Regulation of α-Synuclein Expression by Poly (ADP Ribose) Polymerase-1 (PARP-1) Binding to the NACP-Rep1 Polymorphic Site Upstream of the SNCA Gene (original) (raw)
Abstract
Alleles at NACP-Rep1, the polymorphic microsatellite repeat located ∼10 kb upstream of the α-synuclein gene (SNCA), are associated, in some reports, with differing risks of sporadic Parkinson disease (PD). We showed previously that NACP-Rep1 acts as a negative modulator of SNCA transcription, with an effect that varied threefold among different NACP-Rep1 alleles. Given that duplications and triplications of SNCA have been implicated in familial Parkinson disease (PD), even a 1.5–2-fold increase in α-synuclein expression may, over many decades, contribute to PD. Thus, the association of different NACP-Rep1 alleles with PD may be a consequence of polymorphic differences in transcriptional regulation of SNCA. Here we aimed to identify the factor(s) that bind to NACP-Rep1 and potentially contribute to SNCA transcriptional modulation, by pulling down proteins that bind to NACP-Rep1 and identifying them by mass spectrometry. One of these proteins was poly-(ADP-ribose) transferase/polymerase-1 (PARP-1), a DNA-binding protein and transcriptional regulator. Electrophoresis mobility shift and chromatin immunoprecipitation assays showed specific binding of PARP-1 to NACP-Rep1. Inhibition of PARP-1’s catalytic domain increased the endogenous SNCA mRNA levels in cultured SH-SY5Y cells. Furthermore, PARP-1 binding to NACP-Rep1 specifically reduced the transcriptional activity of the SNCA promoter/enhancer in luciferase reporter assays. This down-regulation effect of PARP-1 depended on NACP-Rep1 being present in the construct and was abrogated by inhibiting PARP-1’s catalytic activity with 3-aminobenzamide. The association of different NACP-Rep1 alleles with PD may be mediated, in part, by the effect of PARP-1, as well as other factors, on SNCA expression.
Introduction
Parkinson disease (PD) (MIM 168600) is the second most common neurodegenerative disease in humans; its etiology is largely unknown. SNCA (MIM 163890), the gene encoding the presynaptic protein α-synuclein, was the first gene in which mutations were found to cause PD. Three missense mutations in the SNCA gene have been shown to cause an early-onset, autosomal dominant PD with, in some families, Lewy body pathology at autopsy (Polymeropoulos et al. 1997; Kruger et al. 1998; Zarranz et al. 2004). Recently, Singleton et al. (2003) reported a genomic triplication of the region containing α-synuclein in affected individuals of another kindred, in which PD presented as an autosomal dominant disease. This triplication resulted in four fully functional copies of SNCA and twofold overexpression of α-synuclein mRNA and protein (Farrer et al. 2004; Miller et al. 2004). Duplications of SNCA in PD have also been found, with even later onset (Chartier-Harlin et al. 2004; Ibanez et al. 2004). Moreover, the toxicity of overexpressed wild-type SNCA has been anticipated in a Drosophila melanogaster model (Feany and Bender 2000) and rodent models (Maries et al. 2003). These observations emphasize the importance of α-synuclein levels in PD by showing that elevated levels of wild-type α-synuclein are sufficient to cause neuronal dysfunction and disease. Although efforts to identify mutations or copy-number changes in the SNCA gene in the more common sporadic forms of PD have failed (Golbe 1999; Johnson et al. 2004), the importance of α-synuclein in sporadic PD was re-enforced by the identification of the protein as a major component of Lewy bodies and Lewy neurites, the pathological hallmark of PD, in patients with PD who had no mutation in the SNCA gene (Spillantini et al. 1997). These findings suggest that even subtle variation in expression of SNCA might play a role in the pathogenesis of the disease in those patients who do not express a mutated protein or have an increase in gene copy number.
