Identification and Characterization of a Novel Adhesin Unique to Oral Fusobacteria (original) (raw)
Abstract
Fusobacterium nucleatum is a gram-negative anaerobe that is prevalent in periodontal disease and infections of different parts of the body. The organism has remarkable adherence properties, binding to partners ranging from eukaryotic and prokaryotic cells to extracellular macromolecules. Understanding its adherence is important for understanding the pathogenesis of F. nucleatum. In this study, a novel adhesin, FadA (_Fusobacterium ad_hesin A), was demonstrated to bind to the surface proteins of the oral mucosal KB cells. FadA is composed of 129 amino acid (aa) residues, including an 18-aa signal peptide, with calculated molecular masses of 13.6 kDa for the intact form and 12.6 kDa for the secreted form. It is highly conserved among F. nucleatum, Fusobacterium periodonticum, and Fusobacterium simiae, the three most closely related oral species, but is absent in the nonoral species, including Fusobacterium gonidiaformans, Fusobacterium mortiferum, Fusobacterium naviforme, Fusobacterium russii, and Fusobacterium ulcerans. In addition to FadA, F. nucleatum ATCC 25586 and ATCC 49256 also encode two paralogues, FN1529 and FNV2159, each sharing 31% identity with FadA. A double-crossover fadA deletion mutant, F. nucleatum 12230-US1, was constructed by utilizing a novel sonoporation procedure. The mutant had a slightly slower growth rate, yet its binding to KB and Chinese hamster ovarian cells was reduced by 70 to 80% compared to that of the wild type, indicating that FadA plays an important role in fusobacterial colonization in the host. Furthermore, due to its uniqueness to oral Fusobacterium species, fadA may be used as a marker to detect orally related fusobacteria. F. nucleatum isolated from other parts of the body may originate from the oral cavity.
Fusobacterium nucleatum is a long filamentous, gram-negative anaerobe associated with various human diseases, including periodontal, peritonsillar, orofacial, brain, chest, lung, abdominal, blood, and obstetrical and gynecological abscesses and infections, existing either as a mixed infection or as the sole infecting agent (3, 6, 8-10, 12, 13, 16, 28, 39, 41, 45, 48, 52). The primary colonization site of F. nucleatum in humans is the oral cavity. It is one of the most abundant gram-negative anaerobes in subgingival plaque and can be isolated in healthy periodontal sites. During periodontal infection, its cell mass increases as much as 10,000-fold, making it one of the most abundant anaerobic species in the diseased sites (48). F. nucleatum is also one of the predominant anaerobes associated with preterm birth, having been frequently isolated from amniotic fluids and placentas of women delivering prematurely (27). F. nucleatum may originate in the oral cavity and be transmitted to the uterus via a hemotogenous route. It has been shown in mice that, once in the bloodstream, F. nucleatum colonizes specifically in the placenta, causing preterm and term stillbirths (25).
Several virulence phenotypes of F. nucleatum have been identified. F. nucleatum is recognized as an “adhesive” organism because it binds to a variety of host mammalian cells, including epithelial and endothelial cells, polymorphonuclear leukocytes, monocytes, erythrocytes, fibroblasts, and HeLa cells, as well as salivary macromolecules, extracellular matrix proteins, and human immunoglobulin G (IgG) (2, 25, 26, 50, 59, 62, 63). It also coaggregates with a wide array of microorganisms in the oral cavity and plays an important role in plaque formation (1, 7, 20, 22, 29, 36-38, 46, 53). Identification of the adhesin molecules on F. nucleatum is thus essential for understanding its pathogenesis. It has been suggested that F. nucleatum possesses both lectin-like and non-lectin-like adhesins (44, 49, 54, 58, 60, 61). Three components, a 40- to 42-kDa major outer membrane porin protein (FomA) and 39.5-kDa and 30-kDa polypeptides, have been suggested as possible adhesins from F. nucleatum that are involved in interbacterial coaggregation (33, 34, 55). FomA was also found to bind to the human IgG Fc fragment (23). A high-molecular-mass component, ranging from 300 to 330 kDa, has been suggested as a galactose-binding agglutinin (49). However, it is unclear if any of these components are involved in F. nucleatum binding to the host cells.
F. nucleatum invades epithelial and endothelial cells in vitro, a mechanism presumably employed for its spreading into deeper tissues (25, 26). Invasion of F. nucleatum into endothelial cells was observed in vivo in the mouse placenta (25). A spontaneous mutant defective in tissue cell attachment and invasion, F. nucleatum 12230 lam, has been isolated, but the nature of its mutational change is unknown (26). The lam mutant exhibited virulence similar to that of the wild type in causing fetal death in the mice (25).
F. nucleatum also induces an array of host cell responses. It is a strong stimulator of the production of interleukin-8 from epithelial cells, indicating its ability to induce inflammation (15, 26). It stimulates apoptosis of human peripheral white blood cells and suppresses T-cell responses (30, 56). The organism also induces production of innate antimicrobial peptides, human β-defensins, in gingival epithelial cells (40). This is presumably a mechanism to suppress the growth of competitive species.
In this study, we report the identification of a novel 13.6-kDa adhesin peptide from F. nucleatum involved in attachment to mammalian cells and the construction of its deletion mutant by a novel sonoporation method.
MATERIALS AND METHODS
Bacterial strains, culture conditions, plasmids, and enzymes.
Bacterial strains and plasmids used in this study are listed in Table 1. All fusobacterial strains were maintained on either Trypticase soy or Columbia agar (BBL) supplemented with 5% defibrinated sheep blood (Cleveland Scientific, OH) or in Trypticase soy or Columbia broth (BBL) and incubated as previously described (25). The Escherichia coli strains were maintained in LB broth (Difco) or on LB agar (Difco) and incubated at 37°C in air. Restriction endonucleases and ligase were purchased from New England BioLabs (Beverly, MA), and PfuUltra high-fidelity DNA polymerase was from Stratagene (La Jolla, CA).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Source of isolation or relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| F. nucleatum 12230 | Transtracheal isolate, working strain in the lab | 26 |
| F. nucleatum 12230-US1 | F. nucleatum 12230 Δ_fadA_::ermF-ermAM | This study |
| F. nucleatum ATCC 10953 | Inflamed gingiva; F. nucleatum subsp. polymorphum | P. E. Kolenbrander |
| F. nucleatum ATCC 23726 | F. nucleatum subsp. nucleatum | P. E. Kolenbrander |
| F. nucleatum ATCC 25586 | Cervicofacial lesion; F. nucleatum subsp. nucleatum | P. E. Kolenbrander |
| F. nucleatum ATCC 49256 | Periodontal pocket; F. nucleatum subsp. vincentii | P. E. Kolenbrander |
| F. nucleatum ATCC 51190 | Sinusitis in upper jaw; F. nucleatum subsp. fusiform | P. E. Kolenbrander |
| F. nucleatum PK 1594 | Periodontal pocket | P. E. Kolenbrander |
| F. nucleatum DUMC1356 | Amniotic fluid; preterm birth | 25 |
| F. nucleatum DUMC2079 | Placenta; preterm birth | 25 |
| F. nucleatum DUMC2929 | Amniotic fluid; preterm birth | 25 |
| F. nucleatum DUMC3156 | Placenta; preterm birth | G. B. Hill |
| F. nucleatum DUMC3349 | Placenta; preterm birth | 25 |
| F. gonidiaformans DUMC CF65-1 | Vaginal tract; bacterial vaginosis | 25 |
| F. gonidiaformans DUMC CF63-1 | Vaginal tract; bacterial vaginosis | 25 |
| F. naviforme DUMC CF108-1 | Vaginal tract; bacterial vaginosis | G. B. Hill |
| F. mortiferum ATCC 25557 | Maxillary abscess | P. E. Kolenbrander |
| F. periodonticum ATCC 33693 | Periodontitis | P. E. Kolenbrander |
| F. russii ATCC 25533 | Infection in a cat | P. E. Kolenbrander |
| F. simiae ATCC 33568 | Monkey dental plaque | P. E. Kolenbrander |
| F. ulcerans ATCC 49185 | Skin ulcer | P. E. Kolenbrander |
| Plasmids | ||
| pLAFR2 | Cosmid vector used for library construction (21.6 kb) | 19 |
| pYWH 1 | pLAFR2 clone containing fadA (48.2 kb) | This study |
| pCR2.1 | Cloning vector (3.9 kb) | Invitrogen |
| pYWH 401 | pCR2.1 carrying 2.4-kb fragment containing fadA (6.3 kb) | This study |
| pVA2198 | Source of the ermF-ermAM cassette (9.2 kb) | 18 |
| pYH1378 | pCR2.1 carrying Δ_fadA_::ermF-ermAM and flanking regions of fadA (7.1 kb) | This study |
| pRL250 | Source of sacB (14.3 kb) | 11 |
| pYH1426 | pYH1378 containing sacB (9.2 kb) | This study |
To construct plasmids pYH1378 and pYH1426, a 526-bp fragment, “up_fadA_,” and a 510-bp fragment, “down_fadA_,” corresponding to the upstream and downstream regions flanking the fadA gene, respectively, were amplified using primer sets _fadA_1f (5′AGGTCAAGAAGCAAAAGG3′)-_fadA_1r (5′TTTTTGGTACCCTTGCTGCATCAGTTGC3′) and _fadA_2f (5′TTTTTGGATCCTCAAGCTTTAAGAGCTGG3′)-_fadA_2r (5′AGGGTTACTTGATTCAGG3′), generating a KpnI site in “up_fadA_” and a BamHI site in “down_fadA_” at ends adjacent to fadA. A KpnI-BamHI fragment containing the ermF-ermAM cassette from pVA2198 (18) was then ligated with the “up_fadA_” and the “down_fadA_” fragments, followed by cloning into pCR2.1 (Invitrogen, Carlsbad, CA). The resulting plasmid, pYH1378, was digested with EcoRV, and a sacB gene from pRL250 (11) was inserted into the EcoRV site to generate pYH1426.
