each primer with the Pfu polymerase (Invitrogen). The PCR product was subcloned in-frame with the GST-tag sequence in the pGEX-KG bacterial expression vector, and the sequence was confirmed by the Applied Biosystems Prism dGTP Big Dye Terminator Ready Reaction kit (PE BioSystems). Expression and purification of recombinant GST-AtIpk1 was exactly as described (11), except that induction of expression occurred at 28°C instead of 37°C. Semiquantitative RT-PCR was performed exactly as described (11).
IPK Assays. Unlabeled IPs were purchased from Cell Signals (Columbus, OH). Tritiated [3H]-I(1,4,5)P3 was purchased from PerkinElmer Life Sciences. 3H-I(1,3,4,6)P4 was synthesized as described (11). 3H-(1,4,5,6)P4 was synthesized by incubating 10 μM [3H]-I(1,4,5)P3 with 1.6 pmol GST-AtIpk2β for 10 min at 37°C. [3H]-inositol 1,3,4,5,6-pentakisphosphate [I(1,3,4,5,6)P5] was synthesized by incubating 10 μM [3H]-I(1,4,5)P3 with 1.6 pmol GST-AtIpk2β for 30 min. The reaction was stopped by heat inactivation at 100°C for 1 min; 1.3 pmol AtIpk1 was then incubated with 10 μM [3H]-I(1,3,4,6)P4, [3H]-inositol 1,4,5,6-tetrakisphosphate [I(1,4,5,6)P4], and [3H]-I(1,3,4,5,6)P5 for 30 min at 37°C in a buffer containing 50 mM Hepes, pH 7.5; 50 mM KCl; 2 mM ATP; and 10 mM MgCl2. The reaction products were separated and analyzed by HPLC over a Partisphere (Whatman strong anion exchange, SAX, column and a linear gradient from 10 mM to 1.7 M NH4H2PO4 (pH 3.5) over 12 min, followed by elution for 25 min with 1.7 M NH4H2PO4. IPs were identified based on comparison to the elution of known IP species.
[3H] _Myo_-Inositol Labeling of Arabidopsis Seedlings and Siliques. Individual wild-type and atipk1-1 seeds were stratified at 4°C for 2 days and then germinated in 25-50 μl of liquid Murashige and Skoog (MS) salts media (GIBCO/BRL Life Technologies) with 0.4 mCi/ml [3H] _myo_-inositol (1 Ci = 37 GBq) for 6 days at 20°C and constant light. The seedlings were washed twice with cold dH2O and then disrupted by agitation twice for 2 min with acid-washed glass beads in the presence of 100 μl of 0.5 M HCl/125 μl 2 M KCl/125 μl of CHCl3/372 μl of CHCl3:MeOH (1:2). Disrupted tissue was centrifuged for 5 min, and the soluble layer was removed and analyzed by Partisphere strong anion exchange HPLC as described above. Siliques were labeled by standing vertically in tubes containing 50 μl of MS salts and 100 μCi [3H] _myo_-inositol for 2 days at 20°C, constant light. IPs were harvested from seeds at the late bent cotyledon stage of development (24) that were dissected from whole siliques exactly as described for seedlings.
Nonradioactive Mass Seed IP Analysis. For each sample, ≈50 μl of seeds were weighed, and 10 volumes of 0.4 M HCl and 50 μl of acid-washed glass beads (425-600 μm) were combined for pulverization by a “bead beater” for 4 min. The seeds were then boiled for 5 min and beaten again for 4 min. The seed extract was then centrifuged for 10 min at maximum speed, and the supernatant was frozen on dry ice, thawed to room temperature, and centrifuged again. The supernatant was loaded onto an IonPac AS7 anion exchange column (Dionex) equilibrated with 10 mM methyl piperazine pH 4.0 (Buffer A). The IPs were eluted with a linear gradient from 0 to 100% buffer B (1 M NaNO3, pH 4.0) with a flow rate of 0.3 ml/min by using an HP190 pump. Eluate was mixed with color reagent [0.015% (wt/vol) FeCl3: 0.15% (wt/vol) sulfosalicylic acid] and detected with a photodiode array detector at an absorbance of 550 nm. IP concentrations were calculated based on standard curves established for IP6, IP5, and IP4 and were linear in the range of 2.5-20 nmol.
Plant Growth Conditions, Handling, and Transformations. Arabidopsis (Ecotype Columbia) seeds were imbibed with water, stratified at 4°C for 2-3 days, and germinated in a subirrigated mixture of vermiculite and sand in a controlled growth chamber with a constant temperature of 21°C. The plants were exposed to a 14-h light (135-150 μmol/m-2·s-1) and 10-h dark cycle. After seedlings were established, they were top-watered with half-strength Hoagland's solution (25) every morning and with dH20 every evening unless otherwise indicated.