NACP-Rep1 is a polymorphic complex repeat site located ∼10 kb upstream of the translational start of SNCA (Xia et al. 1996; Touchman et al. 2001). The basic sequence of NACP-Rep1 is (TC)x(T)2(TC)y(TA)z(CA)w. The repeat is of interest because several association studies have shown that certain alleles of NACP-Rep1 may confer an increased risk of sporadic PD in white (Kruger et al. 1999; Tan et al. 2000; Farrer et al. 2001) and Asian (Mizuta et al. 2002; Tan et al. 2003; Tan et al. 2004) populations, although other studies have failed to replicate these findings (Parsian et al. 1998; Izumi et al. 2001; Khan et al. 2001; Spadafora et al. 2003; Tan et al. 2003). Previously, we attempted to determine if the genetic-association studies correlated with biological function. Using the luciferase reporter assay system for promoter activity, we examined the 10-kb region located upstream of the SNCA gene, including the NACP-Rep1 repeat, in SH-SY5Y cells (Chiba-Falek and Nussbaum 2001). We showed that two segments, a few hundred base pairs in length, flanking the NACP-Rep1 acted in an additive manner to increase promoter function, whereas the NACP-Rep1 itself was shown to have a negative effect on expression (Chiba-Falek and Nussbaum 2001). In addition, we showed that, in SH-SY5Y cells, different alleles had a significant effect on expression levels over a nearly threefold range (Chiba-Falek and Nussbaum 2001; Chiba-Falek et al. 2003). The mechanism by which the polymorphic NACP-Rep1 element affects transcriptional activity of the SNCA promoter remains unknown.
We set out to find proteins that interact with NACP-Rep1 and thereby mediate the effect of the repeat on the activity of the SNCA upstream promoter/enhancer. Here we demonstrate that the DNA-binding protein PARP-1 interacts with the polymorphic NACP-Rep1 element and contributes to the transcriptional regulation of SNCA expression.
Material and Methods
SH-SY5Y Cell Culture and Nuclear Protein Extract
SH-SY5Y, a human neuroblastoma cell line (American Type Culture Collection), was grown in DMEM/F-12 1:1 medium, supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. Cells were maintained at 37°C in a humidified 5% CO2 incubator. Nuclear extracts from this cell line were prepared as described elsewhere (Dignam et al. 1983).
Oligonucleotides and Electrophoretic Mobility Shift Assays
NACP-Rep1 oligonucleotide and a control oligonucleotide (SNCA exon 4) and their matched reverse oligonucleotides were synthesized chemically (Operon), as follows: NACP-oligo, TCTCTCTCTCTCTCTCTCTCTTTCTCTCTCTCTCTCTCTCTCTATATATATATATATACACACACACACACACACACACA; NACP-oligo-r, TGTGTGTGTGTGTGTGTGTGTGTATATATATATATATAGAGAGAGAGAGAGAGAGAGAAAGAGAGAGAGAGAGAGAGAGA; control-oligo, GAGAAGACCAAAGAGCAAGTGACAAATGTTGGAGGAGCAGTGGTGACGGGTGTGACAGCAGTAGCCCAGAAGACAGTGGA; control-oligo-r, TCCACTGTCTTCTGGGCTACTGCTGTCACACCCGTCACCACTGCTCCTCCAACATTTGTCACTTGCTCTTTGGTCTTCTC.
Each single-strand (ss) oligonucleotide (NACP and control) was mixed with the matched reverse ss-oligonucleotide in a molar ratio of 1:1 in TEN-buffer, was heated to 95°C for 10 min, and was slowly cooled (1°C per min) to 15°C, to allow a perfect annealing. NACP-Rep1 and control double stranded (ds) oligonucleotides were 3′ end-labeled with digoxigenin-11-ddUTP (DIG). DIG-labeled NACP-Rep1 and control ds-oligonucleotides were used in electrophoretic mobility shift assays (EMSAs) performed with the DIG Gel Shift kit (Roche) in accordance with the manufacturer’s instructions. In brief, DIG-labeled NACP-Rep1 and control ds-oligonucleotides were incubated with SH-SY5Y nuclear extract or a purified PARP-1 protein (_s_-recombinant human wild-type PARP-1, expressed as 6-histidine–tagged fusion protein in E. coli and purified to >95% homogeneity [Smulson et al. 1998], a kind gift from A. Spoonde) at 25°C for 15 min in the presence of poly[d(I-C)]. For the supershift analysis, 4 μl of monoclonal anti-PARP-1 antibody (PharMingen) was added to the SH-SY5Y nuclear extract or the purified PARP-1 and incubated on ice for 30 min prior to the addition of the DIG-labeled ds-oligonucleotides. Following the binding reaction, the mixtures were subjected to electrophoresis on Novex Retardation Gel (6% polyacrylamide) (Invitrogen) and electroblotted to a positively charged nylon membrane (Roche). The blots were visualized by an enzyme immunoassay using anti-digoxigenin-AP, Fab fragments, and the chemiluminescent substrate CSPD (Tropix) and subsequently were exposed to x-ray films.