Preparation of biotinylated and nonlabeled KB surface proteins.
The human oral mucosal epithelial cell line KB (ATCC CCL-17; American Type Culture Collection, Manassas, VA) was maintained in MEM medium (GibcoBRL, Rockville, MD) supplemented with 10% fetal bovine serum (Mediatech, Herndon, VA). The cultures were grown in four 75-cm2 tissue culture flasks (Fisher Scientific, Pittsburgh, PA) under 5% CO2 at 37°C to near confluence. The cells were detached from the flasks by using enzyme-free cell dissociation buffer (GibcoBRL). Following washes with sterile phosphate-buffered saline (PBS) (Sigma, St. Louis, MO), the cells were incubated in 2 ml of 1 mM sulfo-NHS-LC-biotin (Pierce Chemical Co., Rockford, IL) at 4°C for 2 h. The outer membrane components were extracted with 1% Triton X-100 (Sigma) at room temperature for 1 h, followed by centrifugation. The supernatant was transferred to a Centricon YM-3 column (Millipore, Bedford, MA) and centrifuged at 7,500 × g. The centrifugation was repeated twice, adding 2 ml sterile 10 mM Tris, pH 7.5, each time to the sample reservoir to change the buffer. At the end of centrifugation, a total of approximately 50 μl of concentrated sample was recovered and stored at 4°C. The protein concentration was determined with bicinchoninic acid (BCA) (Pierce, Rockford, IL). Nonlabeled KB surface proteins were prepared following the same procedures except that the KB cells were not incubated with sulfo-NHS-LC-biotin.
Far-Western analysis.
A total of approximately 1 × 108 to 5 × 108 CFU of F. nucleatum 12230 or 10 μg of fractionated F. nucleatum components, unless otherwise indicated, were subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (0.45-μm pore size; Millipore). The membranes were blocked with 1% bovine serum albumin (Sigma), followed by incubation with biotinylated KB surface proteins at a 1:500 dilution in TBST (50 mM Tris, pH 7.5, 0.5 M NaCl, 0.1% Tween 20) at room temperature for 1 h. The membranes were washed with TBST and incubated with avidin-horseradish peroxidase (HRP) conjugate (Bio-Rad, Hercules, CA) at a 1:1,000 dilution. The membranes were developed using 4-chloro-1-naphthol (Bio-Rad) and hydrogen peroxide (Sigma). For controls, the membranes were incubated directly with avidin-HRP conjugate without incubation with biotinylated KB surface proteins. For competitive far-Western analysis, the membrane was preincubated with nonbiotinylated KB surface proteins in 20-fold excess at room temperature for 1 h prior to incubation with biotinylated KB surface proteins.
Preparation of “40P.”
One liter of freshly grown F. nucleatum 12230 culture was centrifuged, and the cell pellet was resuspended in 10 ml sterile PBS, followed by 10 min of ultrasonication in an ice-water bath with a 3-mm microtip at 20-W output pulse setting at a 50% duty cycle (Vibra Cell, model VC250; Sonic and Materials Inc., Danbury, CT). The suspension was then centrifuged at 3000 × g, and the supernatant was centrifuged again at 100,000 × g. The twice-centrifuged supernatant was designated the cell extract, to which ammonium sulfate was added to a final concentration of 40% (wt/vol) and incubated at 4°C with agitation for >4 h. The suspension was centrifuged at 100,000 × g for 2 h, and the pellet was dissolved in 10 mM Tris, pH 7.5, followed by dialysis against 10 mM Tris, pH 7.5, at 4°C. The resulting solution was designated “40P,” and its protein concentration was determined by BCA.
Construction and screening of F. nucleatum 12230 cosmid library.
Chromosomal DNA of F. nucleatum 12230 was purified, partially digested with Tsp509I, and cloned into the EcoRI site of cosmid pLAFR2 (19). The ligation mixture was incubated with Gigapack III XL packaging extract (Stratagene), and the cosmid phage lysate was prepared according to the manufacturer's instructions. The phage lysate was used to transfect JM109, and the cosmid clones were selected on LB plates containing 20 μg/ml tetracycline. The clones were saved in 96-well plates and stored at −80°C. Four degenerate oligonucleotide pools were designed based on the protein N-terminal sequence and the Codon Usage Database (Table 2). The probes were labeled with digoxigenin (DIG) by using the DIG DNA labeling and detection kit (Roche, Indianapolis, IN). They were then used to screen the F. nucleatum 12230 cosmid library by colony hybridization as described previously (21). Putative positive clones were examined by Southern blotting analysis using pool 4 probes.
TABLE 2.
Degenerate oligonuleotide probes used to identify the putative adhesin from an F. nucleatum genomic library
| Probe(s) | Sequenceb |
|---|---|
| FadA N-terminal sequencea | ATDAASLVGELQALDA |
| Pool 1 | GCXACXGATGCTGCTTCATTAGTXGGXGAA |
| Pool 2 | GCXACXGATGCTGCTTCTTTAGTXGGXGAA |
| Pool 3 | GCXACXGATGCTGCTAGTTTAGTXGGXGAA |
| Pool 4 | TTAGTXGGXGAATTACAGGCXTTAGATGC |
fadA sequence analysis.