T3 SALK lines obtained from the Arabidopsis Biological Resource Center were grown, genotyped, and carried through subsequent generations to obtain lines that were homozygous for the T-DNA insertion, which was verified by PCR and Southern blotting. atipk1-1 and atipk2_β_-1 plants that were homozygous for the T-DNA insertion were crossed by using standard procedures and carried through two generations to obtain an atipk1-1atipk2_β_-1 double mutant that was homozygous for each T-DNA insertion.
The cDNA of AtIPK1 was subcloned into pART7 (26) by using standard cloning techniques (27) behind the 35S promoter. The 35S-_AtIPK1_-ocs terminator cassette was then subcloned into pBART27 and subsequently introduced into Agrobacterium strain GV3103 via electroporation. pBART27 is identical to pART27 (26), except that the bar gene replaces the NPTII gene. Wild-type and mutant Arabidopsis plants were transformed via Agrobacterium by floral dipping (28). Transformants were selected with Finale herbicide (Farncom, Phoenix) and verified by PCR.
Pi Assay. Leaf samples were homogenized in 0.4 M HCl, centrifuged, and the supernatant was incubated 1:1 with color reagent (3MH2SO4:2.5%NH4Mo: 10% ascorbate: dH2O; 1:1:1:1). Seed samples were cleared of all nonseed tissue, weighed, and added to 0.4 M HCl to a final concentration of 0.1 mg of seed per ml. The seeds were homogenized for 4 min with a bead beater and glass beads in 0.4 M HCl, boiled for 4 min, and then homogenized again for 4 min. The seed extract was spun at maximum speed in a microfuge for 10 min. The supernatant was frozen and then spun again. The supernatant was analyzed for Pi content and IPs, as described above. A standard curve was generated with K2HPO4 and found to be linear between 0 and 100 nmol phosphate.
Results
AtIPK1 Encodes an IP4 and IP5 2-Kinase. We identified the Arabidopsis orthologs of Ipk2 and Ipk1 through blast sequence homology searches. The IP5 2-kinase amino acid sequences from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida albicans (13) were used as queries against the Arabidopsis genome database. Three candidate genes were identified, two of which appeared to be pseudogenes. The predicted ORF of a gene on chromosome 5 at locus At5g42810 [GeneInfo Identifier (GI): 45680271], designated herein as AtIPK1, had <25% similarity to fungal IP5-2 kinases and 42% similarity to the human IP5 2-kinase (GI:21693577). Multiple sequence alignment of the family of fungal, plant, and metazoan Ipk1 sequences revealed three motifs that define elements present in all Ipk1 members (Fig. 5, which is published as supporting information on the PNAS web site). To identify other plant orthologs, plant genome databases were searched, and full-length cDNAs with high overall similarity (66-82%) to the Arabidopsis sequence were found for rice (OsIpk1; GI: 32988051), maize [ZmIpk1; The Institute for Genomic Research (TIGR) TC222390], and wheat (TaIpk1; TIGR TC152839) (Fig. 5 and Table 2, which is published as supporting information on the PNAS web site). Partial sequences for soybean (TIGR TC184104), barley (TIGR TC222390), and sorghum (TIGR TC97085) were also identified with high homology (not shown). Of note, two other Arabidopsis genes at loci At1g22100 (GI: 18395072) and At1g59312 (GI: 18406350) had deduced amino acid sequences that were ≈70% similar to AtIPK1. Expression of these genes could not be detected, and there are currently no corresponding ESTs in the database, indicating they are either pseudogenes or are expressed at extremely low levels at limited times and places within the plant.