DNA-Affinity Purification of NACP-Rep1-Binding Proteins
The protein(s) binding to the NACP-Rep1 DNA element were purified using a DNA-Binding Protein Purification Kit (Roche) in accordance with the manufacturer’s instructions. In brief, NACP-Rep1 ds-oligonucleotide was ligated to a tethered 16-mer ds-oligonucleotide which was 3′ end labeled with biotin at one site and coupled to streptavidin magnetic particles. SH-SY5Y nuclear extract (100 μg) was applied to the DNA–magnetic particle complex, and the binding reaction was performed at 25°C for 30 min in protein-binding buffer (20 mM Hepes, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween 20, 30 mM KCl). Nonspecific DNA binding was inhibited by the addition of poly[d(CI)] and [d(AT)]. Subsequently, the bound protein(s) were separated from the supernatant by use of a magnetic separator and three washing steps with the protein-binding buffer. Next, the DNA-binding–protein(s) mixture was eluted from the magnetic particles by use of a high-ionic-strength buffer (20 mM Hepes, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween 20, and 2 M KCl), followed by dialysis to remove the high salt concentration. The purified proteins were run on 4%–20% Novex SDS PAGE gel (Invitrogen) and the gel was Commassie stained using GelCode Blue staining reagent (PIERCE).
In Situ Digestion of Proteins in SDS-PAGE Gels
Commassie-stained gel bands were excised with a clean razor blade and subjected to in situ enzymatic digestion in accordance with a modified version of a standard protocol (Stone and Williams 1993). In brief, gel bands were placed in 0.5 ml Eppendorf tubes and destained by the addition of 0.5 ml 50% MeOH:100 mM NH4HCO3 (1:1) for 1 h. The supernatant was decanted and discarded. Gel bands were then washed with 0.5 ml 100 mM NH4HCO3 (1:1) for 1 h, followed by 0.5 ml 50% CH3 CN:100 mM NH4HCO3 (1:1) for 1 h. Gel bands were cut into two or three pieces and then were washed with 150 μL of neat CH3CN for 15 min to cause the gel pieces to shrink. The gel pieces were rehydrated by the addition of 10 μL 100 mM NH4HCO3 containing 10 ng/μL sequencing grade trypsin. Digestion was allowed to proceed overnight at room temperature. Peptides were extracted from the gel pieces by the addition of an equal volume of 50% CH3CN, 1% HCO2H and sonication for 30 min. The extraction procedure was repeated using 80% CH3CN, 1% HCO2H. The pooled supernatants were dried in a vacuum centrifuge and resuspended in 10 μL 5% CH3CN, 1% HCO2H.
LC-MS/MS
The automated HPLC system used was composed of LC-VP series liquid chromatograph components, including three LC-10AD_VP_ solvent delivery pumps, a DGU-14A online degasser, an SIL-10AD_VP_ autosampler, an SCL-10A_VP_ system controller, and an FCV-12AH two-position six-port switching valve (Shimadzu Corporation), with a 0.5 × 2 mm RP trapping column (Peptide CapTrap; Michrom BioResource) mounted on the FCV-12AH switching valve. Sample solutions were injected using the SIL-10AD autosampler. Mobile phase A for sample loading was pumped at 50 μL/min, and, after a 3-min loading and trapping period, the position of the switching valve was changed to an analysis position to elute the trapped peptides onto a 0.3 × 100 mm C18 RP column (BetaBasic-18, 0.3 × 100 mm, 5 μm particle size [Thermo Hypersil-Keystone]), operating at a 10 μL/min flow rate. RP mobile phase A and sample loading buffer was CH3CN:H2O:HCO2H = 5:95:0.1 (v:v:v) and RP mobile phase B, CH3CN:H2O:HCO2H = 80:20:0.1. The chromatograph was developed using a linear gradient from 5% to 65% B in 40 min. The entire chromatographic effluent was introduced into the ESI ion source of an LCQ ion trap mass spectrometer (ThermoFinnigan).
The LCQ was operated in positive ion mode with dynamic exclusion set to repeat count 2, repeat duration 0.5 min, exclusion duration 1 min, exclusion mass width 3 atomic mass units (amu). Spectra were acquired in a data-dependent manner, with the top three most intense ions in the MS scan selected for MS/MS—that is, iterative collection of a survey scan (a full-scan mass spectrum from 400 to 1800 m/z [mass/charge ratio]), followed by MS/MS of the three most abundant ions detected in the previous survey scan. Raw data were submitted to the Mascot cluster maintained by the Center for Information Technology of the National Institutes of Health.