The DNA sequence of the 2.4-kb fragment from pYWH401 was determined (CAMBI Nucleic Acid Facility, Buffalo, NY), first using the M13 reverse primer and the T7 primer and then using synthetic oligonucleotide primers M13ext1 (5′GCTTCCATTTGTTAAAACCACC3′), M13ext2 (5′GCAATTAAACTTACAATCTGAAAGCC3′), M13ext3 (5′CAGTTAGACCAAAGGGTCCTG3′), T7ext1 (5′TCCATAACCAAATAACTTATAC3′), and T7ext2 (5′CTAGCAGCGTCAGCTTGTGCTC3′), derived from the sequence of the fragment. The open reading frames (ORFs) were identified using the National Center for Biotechnology Information ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The fadA genes from F. nucleatum ATCC 49256, DUMC1356, and ATCC 33693 were amplified with oligonucleotide primers _fadA_1Kf (5′CTTTTAAAACCTCTCCAAGC3′) and T7ext1. All other fadA genes were amplified with primers M13ext2 and T7ext1. The PCR conditions were as following: denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 5 min, with repeats of 30 cycles. The PCR products were treated with Exo/SAP-IT (USB, Cleveland, OH) and their nucleotide sequences determined (Molecular Biotechnology Core, Lerner Research Institute, Cleveland, OH) using primers _fadA_for (5′TTAGCTGTTTCTGCTTCAGC3′) and _fadA_rev (5′TTACCAGCTCTTAAAGCTTG3′).
DNA dot blotting.
Chromosomal DNA of Fusobacterium species was denatured by heating at 95°C for 10 min. A total of 0.5 μg denatured DNA was spotted onto an Immobilon-NY+ membrane (Millipore). A DIG-labeled 359-bp fadA fragment was used as the probe. It was amplified by PCR using primers _fadA_for and _fadA_rev with chromosomal DNA of F. nucleatum 12230 as the template.
Construction of fadA mutant of F. nucleatum 12230 via sonoporation.
Log-phase F. nucleatum 12230 cells were washed and resuspended to a final concentration of 1 × 1010 CFU/ml in PBS supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2. A total of 100 μl of the bacterial suspension was mixed with 50 μg plasmid DNA and 50 μl Optison (Perflutren protein-type A microspheres for injection, USP; Amersham, Princeton, NJ) in a 96-well plate and subjected to ultrasound (US) treatment. A custom-made regular planar piezoelectric lead-zirconate-titanate US transducer of a circular aperture with a diameter of 5.1 cm (center frequency of 0.96 MHz) was vertically directed upward to irradiate the bacteria in the 96-well plate. A signal generator (33250A; Agilent Technologies, Palo Alto, CA) controlled the duty cycle and initial amplitude of the input signal, which was amplified using a 75-W power amplifier (75A250; Amplifier Research, Souderton, PA). The amplified signal was connected to the US transducer to generate the desired US field. Pulsed US exposures at a duty cycle of 50% and a pulse repetition frequency of 1 Hz were used for a total duration of 90 s. The US beam profile was measured using a calibrated hydrophone system (HPM04/1; Precision Acoustics, United Kingdom), and the effective US output powers were calibrated using a US power meter (UPM-DT-10; Ohmic Instrument Co, Easton, MD). The acoustic pressure of US exposure was 0.5 MPa (corresponding to an initial input signal at 130 mV). Following US treatment, the suspension was plated onto Columbia blood agar plates and incubated under anaerobic conditions at 37°C for 24 h. The bacteria were then replicated onto Columbia blood agar plates containing 0.4 μg/μl clindamycin and incubated for 3 additional days. The clindamycin-resistant colonies were purified on plates before being inoculated in Columbia broth containing 0.4 μg/μl clindamycin. The genetic nature of the mutants was verified by PCR, using primers _fadA_for and _fadA_rev, and by Southern blot analysis, using the same 359-bp DIG-labeled fadA probe used for DNA dot blotting.
Western blot analysis.
Whole-cell F. nucleatum was boiled for 3 min in Laemmli sample buffer, subjected to 15% SDS-PAGE, and blotted onto a PVDF membrane. The membrane was incubated overnight with polyclonal anti-FadA serum (unpublished results) at a 1:1,000 dilution at 4°C. After washing, the membrane was incubated with goat anti-rabbit IgG-HRP at a 1:1,000 dilution at room temperature for 1 h, followed by color development as described above.
Northern blot analysis.
RNA was prepared from mid-log-phase F. nucleatum by phenol extraction. A total of 10 μg RNA/lane was loaded onto a 1.5% agarose-formaldehyde gel, alongside a 0.16- to 1.77-kb RNA ladder (Invitrogen), followed by electrophoresis at 50 V for 1.5 h. The RNA was then transferred onto a Zeta-Probe GT blotting membrane (Bio-Rad) by alkaline blotting for 4 h. The above-mentioned 359-bp fadA fragment was used as a probe, using the ECL direct nucleic acid labeling and detection system (Amersham Biosciences) according to the manufacturer's instructions. The membrane was washed, exposed on an X-ray film, and developed. The experiment was repeated at least twice.
RT-PCR.
RNA was prepared from mid-log-phase F. nucleatum by using the RNeasy minikit (QIAGEN, Valencia, CA), followed by treatment with RNase-free DNase (QIAGEN). DNA contamination in the RNA samples was determined by PCR amplification of ORF2, fadA, and ORF3 with primers Orf2-F (5′GGAGGGGAAGATGGAAGAAG3′) and Orf2-R (5′TCTTCTGCTATTGCTGGATGAA3′), _fadA_for and _fadA_rev, and Orf3-F (5′AAGGGTTACTTGATTCAGGAATTG3′) and Orf3-R (5′CAATTCCTGAATCAAGTAACCCTT3′), respectively. Samples with no detectable DNA contamination were used for reverse transcription-PCR (RT-PCR). Reverse transcription was performed using SuperScript II (Invitrogen) with 1 μg DNA-free RNA and 10 pmol of the forward primer for each gene in a final volume of 50 μl per reaction. An aliquot of 2 μl of the RT reaction mix was then used for PCR amplification of 25 cycles (94°C for 45 s, 55°C for 30 s, and 72°C for 1 min, followed by a 7-min extension at 72°C), using both the forward and reverse primers described above. The PCR products were subjected to electrophoresis on a 1.0% agarose gel. Each experiment was repeated at least twice.
Bacterial growth curve.
Fresh broth cultures of F. nucleatum were transferred into fresh medium at a 1:4 dilution. An aliquot was taken out every hour, and its optical density at 600 nm was measured using a Genesys 5 UV-visible spectrophotometer (Thermo Electron, Waltham, MA). The experiment was repeated twice.
Tissue culture cell attachment assay.
KB cells were cultured as described above. Chinese hamster ovary (CHO) cells were maintained in F12K medium (Mediatech) supplemented with 10% fetal bovine serum. The attachment assays were carried out as previously described (26). Briefly, KB or CHO cells were seeded into 24-well trays and allowed to grow to near confluence. Immediately before the assay, the spent medium was replaced with fresh nonsupplemented medium. F. nucleatum strains were harvested and resuspended in PBS to a density of 5 × 108 cells/ml. Approximately 5 × 106 CFU was added into each well and incubated at 37°C under 5% CO2 for 1 h. The monolayers were then washed four times with PBS and lysed with water. F. nucleatum attached to the cells was enumerated on blood agar plates. Attachment values were expressed as the percentage of bacteria associated with the host cells relative to the total number of bacteria initially added.
Nucleotide sequence accession number.
The nucleotide sequence of the 2.4-kb fragment from F. nucleatum 12230 containing the fadA gene has been deposited in the GenBank database with an assigned accession number AY850357. The accession numbers for the FadA sequences from other fusobacterial strains and species are as follows: DQ012969 for F. nucleatum ATCC 10953, DQ012970 for F. nucleatum ATCC 23726, DQ012971 for F. nucleatum ATCC 25586, DQ012972 for F. nucleatum ATCC 49256, DQ012973 for F. nucleatum ATCC 51190, DQ012974 for F. nucleatum DUMC1356, DQ012975 for F. nucleatum DUMC2079, DQ012976 for F. nucleatum DUMC2929, DQ012977 for F. nucleatum DUMC3156, DQ012978 for F. nucleatum DUMC3349, DQ012979 for F. nucleatum PK1594, DQ012980 for F. periodonticum ATCC 33693, and DQ012981 for F. simiae ATCC 33568.
RESULTS
Identification of a putative adhesin molecule from F. nucleatum 12230.