Recombinant AtIpk1 and AtIpk2_β (11) were analyzed for functional properties and the ability to reconstitute IP6 synthesis in vitro. In the presence of ATP, recombinant AtIpk1 phosphorylated I(1,3,4,5,6)P5 to IP6 with a K_m and V_max of 7.6 μM and 0.1 μmol/min/mg (Fig. 6_A and Table 3, which are published as supporting information on the PNAS web site). AtIpk1 also efficiently phosphorylates I(1,3,4,6)P4 and I(1,4,5,6)P4 to produce inositol 1,2,3,4,6-pentakisphosphate [I(1,2,3,4,6)P5] and inositol 1,2,4,5,6-pentakisphosphate [I(1,2,4,5,6)P5]. To reconstitute Pathway (I)-dependent synthesis of IP6 in vitro, we incubated nanogram quantities of recombinant AtIpk2β (11) and AtIpk1 with ATP and I(1,4,5)P3 and found that IP6 was produced (Fig. 6_B). Of interest, by altering the molar ratio of AtIpk2β and AtIpk1 in our in vitro assays, we observed several new IP4 and IP5 species consistent with a number of minor IP species reported from metabolic labeling of plant cells (Fig. 6_B). When AtIpk2β and AtIpk1 were incubated with I(1,3,4)P3 and ATP, no product formation of any kind was observed; however, addition of recombinant I(1,3,4,)P3 5/6-kinase, AtIpk2β, and AtIpk1 enabled IP6 production from I(1,3,4)P3 (Fig. 6_C_). Incubation of I(1,3,4)P3, ATP, recombinant I(1,3,4,)P3 5/6-kinase, and AtIpk1 resulted in the formation of I(1,2,3,4,6)P5. Incubation of I(1,3,4)P3, ATP, recombinant I(1,3,4,)P3 5/6-kinase, and AtIpk2β resulted in the formation of I(1,3,4,5,6)P5. These data demonstrate that plant Ipk2 and Ipk1 define pathway (I) and are able to participate in the last two steps of pathway (II).
AtIpk1 and AtIpk2β Are Necessary for IP6 Synthesis in Vivo. To assess the roles of AtIPK1 and _AtIPK2_β in IP6 production in plants, we obtained loss-of-function mutants of Arabidopsis carrying T-DNA insertions that disrupted either AtIPK1 or AtIPK2_β. We identified an AtIPK1 mutant (SALK_065337, designated here as atipk1-1) and an AtIPK2_β mutant (SALK_104995, designated as atipk2_β_-1) from the Salk Institute Genome Analysis Laboratory population of mapped insertions. The T-DNA insertion in the atipk1-1 mutant is 77 nucleotides upstream of the stop codon in the last exon of the ORF (Fig. 7_A, which is published as supporting information on the PNAS web site). atipk1-1 mutant tissue exhibits >70% reduction in AtIPK1 expression, as indicated by Northern blot analysis (Fig. 7_A). The T-DNA insertion in the atipk2_β_-1 mutant is 400 nucleotides downstream of the start codon and appears to completely ablate _AtIPK2_β expression without altering AtIPK1 or AtIPK2_α expression (Fig. 7_B). Southern blot analysis confirmed that atipk1-1 and atipk2_β_-1 mutants contain a T-DNA insertion at a single locus (data not shown).
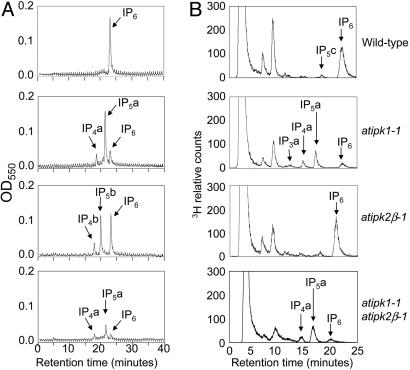
We next measured the mass of IPs, including IP6, in seed extracts from normal and mutant plants by using a quantitative nonradioactive method. Soluble extracts made from mature desiccated wild-type seeds reveal that IP6 comprises 100% of the detectable IPs (Fig. 2_A_), corresponding to an average of 22 nmol IP6/mg seed, ≈1.4% by weight (Table 1). By comparison, the seed phytate levels are reduced by 83% in the atipk1-1, 35% in the atipk2_β_-1, and >95% in the atipk1-1 atipk2_β_-1 double mutant extracts. Interestingly, in both the atipk1-1 and atipk2_β_-1 single mutants, there is a significant increase in IP6 precursors, including IP4 and IP5, such that the total amount of IPs produced is similar to the amount of IP6 observed in wild-type seed extracts (Fig. 2 A). Importantly, the accumulation of IP precursors and IP6 is nearly ablated in the double mutant (Fig. 2 A). Similar results were obtained by using a silique radiolabeling strategy that we developed to measure IP levels in developing embryos (data not shown).
Fig. 2.