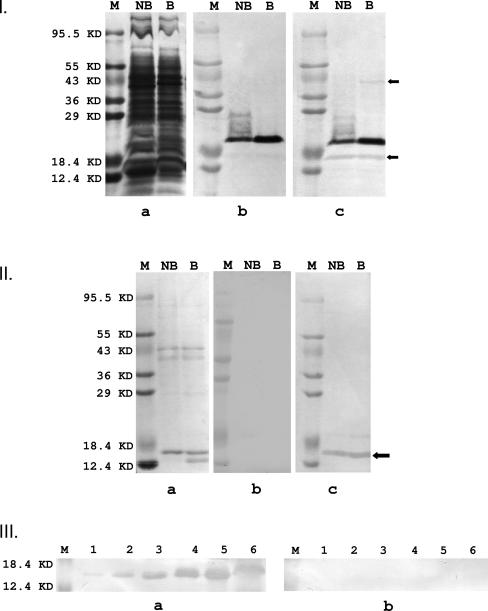
It was shown previously that F. nucleatum binds to both KB cells and normal human gingival epithelial cells (26). Therefore, KB cells were used in this study for ease of manipulation. Sulfo-NHS-LC-biotin is a nonspecific biotinylating agent which does not penetrate mammalian cell membranes. Thus, upon incubation with the KB cells, it nonspecifically labeled proteins on the KB cell surface. Triton X-100 extraction of the KB cells produced a “cocktail” of biotinylated KB surface proteins. When this “cocktail” was incubated with F. nucleatum components immobilized on PVDF membranes, binding between F. nucleatum adhesins and their receptors on the KB cells could occur. When whole-cell F. nucleatum 12230 was tested, one dark and two light bands were identified (Fig. 1I, panel c). When the same components were incubated directly with avidin-HRP, the two light bands were not detected, while the dark band remained strongly visible (Fig. 1I, panel b). This observation indicates that the components identified by the two light bands bound KB surface proteins, while the dark band represents an F. nucleatum component(s) naturally bound with biotin, such as a carboxylase. The two light bands had apparent molecular masses of 40 kDa and 16 kDa. The 40-kDa component was detected only in the boiled F. nucleatum and not in the nonboiled sample, indicating it likely formed oligomers too large to migrate into the gel (Fig. 1I, panel c). The size of the monomer and its oligomerization suggest that this component could be the trimer-forming major outer membrane porin protein FomA (35). Since the 16-kDa component was more prominent, it was analyzed further.
FIG. 1.
I and II. Identification of F. nucleatum adhesins by far-Western analysis. a. F. nucleatum 12230 (I) or 40P (II) components stained with Coomassie blue following 12% SDS-PAGE. b. F. nucleatum 12230 (I) or 40P (II) components immobilized on PVDF membranes were incubated with streptavidin-HRP conjugate, followed by chemiluminescence reaction. c. F. nucleatum 12230 (I) or 40P (II) components immobilized on PVDF membranes were first incubated with biotinylated KB surface proteins, followed by incubation with streptavidin-HRP. “M,” protein size marker, with sizes indicated on the left; “NB,” nonboiled whole-cell F. nucleatum 12230 (I) or 40P (II); “B,” boiled whole-cell F. nucleatum 12230 (I) or 40P (II). The arrows point to bands visible in c but not in b, indicating binding to biotinylated KB proteins. The bottom arrow in Ic and the arrow in IIc indicate FadA. III. Competitive far-Western analysis. 40P or F. nucleatum 12230 whole-cell components immobilized on PVDF membranes were incubated directly with biotinylated KB surface proteins (a) or preincubated with 20× nonlabeled KB surface proteins prior to incubation with biotinylated KB surface proteins (b). “M,” protein size markers, as indicated on the left; lanes 1 to 5, 40P in increasing amounts (0.375, 0.625, 1.25, 2.5, and 5 μg, respectively); lanes 6, F. nucleatum 12230.
Incubation of F. nucleatum 12230 cell extract with 40% (wt/vol) (NH4)2SO4 resulted in the precipitation of a limited number of F. nucleatum components, designated “40P” (Fig.1II, panel a). Far-Western blot analysis of 40P using biotinylated KB surface proteins identified one major band (Fig. 1II, panel c), which was not detected if the membrane was incubated only with avidin-HRP (Fig.1II, panel b). The component in Fig.1II, panel a, corresponding to that in Fig.1II, panel c, was identified by aligning the two PVDF membranes. This component was designated “FadA,” for _Fusobacterium ad_hesin A, and its N-terminal amino acid sequence was determined by protein microsequencing (ProSeq, Boxford, MA) (Table 2). Binding between FadA and the biotinylated KB surface proteins increased as the FadA quantity increased (Fig.1III, panel a). Furthermore, the binding was inhibited by preincubation with nonlabeled KB surface proteins prior to incubation with the biotinylated proteins (Fig.1III, panel b). These results further indicate that FadA bound specifically to a component(s) on the KB cell surface.
Identification of the fadA gene.
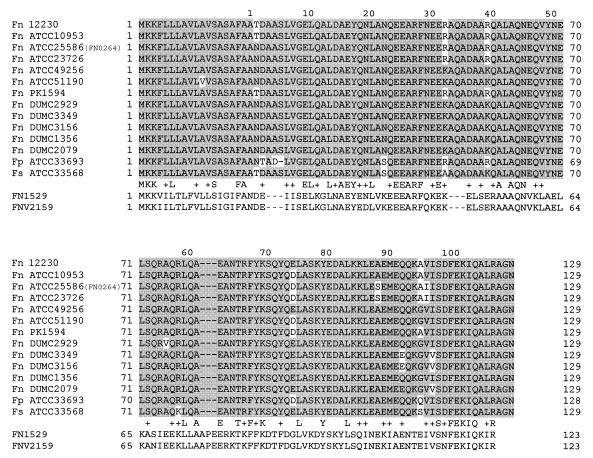
A genomic library of F. nucleatum 12230 was constructed by cloning the bacterial chromosomal DNA into the cosmid vector pLAFR2. A total of 576 cosmid clones were saved in six 96-well plates. AseI digestion of 10 randomly picked clones showed that all were independent clones (data not shown). With a mean insert size of approximately 20 kb, and assuming that all clones were independent, this library should have covered the entire F. nucleatum 12230 genome four to five times. A total of four different degenerate oligonucleotide pools were used to screen the library (Table 2). Pools 1 to 3 correspond to the first 10 amino acids of the FadA N-terminal sequence. The only difference between these three pools was the codon used for the serine residue at position 6. Pool 4 corresponds to the amino acid sequence from position 7 through 16. Since the F. nucleatum genome consists of more than 70% AT, the pools were designed such that only the third position in selected codons carried a mixture of A and T. This design reduced the degeneracy of the oligonucleotide pools. Through repeated colony hybridization and Southern blot analyses, one true positive clone was identified and designated YWH1 (data not shown). Cosmid pYWH1 was purified and digested with different restriction endonucleases. A 6.2-kb EcoRV fragment, a 1.3-kb EcoRI fragment, and a 2.4-kb Sau3AI fragment were identified through Southern blot analysis using pool 4 oligonucleotides as probes (data not shown). The 2.4-kb Sau3AI fragment was subcloned into the BamHI site of pCR2.1 to generate pYWH401, and its DNA sequence was determined (data not shown). A total of four ORFs were identified. The smallest ORF encodes 129 amino acids, with the first eighteen residues corresponding to a typical signal peptide, which should be absent in the secreted form. The next 16 residues perfectly matched the N-terminal peptide sequence of FadA (Table 2), indicating that the component identified by far-Western analysis was the secreted form. FadA is alanine (20%) and leucine (10%) rich. It shares no homology with any known adhesins. Secondary structure analysis preformed by the Ph.D. method at the European Molecular Biology server indicated that it was composed almost exclusively of α-helix. The intact FadA had a calculated molecular mass of 13.6 kDa, while the secreted form was 12.6 kDa, smaller than the apparent molecular mass of 16 kDa identified by SDS-PAGE.
Conservation of FadA among fusobacteria.
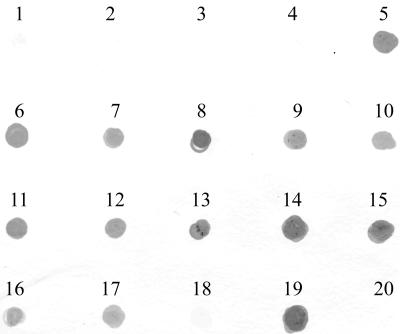
The presence of fadA among other species and strains of fusobacteria was examined by DNA dot blotting with 12 strains of F. nucleatum, 2 strains of F. gonidiaformans, and one strain each of F. mortiferum, F. naviforme, F. periodonticum, F. russii, F. simiae, and F. ulcerans (Fig. 2). The fadA gene appeared to exist in the three most closely related species, F. nucleatum, F. periodonticum, and F. simiae, but was absent in the other species (Fig. 2). DNA sequence analysis showed that FadA was highly conserved among the _fadA_-positive species (Fig. 3).
FIG. 2.
DNA dot blot analysis of the fadA gene in different Fusobacterium species. 1, F. gonidiaformans DUMC CF65-1; 2, F. gonidiaformans DUMC CF63-1; 3, F. mortiferum ATCC 25557; 4, F. naviforme DUMC CF108-1; 5, F. nucleatum ATCC 10953; 6, F. nucleatum ATCC 25586; 7, F. nucleatum ATCC 23726; 8, F. nucleatum 12230; 9, F. nucleatum ATCC 49256; 10, F. nucleatum ATCC 51190; 11, F. nucleatum PK1594; 12, F. nucleatum DUMC2929; 13, F. nucleatum DUMC3349; 14, F. nucleatum DUMC3156; 15, F. nucleatum DUMC1356; 16, F. nucleatum DUMC2079; 17, F. periodonticum ATCC 33693; 18, F. russii ATCC 25533; 19, F. simiae ATCC 33568; 20, F. ulcerans ATCC 49185.
FIG. 3.
Amino acid sequence alignment of FadA and its paralogues. Highlighted in gray are the identical residues shared among FadA proteins. The sequences of two FadA paralogues, FN1529 from F. nucleatum ATCC 25586 and FNV2159 from F. nucleatum ATCC 49256, are listed below FadA. The conserved and identical residues between FadA and the paralogues are indicated. Fn, F. nucleatum; Fp, F. periodonticum; Fs, F. simiae. The numbers above the sequence indicate amino acid positions in the secreted form of FadA, and the numbers beside the sequence indicate positions in the intact form.
Following identification of the fadA gene, the genomic sequence of F. nucleatum ATCC 25586 became available (31). A BLAST search showed that the 2.4-kb fragment containing the fadA gene was highly conserved between F. nucleatum 12230 and ATCC 25586 (data not shown). The ORF corresponding to fadA in F. nucleatum ATCC was FN0264. The three additional ORFs near fadA were identified as follows: ORF1 (FN0262), which transcribes in the opposite direction relative to fadA, encodes a formate acetyltransferase; ORF2 (FN0263), immediately upstream of fadA, encodes a peptidyl-prolyl _cis_-_trans_-isomerase; and ORF3 (FN0265) is downstream of fadA and encodes a cell division protein, FtsX, with 29% identity to the ABC transporter permease cell division protein FtsX of Listeria monocytogenes (data not shown). In addition, a paralogue (i.e., homologue on the same chromosome), FN1529, is present in F. nucleatum ATCC 25586, sharing 31% identity with FadA (Fig. 3).
More recently, the genome sequence of another strain, F. nucleatum subsp. vincentii ATCC 49256, became available (32). DNA dot blot and PCR analyses indicate the presence of fadA in this strain (Fig. 2 and 3). However, a BLAST search failed to identify fadA in the gapped genome sequence (data not shown). Instead, a FadA paralogue, FNV2159, which is 31% identical to FadA and 98% identical to FN1529, was identified (Fig. 3).
Construction of a fadA deletion mutant of F. nucleatum 12230.
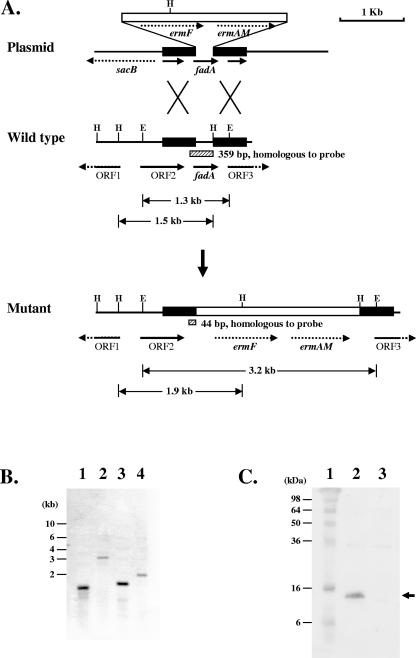
To the best of our knowledge, currently no report is available on the construction of genetic knockout mutants of F. nucleatum. Since fadA is made up of fewer than 400 bases, it would be difficult to construct a knockout mutant by integrating a suicide plasmid containing an internal fragment of fadA. Following unsuccessful attempts to generate a correct fadA deletion mutant of F. nucleatum 12230 by either electroporation or conjugation, DNA delivery via sonoporation, i.e., transient membrane permeabilization by ultrasound, was tested. Plasmid pYH1426, which contains a homologous fragment of approximately 500 bp at either end of fadA and a 2.1-kb ermF-ermAM cassette replacing fadA, was used (Fig. 4A). The ermF-ermAM cassette confers erythromycin and clindamycin resistance (24). The plasmid also carries a 2.1-kb fragment containing a sacB gene, conferring sucrose sensitivity (11). Intact pYH1426 was mixed with F. nucleatum 12230 and Optison, followed by a 90-s (pulse repetition frequency, 1 Hz; duty cycle, 50%) ultrasonic treatment. Optison is a Food and Drug Administration-approved contrast agent consisting of albumin-coated perfluoropropane (C3F8) gas bubbles and is routinely used in ultrasound imaging for cardiac diagnosis. It has been used to facilitate sonoporation in mammalian cells (47). Under these experimental conditions, the viability of F. nucleatum 12230 was not affected; nor was there any detectable DNA damage when examined by agarose gel electrophoresis (data not shown). Ultrasonic delivery of pYH1426 into F. nucleatum 12230 produced more than 30 independent transformants, at an efficiency of approximately 0.05 transformant/μg DNA. All transformants were genetically identical double-crossover fadA deletion mutants, as determined by PCR (data not shown) and Southern blot analyses (Fig. 4B). Loss of FadA in these mutants was verified by Western blotting using anti-FadA polyclonal antibodies (Fig. 4C). One of the mutants was designated F. nucleatum 12230-US1 (Table 1).
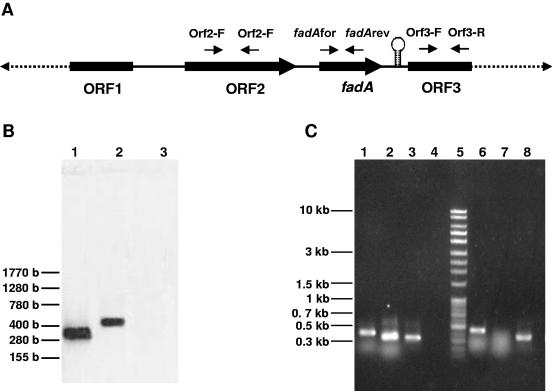
FIG. 4.