IP metabolism of IPK mutants. (A) Nonradioactive mass IP analysis of mature dessicated seed extracts. Seed extracts were separated by an IonPac AS7 anion-exchange column, detected by metal-dye chelation, and measured at 550 nm. Data points are plotted as negative absorbance values. (B) Wild-type and IPK mutant seeds were germinated in liquid MS salts with 400 μCi/ml [3H]-_myo_-inositol for 6 days, and IPs were harvested from seedlings and analyzed by Partisphere strong anion exchange HPLC. Representative chromatograms for each genotype are shown and indicated at the right. The chemical identity of IP3 a is currently unknown. Eighty percent of IP4 a is I(3,4,5,6)P4, and 20% is I(1,3,4,6)P4 and/or I(1,4,5,6)P4. IP5 a is I(1,3,4,5,6)P5. IP4b is I(1,3,4,6)P4 and/or I(1,4,5,6)P4. IP5b is I(1,2,3,4,6)P5. IP5c is either I(1,2,4,5,6)P5 or its enantiomer I(2,3,4,5,6)P5. Sometimes slight alterations in IP migration were observed and are due to different strong anion exchange columns. In all cases, the IPs are compared with known standards.
Table 1. Inositol phosphate, free phosphate, and seed yield analysis.
| Genotype | IP4, nmol/mg | IP5, nmol/mg | IP6, nmol/mg | Pi, nmol/mg | Weight, mg/200 seeds |
|---|---|---|---|---|---|
| Wild type | ND | ND | 22.3 ± 0.4 | 13.7 ± 5.3 | 3.3 ± 0.1 |
| atipk1-1 | 3.3 ± 0.3 | 29.7 ± 0.6 | 3.9 ± 0.9 | 23.9 ± 0.3 | 3.0 ± 0.4 |
| atipk2β-1 | 2.5 ± 0.9 | 15.3 ± 1.4 | 14.5 ± 0.2 | 23.7 ± 0.9 | 3.3 ± 0.3 |
| atipk1-1 atipk2β-1 | <1 | 6.7 ± 1.1 | <1 | 127.3 ± 6.0 | 3.1 ± 0.2 |
The effects of IPK disruption on IP metabolism in plant tissue were distinct from those observed in seeds for _atipk2_β-1 but not atipk1-1. Radiolabeled seedlings from atipk1-1, atipk2_β_-1, and atipk1-1 atipk2_β_-1 double mutant plants were analyzed (Fig. 2_B_). The IP6 and inositol content of wild-type seedlings was 9.6% and 87% of the total recovered labeled soluble inositol species. Phytate production in atipk2_β_-1 mutant seedlings looked normal compared with wild-type, indicating that _AtIPK2_α was able to fully compensate in tissue but not seeds. Conversely, the IP6 content of atipk1-1 seedlings was 0.71% of the total labeled inositol species, representing a 93% reduction from wild type. These seedlings also exhibited IP6 precursor accumulation, including IP5, IP4, and inositol trisphosphate (IP3) (representing 3.4%, 1.3%, and 1.3%, respectively). The minor I(1,2,4,5,6)P5 peak, which is typically seen in wild-type seedlings, was no longer detectable in the atipk1-1 seedlings, consistent with the in vitro data that I(1,2,4,5,6)P5 synthesis is _AtIPK1_-dependent (data not shown). The atipk1-1 atipk2_β_-1 double mutant seedlings had an IP profile similar to the atipk1-1 single mutant, which also verified the dispensability of AtIpk2β in IP6 synthesis in seedling tissue.
Seed Yield and Free Phosphate Analysis. As a measure of seed yield, we quantified the number of seeds per silique and average seed weight from our mutant plants. Analysis of >20 mature siliques obtained from several independent plants from wild-type and mutant plants had similar average seed numbers per silique (48 ± 2). Two hundred mature desiccated seeds were weighed and varied from 14 to 16 μg per seed without any statistical differences among the wild-type, single, or double mutants (Table 1). Because previously described low-phytate grains are characterized by having above-average inorganic phosphate levels, we measured the amount of free inorganic phosphate in the IPK mutants. As shown in Table 1, whereas the atipk1-1 and atipk2_β_-1 mutants had an approximate 2-fold increase, the double mutant had a 10-fold increase in inorganic phosphate. Therefore, despite significant changes in IP and phosphate levels in seeds, the yield remained largely unaffected in the IPK mutants.
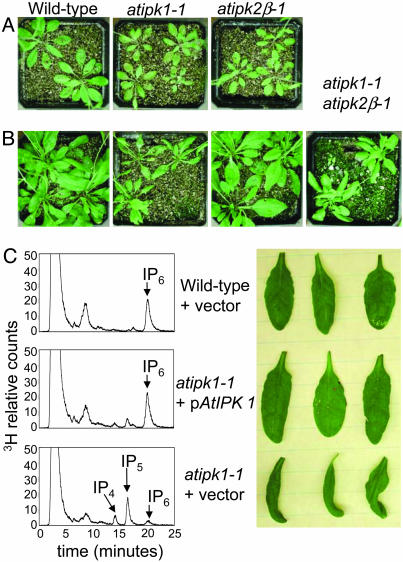
A Requirement for IP4/IP52-Kinase Activity in Phosphate Sensing and Signaling. During the course of breeding, we observed an obvious but conditional phenotype in atipk1-1 mutant plants. Given the proposed roles for seed phytate in germination, in no case did homozygous mutation in either AtIPK1 or _AtIPK2_β result in altering the timing of seed germination or appearance of cotyledons. Furthermore, no alterations were observed in timing of development of the mature plant. Wild-type and mutant plants, when grown on a mixture of sand and vermiculite, watered daily with tenth-strength Hoagland's solution (0.1×), appeared similar in size, with only a slight defect observed in the atipk1-1 mutant plants (Fig. 3_A_). However, atipk1-1 mutant plants, when watered daily with half-strength Hoagland's solution (0.5×), appeared significantly smaller than wild-type or atipk2_β_-1 plants and were typified by abaxially curled (epinastic) rosette leaves. The epinasty became more pronounced over time, and by 4 weeks, the mutant leaves exhibited necrotic margins and were approximately one-half to one-third of the mass of wild-type or atipk2_β_-1 plants (Fig. 3 B and C). Both the epinasty and inositol metabolism defects were rescued by complementing the expression of AtIPK1 (Fig. 3_C_). The phenotype of the atipk1-1 atipk2_β_-1 double mutant was similar to atipk1-1, indicating this effect depends on IP4/IP5 2-kinase activity alone.
Fig. 3.
Phenotypic analysis of IPK mutants. Seeds were germinated on a mixture of sand and vermiculite through subirrigation until the cotyledons had emerged. Plants were then top-watered with either 0.1 × or 0.5 × Hoagland's solution daily and were grown at 20°C under a 14-hr light (150 μmol/m2·s-1) and 10-hr dark cycle in the Duke University Phytotron. Top view of 5-week-old plants watered with 0.1 × Hoagland's solution (A) or 0.5 × Hoagland's solution (B). Genotypes are indicated. (C) Complementation of IP synthesis and growth phenotype in atipk1-1 by constitutive expression of AtIPK1 through a 35S promoter. (Left) HPLC chromatograms of IP profiles from [3H]-inositol-labeled 5-day-old seedlings. (Right) Leaves from 5-week-old plants grown in half-strength Hoagland's solution as described. Upper and leaf row are wild-type plants transformed with the empty expression vector. Middle and leaf row are atipk1-1 plants transformed with the empty expression vector. Bottom and leaf row are atipk1-1 plants transformed with the expression vector harboring the AtIPK1 gene.
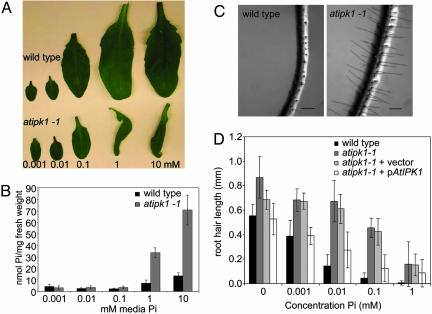
To further probe the negative effects of Hoagland's solution on atipk1-1 plants, we (i) systematically removed each nutrient individually from the Hoagland's solution or (ii) added individual nutrients to water. Only the removal of phosphate was able to alleviate the leaf epinasty and necrosis (not shown). Through additional experiments, we found that 0.5× Hoagland's solution containing Pi at concentrations of 0.1 mM or below did not induce epinasty, and the leaf size difference between wild type and atipk1-1 was minimal (Fig. 4_A_). In contrast, at Pi concentrations of 1 mM or higher, the leaves of the atipk1-1 plants were epinastic and were one-third to one-quarter the size of wild-type (Fig. 4_A_). The leaf epinasty also corresponded with an increased intracellular concentration of Pi in the atipk1-1 mutants (Fig. 4_B_). Although the leaf mass is much smaller in the atipk1-1 mutants compared with wild type, the increased intracellular Pi could not be explained by a more concentrated cytosol, because the gross measurements of nucleic acids (A260) and proteins (A280) varied by <13% between the atipk1-1 and wild-type leaves (not shown). Thus, the atipk1-1 plants are unable to maintain normal intracellular phosphate concentrations and appear to suffer from phosphate toxicity when administered phosphate levels that are typically optimal for plant growth.
Fig. 4.