Inactivation of the fadA gene of F. nucleatum 12230. A. Schematic diagram of construction of the Δ_fadA_::erm mutant by double-crossover allelic exchange. The erythromycin resistance cassette ermF-ermAM was inserted between bp 71 and 365 of the fadA gene. The shaded boxes represent regions hybridizing with the probe during Southern blotting. The sizes of the fragments hybridized with the probe are indicated. H, HindIII cleavage sites; E, EcoRI cleavage sites. B. Southern blot analysis of F. nucleatum 12230 and F. nucleatum 12230-US1, using a 359-bp fadA fragment as a probe. Lanes: 1, F. nucleatum 12230 digested with EcoRI; 2, F. nucleatum 12230-US1 digested with EcoRI; 3, F. nucleatum 12230 digested with HindIII; 4, F. nucleatum 12230-US1 digested with HindIII. The DNA size markers are indicated on the left. C. Western blot analysis of F. nucleatum 12230 and 12230-US1, using anti-FadA antibodies. Lanes: 1, protein size markers, with molecular masses shown on the left; 2, F. nucleatum 12230; 3, F. nucleatum 12230-US1. The arrow indicates FadA.
In order to determine if insertional inactivation had any polar effects on the downstream gene ORF3, Northern blotting and RT-PCR were performed (Fig. 5). Northern blotting using fadA as a probe revealed a transcript of approximately 400 bases, indicating that fadA was transcribed monocistronically (Fig. 5A). DNA sequence analysis indicated the likely existence of a rho-independent transcription terminator immediately downstream of fadA from position 1893 to 1941 (data not shown). Transcription of both neighboring genes of fadA, the upstream ORF2 and the downstream ORF3, was unaffected in F. nucleatum 12230-US1 as indicated by RT-PCR (Fig. 5C).
FIG. 5.
RT-PCR and Northern blot analyses of F. nucleatum 12230 and F. nucleatum 12230-US1. A. Schematic diagram showing locations of primers used for RT-PCR. The 2.4-kb _fadA_-containing fragment from F. nucleatum 12230 is presented as solid lines. The hairpin indicates the location of a putative transcription terminator. B. Northern blot analysis of F. nucleatum 12230 (lane 2) and F. nucleatum 12230-US1 (lane 3), using the 359-bp fadA fragment as a probe. Lane 1, 359-bp fadA fragment (positive control). C. RT-PCR analysis of expression of ORF2 (lanes 1 and 6), fadA (lanes 2 and 7), and ORF3 (lanes 3 and 8) in F. nucleatum 12230 (lanes 1 to 3) and F. nucleatum 12230-US1 (lanes 6 to 8). Lane 4, negative control without RNA; lane 5, 1.0-kb Plus DNA ladder.
Characterization of the fadA deletion mutant.
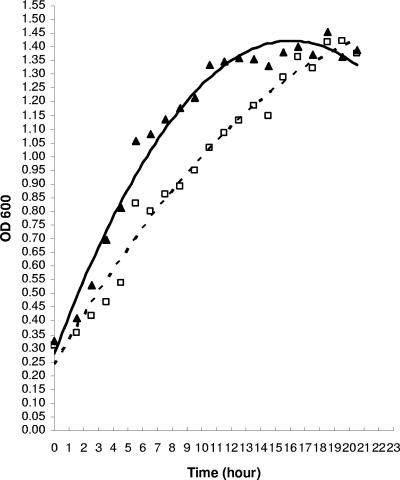
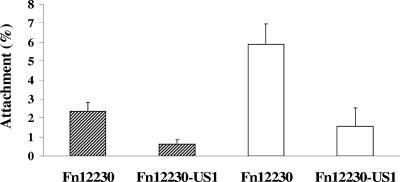
F. nucleatum 12230-US1 was characterized by its growth rate, aerotolerance, and ability to bind to KB and CHO cells, each in comparison with its parental strain F. nucleatum 12230. The mutant consistently grew at a lower rate, with a doubling time in the exponential phase of approximately 6 h, compared to 5 h for the parental strain (Fig. 6). The mutant exhibited aerotolerance similar to that of the wild type (data not shown). When tested for binding to KB and CHO cells, F. nucleatum 12230-US1 was found to be severely defective. Although the percent attachment levels varied when KB or CHO cells were used, the difference between the wild type and the mutant remained consistent, with the mutant exhibiting a reduction of approximately 70 to 80% (Fig. 7). These observations indicate that FadA is nonessential for bacterial integrity but is required for its binding to host cells.
FIG. 6.
Growth of F. nucleatum 12230 (solid triangles and solid line) and F. nucleatum 12230-US1 (open squares and dashed line) in Columbia broth. OD 600, optical density at 600 nm.
FIG. 7.
Attachment of F. nucleatum 12230 and F. nucleatum 12230-US1 to KB (hatched bars) and CHO (open bars) cells. The levels of attachment are means and standard deviations from three separate experiments, each performed in triplicate.
DISCUSSION
F. nucleatum binds to a wide variety of partners, including both eukaryotic and prokaryotic cells. Although a few putative adhesins have been suggested to be involved in interbacterial coaggregation or agglutinination of red blood cells, it is unclear if they are also required for F. nucleatum binding to other host cells. Known for its wide-ranging adherence properties, F. nucleatum may possess multiple adhesins, some of which may be partner specific while others may have multiple binding substrates. In this study, a novel peptide, FadA, was identified, with the α-helix predicted as the predominant secondary structure. The secreted form of FadA has a larger apparent molecular mass than its calculated molecular mass (Fig. 1). As a putative adhesin, FadA is likely associated with the outer membrane. It is not unusual for a membrane protein to have aberrant migration on SDS-PAGE. For instance, the 40-kDa FomA protein has been reported to migrate as 37-kDa, 40-kDa, 42-kDa, and 62-kDa proteins (5, 34, 35).
Loss of FadA resulted in a 70 to 80% reduction of the organism's ability to bind to KB and CHO cells (Fig. 7). Several possibilities exist: (i) the loss of attachment was due to the lower growth rate of F. nucleatum 12230-US1, (ii) the defect was due to a polar effect on the downstream gene(s), (iii) FadA serves as an accessory protein for binding, or (iv) FadA is a major adhesin directly involved in F. nucleatum binding to host cells. The first three possibilities are unlikely for the following reasons: (i) the incubation time during the attachment assay was 1 hour, during which the bacterial growth was minimal; (ii) Northern blot and RT-PCR analyses indicated that fadA was transcribed monocistronically and that transcription of ORF3 was unaffected by the mutational change in fadA (Fig. 5) (these observations were also supported by the detection of a putative transcription terminator immediately downstream of fadA), and (iii) FadA was identified by far-Western analysis as directly and specifically bound by biotinylated KB surface proteins (Fig. 1). Taken together, the most reasonable explanation would be that FadA is directly involved in binding. Further supporting this notion is that expression of FadA in E. coli enhanced the ability of E. coli to bind to mammalian cells (unpublished results). Since F. nucleatum 12230-US1 was defective in binding to both KB and CHO cells, it is likely that FadA binds to a receptor(s) common to both types of cells. It should be pointed out that although FadA appears to be a significant adhesin for F. nucleatum to bind to host cells, an additional adhesin(s) exists, likely accounting for the remaining binding activities observed in F. nucleatum 12230-US1.
Although BLAST searches failed to identify FadA in the gapped genome of F. nucleatum ATCC 49256, DNA hybridization, PCR, and sequence analyses indicated that FadA is highly conserved among F. nucleatum, F. periodonticum, and F. simiae yet is absent in F. mortiferum, F. gonidiaformans, F. naviforme, F. mortiferum, F. russii, and F. ulcerans (Fig. 2 and 3). F. nucleatum, F. periodonticum, and F. simiae have been reported as three closely related oral species, forming a distinct group within the genus (42, 51). The presence of FadA in these three species and its absence in others are consistent with the previously described genetic relatedness within the group. Therefore, fadA may be used as a marker for identification of orally related fusobacteria. The conservation of fadA in F. nucleatum isolated from intrauterine infections and its absence in the vaginal species F. gonidiaformans and F. naviforme further support the hypothesis that intrauterine F. nucleatum originates from the oral cavity rather than the vaginal tract (25). BLAST searches also identified two paralogues of FadA, FN1529 from F. nucleatum ATCC 25586 and FNV2159 from F. nucleatum ATCC 49256, which share 31% identity with FadA and 98% identity with each other. The conservation of the FadA paralogue among fusobacteria and its role in adherence are currently under investigation.