Aberrant phosphate sensing and root hair growth in atipk1-1 mutants. (A) Leaves from 5-week-old wild-type and atipk1-1 plants grown as described above in half-strength Hoagland's solution with variable phosphate concentrations as indicated. (B) Inorganic phosphate concentrations in the leaves from plants grown at various phosphate concentrations. (C) Representative images of main root and root hairs from wild-type (Left) and atipk1-1 (Right) plants grown vertically on MS-agar plates containing 10 μM phosphate. (Bar, 0.2 mm.) (D) Root hair lengths of the indicated genotypes grown vertically on MS-agar plates containing various phosphate concentrations as indicated. n = 12 for each genotype.
Because the atipk1-1 mutant appears to have altered phosphate homeostasis, we tested to see whether it also had an altered sensitivity to extracellular Pi levels. Plants respond to a sensed change in phosphate by increasing or decreasing the surface area of the roots to alter Pi availability and uptake. One of the ways that a change in surface area is achieved is by altering the length of the root hairs. Thus, for instance, when phosphate is limiting, Arabidopsis seedlings produce longer root hairs to scavenge more Pi (29). Seeds from wild-type, atipk1-1, and atipk1-1 plants transformed with p_AtIPK1_ were plated onto MS agar containing variable concentrations of Pi and were grown in a vertical position under constant light. After 12 days, primary roots were imaged, and the root hair length was measured. We found that the atipk1-1 roots had longer root hairs than wild-type at lower concentrations of Pi and an attenuated ability to sense the increase in Pi, as indicated by the inability to form shorter root hairs at higher Pi concentrations (Fig. 4 C and D). The Pi -insensitive root hair growth could be rescued in the atipk1-1 mutant by complementation of AtIPK1, indicating that the effect is due to the loss of AtIPK1. This result suggests an inability of the mutants to sense the extracellular phosphate levels as readily as wild-type.
Discussion
Molecular definition of the later stages of phytate synthesis in plants has important ramifications in agricultural and biotechnology fields. We suggest that seed-specific disruption of IPK2 and IPK1 in cereal grains represents an excellent strategy for the generation of nutritionally improved crops, especially because IPK mutants do not exhibit defects in seed yield. IPK mutants meet other criteria for improving the nutritional and agricultural quality of grains: (i) they do not quantitatively accumulate phytate precursors, which could counterbalance the benefits of an IP6 reduction; and (ii) they exhibit increased levels of bioavailable free phosphorus, so that animal dietary needs can be achieved without supplementation. Of note, whereas the seed yield of IPK mutants does not decrease significantly, the reduced size of the at_ipk1-1_ mutant plants, especially when grown under higher phosphate conditions, may be problematic for commercialization. We suggest this is likely due to a role for IPK1 in tissue signaling, which would easily be overcome through seed-selective disruption strategies. This is supported by the lack of growth defect observed in atIpk2_β_-1 mutants, which have normal tissue IP metabolism but compromised seed phytate levels.
Our study provides insights into roles for higher IP messenger pathways downstream of IP3 in the regulation of phosphate biology. The phosphate-dependent root hair elongation defects observed in the atipk1-1 mutants indicate that products of Ipk1 regulate phosphate acquisition machinery. Despite elevated intracellular phosphate concentrations and adequate external phosphate, the atipk1-1 plants behave as if they are in a phosphorus-limiting environment, indicating an apparent defect in the rheostat that monitors the Pi status of the leaves and roots. Perhaps IP signals are required to integrate these processes through regulating the activity or expression of genes involved in phosphate uptake and acquisition, such as phosphate transporters or secreted acid phosphatases. Precedence for such a hypothesis comes from studies in yeast (30), where it has been shown that proper control of transcription and chromatin remodeling of the PHO5 locus depend on IPKs. Many phosphate-responsive genes have been identified in Arabidopsis. The Arabidopsis mutant pho2 is a phosphate accumulator that suffers from phosphate toxicity (31). Like atipk1-1, pho2 mutants accumulate phosphate by 2-5-fold over wild type in leaf tissue. Although the molecular identity of the pho2 mutation is not known, it is presumed to be a defect in a regulator of the distribution of Pi from the roots to the shoots or to otherwise regulate shoot Pi concentrations (32). It will be interesting to explore a possible relationship between pho2 and atipk1-1.
Root hair elongation requires the biosynthesis of new cell wall components, cell wall loosening, and targeted vesicle transport to the growing tip of the cell. Several mutants have been isolated that affect root hair elongation and potentially disrupt these processes (33-36). Thus, the elongated root hairs of the atipk1-1 plants may result from the aberrant regulation of one of these processes. Additionally, ethylene and auxin have been implicated as diffusible signals that alter root growth. The ethylene and auxin response mutants, axr1, aux1, etr1, and ein2, have shorter root hairs than wild type, and exogenously applied hormone precursors encourage root hair elongation (37). The atipk1-1 mutation may enhance ethylene and/or auxin production or responses and therefore cause increased root hair growth and leaf epinasty, although mutant seedlings do not display the classical deetiolated phenotype of constitutive ethylene responses in the dark, suggesting that ethylene production and responses are normal, at least during initial seedling development.