Genetic manipulation of F. nucleatum has been difficult, presumably due, in part, to its diversified restriction endonuclease systems, which differ between strains and cleave DNA irrespective of the extent of methylation (43). Attempts to construct a fadA deletion mutant of F. nucleatum 12230 by either electroporation or conjugation were unfruitful. This could be attributed to one or more of the following: (i) inefficient DNA delivery by electroporation or conjugation, (ii) inefficient homologous recombination between the exogenous plasmid and the bacterial chromosome, (iii) exogenous DNA being digested by a restriction endonuclease(s) before recombination could occur, or (iv) killing of the bacteria by electroporation. DNA delivery via ultrasound has been employed with mammalian cells (4, 14, 47, 57, 64). It has been suggested that ultrasonic treatment of mammalian cells induces transient membrane permeability, allowing uptake of extracellular compounds, such as chemotherapeutic agents, genetic materials, and fluorescence markers, which normally do not permeate the cell membrane (17). Although ultrasound treatment in the presence of Optison enhances sonoporation, its mechanism is not clearly understood (47). Our results demonstrate that the same technology could also be applied to bacteria, even though the bacterial cell envelope is quite different from that of mammalian cells. Unlike electroporation, which kills the majority of the bacteria, ultrasonic treatment under the testing conditions used did not affect the viability of F. nucleatum. By mixing F. nucleatum 12230 with intact pYH1426, we intended to first obtain a single-crossover merodiploid construct through sonoporation and then utilize the sacB gene on pYH1426 to select for a double-crossover mutant on sucrose medium (11). Surprisingly, all transformants obtained were fadA deletion mutants. The mechanism of this one-step double-crossover allelic exchange is unclear and is currently under investigation. It should be pointed out that, as a preliminary test, the concentrations and ratios of the bacteria, plasmid, and Optison were empirically determined and thus may be far from optimal. Additional work is needed to understand the sonoporation mechanism and to optimize its conditions.
In summary, a novel adhesin, FadA, which is unique to oral fusobacteria, was identified. It was required for F. nucleatum attachment to epithelial cells and thus may play an important role in Fusobacterium colonization in the host.
Acknowledgments
We are indebted to Gale B. Hill and Paul E. Kolenbrander for generously providing fusobacterial strains.
This work was supported in part by NIH grants DE 14924 and DE 14447 and Philip Morris External Research grant to Y.W.H., NIH grant DE 09821 to H.K.K., and start-up funds from the Department of Biomedical Engineering, Case Western Reserve University, to C.X.D.
REFERENCES
- 1.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13**:**51-54. [DOI] [PubMed] [Google Scholar]
- 2.Babu, J. P., J. W. Dean, and M. J. Pabst. 1995. Attachment of Fusobacterium nucleatum to fibronectin immobilized on gingival epithelial cells or glass coverslips. J. Periodontol. 66**:**285-290. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, C., D. Schoonbroodt, C. Wagner, and Y. Horsmans. 2000. Liver abscesses due to Fusobacterium species. Liver 20**:**267-268. [DOI] [PubMed] [Google Scholar]
- 4.Bekeredjian, R., S. Chen, P. A. Frenkel, P. A. Grayburn, and R. V. Shohet. 2003. Ultrasound-targeted microbubble destruction can repeatedly direct highly specific plasmid expression to the heart. Circulation 108**:**1022-1026. [DOI] [PubMed] [Google Scholar]
- 5.Bolstad, A. I., B. T. Hogh, and H. B. Jensen. 1995. Molecular characterization of a 40-kDa outer membrane protein, FomA, of Fusobacterium periodonticum and comparison with Fusobacterium nucleatum. Oral Microbiol. Immunol. 10**:**257-264. [DOI] [PubMed] [Google Scholar]
- 6.Botha, S. J., R. Senekal, P. L. Steyn, and W. J. Coetzee. 1993. Anaerobic bacteria in orofacial abscesses. J Dent. Assoc. S. Afr. 48**:**445-449. [PubMed] [Google Scholar]
- 7.Bradshaw, D. J., P. D. Marsh, G. K. Watson, and C. Allison. 1998. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66**:**4729-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook, I. 1990. Bacteremia due to anaerobic bacteria in newborns. J. Perinatol. 10**:**351-356. [PubMed] [Google Scholar]
- 9.Brook, I., and E. H. Frazier. 2000. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J. Med. Microbiol. 49**:**827-830. [DOI] [PubMed] [Google Scholar]
- 10.Bultink, I. E., J. W. Dorigo-Zetsma, M. G. Koopman, and E. J. Kuijper. 1999. Fusobacterium nucleatum septicemia and portal vein thrombosis. Clin. Infect. Dis. 28**:**1325-1326. [DOI] [PubMed] [Google Scholar]
- 11.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172**:**3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaim, W., and M. Mazor. 1992. Intraamniotic infection with fusobacteria. Arch. Gynecol. Obstet. 251**:**1-7. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhry, R., B. Dhawan, B. V. Laxmi, and V. S. Mehta. 1998. The microbial spectrum of brain abscess with special reference to anaerobic bacteria. Br. J. Neurosurg. 12**:**127-130. [DOI] [PubMed] [Google Scholar]
- 14.Christiansen, J. P., B. A. French, A. L. Klibanov, S. Kaul, and J. R. Lindner. 2003. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med. Biol. 29**:**1759-1767. [DOI] [PubMed] [Google Scholar]
- 15.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66**:**1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debelian, G. J., I. Olsen, and L. Tronstad. 1998. Anaerobic bacteremia and fungemia in patients undergoing endodontic therapy: an overview. Ann. Periodontol. 3**:**281-287. [DOI] [PubMed] [Google Scholar]
- 17.Deng, C. X., F. Sieling, H. Pan, and J. Cui. 2004. Ultrasound-induced cell membrane porosity. Ultrasound Med. Biol. 30**:**519-526. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63**:**1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18**:**289-296. [DOI] [PubMed] [Google Scholar]
- 20.George, K. S., and W. A. Falkler, Jr. 1992**. Coaggregation studies of the Eubacterium species.** Oral Microbiol. Immunol. 7**:**285-290. [DOI] [PubMed] [Google Scholar]
- 21.Gergen, J. P., R. H. Stern, and P. C. Wensink. 1979. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 7**:**2115-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimaudo, N. J., and W. E. Nesbitt. 1997. Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol. Immunol. 12**:**168-173. [DOI] [PubMed] [Google Scholar]
- 23.Guo, M., Y. W. Han, A. Sharma, and E. De Nardin. 2000. Identification and characterization of human immunoglobulin G Fc receptors of Fusobacterium nucleatum. Oral Microbiol. Immunol. 15**:**119-123. [DOI] [PubMed] [Google Scholar]
- 24.Haake, S. K., S. C. Yoder, G. Attarian, and K. Podkaminer. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182**:**1176-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, Y. W., R. W. Redline, M. Li, L. Yin, G. B. Hill, and T. S. McCormick. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72**:**2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, Y. W., W. Shi, G. T. Huang, S. Kinder Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68**:**3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3**:**222-232. [DOI] [PubMed] [Google Scholar]
- 28.Hockensmith, M. L., D. L. Mellman, and E. L. Aronsen. 1999. Fusobacterium nucleatum empyema necessitans. Clin. Infect. Dis. 29**:**1596-1598. [DOI] [PubMed] [Google Scholar]
- 29.Jabra-Rizk, M. A., W. A. Falkler, Jr., W. G. Merz, J. I. Kelley, A. A. Baqui, and T. F. Meiller. 1999. Coaggregation of Candida dubliniensis with Fusobacterium nucleatum. J. Clin. Microbiol. 37**:**1464-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jewett, A., W. R. Hume, H. Le, T. N. Huynh, Y. W. Han, G. Cheng, and W. Shi. 2000. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect. Immun. 68**:**1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184**:**2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapatral, V., N. Ivanova, I. Anderson, G. Reznik, A. Bhattacharyya, W. L. Gardner, N. Mikhailova, A. Lapidus, N. Larsen, M. D'Souza, T. Walunas, R. Haselkorn, R. Overbeek, and N. Kyrpides. 2003. Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Res. 13**:**1180-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman, J., and J. M. DiRienzo. 1989. Isolation of a corncob (coaggregation) receptor polypeptide from Fusobacterium nucleatum. Infect. Immun. 57**:**331-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinder, S. A., and S. C. Holt. 1993. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J. Bacteriol. 175**:**840-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleivdal, H., R. Benz, and H. B. Jensen. 1995. The Fusobacterium nucleatum major outer-membrane protein (FomA) forms trimeric, water-filled channels in lipid bilayer membranes. Eur. J. Biochem. 233**:**310-316. [DOI] [PubMed] [Google Scholar]
- 36.Kolenbrander, P. E., and R. N. Andersen. 1989. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect. Immun. 57**:**3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57**:**3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolenbrander, P. E., K. D. Parrish, R. N. Andersen, and E. P. Greenberg. 1995. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect. Immun. 63**:**4584-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koornstra, J. J., D. Veenendaal, G. A. Bruyn, and H. de Graaf. 1998. Septic arthritis due to Fusobacterium nucleatum. Br. J. Rheumatol. 37**:**1249. [DOI] [PubMed] [Google Scholar]
- 40.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68**:**2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lark, R. L., S. A. McNeil, K. VanderHyde, Z. Noorani, J. Uberti, and C. Chenoweth. 2001. Risk factors for anaerobic bloodstream infections in bone marrow transplant recipients. Clin. Infect. Dis. 33**:**338-343. [DOI] [PubMed] [Google Scholar]
- 42.Lawson, P. A., S. E. Gharbia, H. N. Shah, D. R. Clark, and M. D. Collins. 1991. Intrageneric relationships of members of the genus Fusobacterium as determined by reverse transcriptase sequencing of small-subunit rRNA. Int. J. Syst. Bacteriol. 41**:**347-354. [DOI] [PubMed] [Google Scholar]
- 43.Liu, A., B. McBride, G. Vovis, and M. Smith. 1979. Site specific endonuclease from Fusobacterium nucleatum. Nucleic Acids Res. 6**:**1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangan, D. F., M. J. Novak, S. A. Vora, J. Mourad, and P. S. Kriger. 1989. Lectinlike interactions of Fusobacterium nucleatum with human neutrophils. Infect. Immun. 57**:**3601-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martius, J., and D. A. Eschenbach. 1990. The role of bacterial vaginosis as a cause of amniotic fluid infection, chorioamnionitis and prematurity—a review. Arch. Gynecol. Obstet. 247**:**1-13. [DOI] [PubMed] [Google Scholar]
- 46.Metzger, Z., L. G. Featherstone, W. W. Ambrose, M. Trope, and R. R. Arnold. 2001. Kinetics of coaggregation of Porphyromonas gingivalis with Fusobacterium nucleatum using an automated microtiter plate assay. Oral Microbiol. Immunol. 16**:**163-169. [DOI] [PubMed] [Google Scholar]
- 47.Miller, D. L., S. V. Pislaru, and J. E. Greenleaf. 2002. Sonoporation: mechanical DNA delivery by ultrasonic cavitation. Somat. Cell Mol. Genet. 27**:**115-134. [DOI] [PubMed] [Google Scholar]
- 48.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontology 2000 5**:**66-77. [DOI] [PubMed] [Google Scholar]
- 49.Murray, P. A., D. G. Kern, and J. R. Winkler. 1988. Identification of a galactose-binding lectin on Fusobacterium nucleatum FN-2. Infect. Immun. 56**:**1314-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozaki, M., Y. Miyake, M. Shirakawa, T. Takemoto, H. Okamoto, and H. Suginaka. 1990. Binding specificity of Fusobacterium nucleatum to human erythrocytes, polymorphonuclear leukocytes, fibroblasts, and HeLa cells. J. Periodontal Res. 25**:**129-134. [DOI] [PubMed] [Google Scholar]
- 51.Potts, T. V., L. V. Holdeman, and J. Slots. 1983. Relationships among the oral fusobacteria assessed by DNA-DNA hybridization. J. Dent. Res. 62**:**702-705. [DOI] [PubMed] [Google Scholar]
- 52.Roberts, G. L. 2000. Fusobacterial infections: an underestimated threat. Br. J. Biomed. Sci. 57**:**156-162. [PubMed] [Google Scholar]
- 53.Rosen, G., I. Nisimov, M. Helcer, and M. N. Sela. 2003. Actinobacillus actinomycetemcomitans serotype b lipopolysaccharide mediates coaggregation with Fusobacterium nucleatum. Infect. Immun. 71**:**3652-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaniztki, B., N. Ganeshkumar, and E. I. Weiss. 1998. Characterization of a novel N-acetylneuraminic acid-specific Fusobacterium nucleatum PK1594 adhesin. Oral Microbiol. Immunol. 13**:**47-50. [DOI] [PubMed] [Google Scholar]
- 55.Shaniztki, B., D. Hurwitz, N. Smorodinsky, N. Ganeshkumar, and E. I. Weiss. 1997. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect. Immun. 65**:**5231-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shenker, B. J., and S. Datar. 1995. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect. Immun. 63**:**4830-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shohet, R. V., S. Chen, Y. T. Zhou, Z. Wang, R. S. Meidell, R. H. Unger, and P. A. Grayburn. 2000. Echocardiographic destruction of albumin microbubbles directs gene delivery to the myocardium. Circulation 101**:**2554-2556. [DOI] [PubMed] [Google Scholar]
- 58.Takemoto, T., T. Hino, M. Yoshida, K. Nakanishi, M. Shirakawa, and H. Okamoto. 1995. Characteristics of multimodal co-aggregation between Fusobacterium nucleatum and streptococci. J. Periodontal Res. 30**:**252-257. [DOI] [PubMed] [Google Scholar]
- 59.Tuttle, R. S., and D. F. Mangan. 1990. Interaction of Fusobacterium nucleatum 191 with human peripheral blood lymphocytes. J. Periodontal Res. 25**:**364-371. [DOI] [PubMed] [Google Scholar]
- 60.Tuttle, R. S., N. A. Strubel, J. Mourad, and D. F. Mangan. 1992. A non-lectin-like mechanism by which Fusobacterium nucleatum 10953 adheres to and activates human lymphocytes. Oral Microbiol. Immunol. 7**:**78-83. [DOI] [PubMed] [Google Scholar]
- 61.Weiss, E. I., B. Shaniztki, M. Dotan, N. Ganeshkumar, P. E. Kolenbrander, and Z. Metzger. 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol. Immunol. 15**:**371-377. [DOI] [PubMed] [Google Scholar]
- 62.Winkler, J. R., S. R. John, R. H. Kramer, C. I. Hoover, and P. A. Murray. 1987. Attachment of oral bacteria to a basement-membrane-like matrix and to purified matrix proteins. Infect. Immun. 55**:**2721-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie, H., R. J. Gibbons, and D. I. Hay. 1991. Adhesive properties of strains of Fusobacterium nucleatum of the subspecies nucleatum, vincentii and polymorphum. Oral Microbiol. Immunol. 6**:**257-263. [DOI] [PubMed] [Google Scholar]
- 64.Zarnitsyn, V. G., and M. R. Prausnitz. 2004. Physical parameters influencing optimization of ultrasound-mediated DNA transfection. Ultrasound Med. Biol. 30**:**527-538. [DOI] [PubMed] [Google Scholar]