Conclusion
Understanding the roles of higher phosphorylated IP messengers, such as IP5 and IP6, has been a long-standing effort in the field of biology. We have now defined a molecular basis for their production in plants and have generated mutants compromised in their ability to produce these messengers. We link IPKs, and presumably their IP products, to phosphate signaling biology and phytate production in seeds, opening up several new areas of study. Which IP messengers are involved in these important cellular pathways? The most likely candidates are IP5, IP6, and/or inositol pyrophosphates, which are all downstream of IP3 signaling. This model is supported by the excellent correlation between the dosage of Ipk1 and Ipk2 and the cellular levels of IP products. However, given our data indicating that AtIpk1 and AtIpk2 are promiscuous enzymes that use/generate several IPs through multiple routes, such as pathway (I) or (II), we cannot rule out other species. The future challenge will be to dissect the specific defects in phosphate sensing and homeostasis of the atipk1-1 mutants and to define the corresponding IP receptors involved.
Supplementary Material
Supporting Information
Acknowledgments
We thank June dela Cruz for technical assistance, the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and the Arabidopsis Biological Resource Center for providing these materials. This work is supported by funds from the Howard Hughes Medical Institute and from National Institutes of Health Grants R01 HL-55672 and R33 DK-070272.
Author contributions: J.S.-P., S.-T.C., and J.D.Y. designed research; J.S.-P., R.J.B., S.-T.C., R.A.F., and J.D.Y. performed research; J.S.-P. contributed new reagents/analytic tools; J.S.-P., R.J.B., S.-T.C., and J.D.Y. analyzed data; and J.S.-P. and J.D.Y. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IP, inositol polyphosphate; IP6, inositol hexakisphosphate; IP3, inositol trisphosphate; IP4, inositol tetrakisphosphate; IP5, inositol pentakisphosphate; I(1,4,5)P3, inositol 1,4,5-trisphosphate; I(1,3,4)P3, inositol 1,3,4-trisphosphate; I(1,4,5,6)P4, inositol 1,4,5,6-tetrakisphosphate; I(1,3,4,6)P4, inositol 1,3,4,6-tetrakisphosphate; I(1,3,4,5,6)P5, inositol 1,3,4,5,6-pentakisphosphate; I(1,2,3,4,6)P5, inositol 1,2,3,4,6-pentakisphosphate; I(1,2,4,5,6)P5, inositol 1,2,4,5,6-pentakisphosphate; IPK, IP kinase; MS, Murashige and Skoog; TIGR, The Institute for Genomic Research.
References
- 1.Bewley, J. D. (1997) Plant Cell 9**,** 1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raboy, V. (2001) Trends Plant Sci. 6**,** 458-462. [DOI] [PubMed] [Google Scholar]
- 3.Brinch-Pedersen, H., Sorensen, L. D. & Holm, P. B. (2002) Trends Plant Sci. 7**,** 118-125. [DOI] [PubMed] [Google Scholar]
- 4.Dorsch, J. A., Cook, A., Young, K. A., Anderson, J. M., Bauman, A. T., Volkmann, C. J., Murthy, P. P. & Raboy, V. (2003) Phytochemistry 62**,** 691-706. [DOI] [PubMed] [Google Scholar]
- 5.Hitz, W. D., Carlson, T. J., Kerr, P. S. & Sebastian, S. A. (2002) Plant Physiol. 128**,** 650-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilu, R., Panzeri, D., Gavazzi, G., Rasmussen, S. K., Consonni, G. & Nielsen, E. (2003) Theor. Appl. Genet. 107**,** 980-987. [DOI] [PubMed] [Google Scholar]
- 7.Fujii, M. & York, J. D. (2005) J. Biol. Chem. 280**,** 1156-1164. [DOI] [PubMed] [Google Scholar]
- 8.Odom, A. R., Stahlberg, A., Wente, S. R. & York, J. D. (2000) Science 287**,** 2026-2029. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi, A., Caffrey, J. J., Snyder, S. H. & Shears, S. B. (2000) FEBS Lett. 468**,** 28-32. [DOI] [PubMed] [Google Scholar]
- 10.Seeds, A. M., Sandquist, J. C., Spana, E. P. & York, J. D. (2004) J. Biol. Chem. 279**,** 47222-47232. [DOI] [PubMed] [Google Scholar]
- 11.Stevenson-Paulik, J., Odom, A. R. & York, J. D. (2002) J. Biol. Chem. 277**,** 42711-42718. [DOI] [PubMed] [Google Scholar]
- 12.Xia, H. J., Brearley, C., Elge, S., Kaplan, B., Fromm, H. & Mueller-Roeber, B. (2003) Plant Cell 15**,** 449-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ives, E. B., Nichols, J., Wente, S. R. & York, J. D. (2000) J. Biol. Chem. 275**,** 36575-36583. [DOI] [PubMed] [Google Scholar]
- 14.Verbsky, J. W., Wilson, M. P., Kisseleva, M. V., Majerus, P. W. & Wente, S. R. (2002) J. Biol. Chem. 277**,** 31857-31862. [DOI] [PubMed] [Google Scholar]
- 15.York, J. D., Odom, A. R., Murphy, R., Ives, E. B. & Wente, S. R. (1999) Science 285**,** 96-100. [DOI] [PubMed] [Google Scholar]
- 16.Chang, S. C., Miller, A. L., Feng, Y., Wente, S. R. & Majerus, P. W. (2002) J. Biol. Chem. 277**,** 43836-43843. [DOI] [PubMed] [Google Scholar]
- 17.Shears, S. B. (1998) Biochim. Biophys. Acta 1436**,** 49-67. [DOI] [PubMed] [Google Scholar]
- 18.Verbsky, J. W., Chang, S. C., Wilson, M. P., Mochizuki, Y. & Majerus, P. W. (2005) J. Biol. Chem. 280**,** 1911-1920. [DOI] [PubMed] [Google Scholar]
- 19.Wilson, M. P. & Majerus, P. W. (1997) Biochem. Biophys. Res. Commun. 232**,** 678-681. [DOI] [PubMed] [Google Scholar]
- 20.Shi, J., Wang, H., Wu, Y., Hazebroek, J., Meeley, R. B. & Ertl, D. S. (2003) Plant Physiol. 131**,** 507-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brearley, C. A. & Hanke, D. E. (1996) Biochem. J. 314 (Pt 1), 227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irvine, R. F. & Schell, M. J. (2001) Nat. Rev. Mol. Cell. Biol. 2**,** 327-338. [DOI] [PubMed] [Google Scholar]
- 23.Shi, J., Wang, H., Hazebroek, J., Ertl, D. S. & Harp, T. (2005) Plant J. 42**,** 708-719. [DOI] [PubMed] [Google Scholar]
- 24.Otegui, M. S., Capp, R. & Staehelin, L. A. (2002) Plant Cell 14**,** 1311-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoagland, D. R. & Arnon, D. I. (1938) The Water-Culture Method for Growing Plants Without Soil (Univ. of California College of Agriculture Agricultural Experiment Station, Berkeley, CA).
- 26.Gleave, A. P. (1992) Plant Mol. Biol. 20**,** 1203-1207. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Clough, S. J. & Bent, A. F. (1998) Plant J. 16**,** 735-743. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Bucio, J., Cruz-Ramirez, A. & Herrera-Estrella, L. (2003) Curr. Opin. Plant Biol. 6**,** 280-287. [DOI] [PubMed] [Google Scholar]
- 30.Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R. & O'Shea, E. K. (2003) Science 299**,** 114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delhaize, E. & Randall, P. J. (1995) Plant Physiol. 107**,** 207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong, B., Rengel, Z. & Delhaize, E. (1998) Planta 205**,** 251-256. [DOI] [PubMed] [Google Scholar]
- 33.Vincent, P., Chua, M., Nogue, F., Fairbrother, A., Mekeel, H., Xu, Y., Allen, N., Bibikova, T. N., Gilroy, S. & Bankaitis, V. A. (2005) J. Cell Biol. 168**,** 801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, X., Cnops, G., Vanderhaeghen, R., De Block, S., Van Montagu, M. & Van Lijsebettens, M. (2001) Plant Physiol. 126**,** 575-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilliland, L. U., Kandasamy, M. K., Pawloski, L. C. & Meagher, R. B. (2002) Plant Physiol. 130**,** 2199-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiefelbein, J. W. & Somerville, C. (1990) Plant Cell 2**,** 235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitts, R. J., Cernac, A. & Estelle, M. (1998) Plant J. 16**,** 553-560. [DOI] [PubMed] [Google Scholar]
- 38.Stephens, L. R. & Irvine, R. F. (1990) Nature 346**,** 580-583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information