Chromatin Immunoprecipitation (ChIP) and PCR Analysis
For each ChIP reaction, 1 × 107 SH-SY5Y cells were fixed by adding formaldehyde to a final concentration of 1% directly to growth media at 37°C for 7 min. The crosslinking reaction was quenched with glycine at a final concentration of 0.125 M. Cells were washed with ice-cold PBS, were lysed in lysis buffer 1 (0.25% Triton X-100, 0.5 mM EGTA, 1 mM EDTA, 10 mM Tris-HCl pH 8 and an EDTA-free tablet of protease inhibitor cocktail [Roche]) for 10 min, rotating at 4°C. Nuclei were pelleted at low speed; were lysed in lysis buffer 2 (200 mM NaCl, 0.5 mM EGTA, 1 mM EDTA, 10 mM Tris-HCl pH 8 and an EDTA-free tablet of protease inhibitor cocktail [Roche]) for 10 min, rotating at 4°C; and were diluted to 5 ml in dilution buffer (1 mM EDTA, 10 mM Tris-HCl pH 8, 0.5 mM EGTA, and an EDTA-free tablet of protease inhibitor cocktail [Roche]). Chromatin was sonicated to ∼500-bp fragments and was centrifuged at 12,000 rpm for 10 min at 4°C to remove debris. The chromatin sample was removed (total ∼4 ml in dilution buffer) and was split into aliquots. Nonspecific background was precleared with 20 μl of protein A agarose beads (Amersham) for 8 h at 4°C. The chromatin supernatant was aliquoted and either 5 μl of rabbit polyclonal anti-PARP-1 antibody (Roche), rabbit polyclonal IgG (nonspecific control), or no antibody (the “No Antibody” sample) were added to each aliquot and incubated overnight at 4°C with rotation. The protein A agarose beads were blocked with 0.5 mg/ml acetylated BSA (Promega) and 23 μg/ml yeast DNA for >1 h at 4°C. Before immunocomplexes were collected with the blocked beads, a portion (10%) from the “No Antibody” sample was set aside to measure input DNA. The immunocomplexes were precipitated with preblocked protein A agarose beads for 3 h at 4°C. Beads were washed four times each with high-salt RIPA buffer (0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 10 mM Tris pH 8, 500 mM NaCl, and an EDTA-free tablet of protease inhibitor cocktail [Roche]), once each with LiCl wash buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris pH 8.0), and then twice with TE buffer. Pellets were resuspended in 150 μl prewarmed elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0) and were rotated at 65°C for 15 min. Samples were centrifuged and eluates removed. Input DNA (10% of total sample previously set aside) was diluted to 150 μl in elution buffer, and all samples were diluted to a final volume of 300 μl in double deionized water. Cross-links were reversed in all samples, and proteins were removed with the addition of 500 μg/ml proteinase K and incubation at 37°C for 8 h, followed by overnight at 65°C. Samples were extracted with 1:1 phenol/chloroform/isoamyl-alcohol and were precipitated in cold ethanol with 33 μl of 3 M NaAcetate and 20 μg glycogen at −20°C for 2 h. Samples were washed with 70% ethanol and resuspended in ultrapure water. DNA enrichment was semiquantified using PCR. The following primers were used to detect the enrichment of the NACP-Rep1 DNA sequence: Rep1 (CCTGGCATATTTGATTGCAA) and Rep2 (GACTGGCCCAAGATTAACCA). For the detection of the SNCA 3′ UTR, the following primers were used: SNCA_3UTRf (TCTGAGGAAGGGTATCAAGACTACGAA) and SNCA_3UTRr (GCACCGAAATGCTGAGTGG). For the INPP5B 3′ UTR, the following primers were used: INPP5B F (TTGATATGACAGAGAAGAAGAAGG) and INPP5B R (ATGTCAGTTTTATTATGGTGGATT).
3-Aminobenzamide (3AB) Treatment of SH-SY5Y and RNA Extraction
SH-SY5Y cells were plated on six-well dishes coated with poly-L-lysine the day before treatment. DMEM/F-12 1:1 medium was prepared with 1% DMSO and either 0, 3, 10, 15, 30, or 60 mM 3-aminobenzamide (Sigma). Medium containing different concentrations of 3-AB was added in duplicate to six pairs of cell cultures and was incubated for 48 h. Then, the cells were washed with phosphate buffered saline, and total RNA was extracted using TRIzol reagent (Invitrogen).
cDNA Synthesis and Real-Time PCR
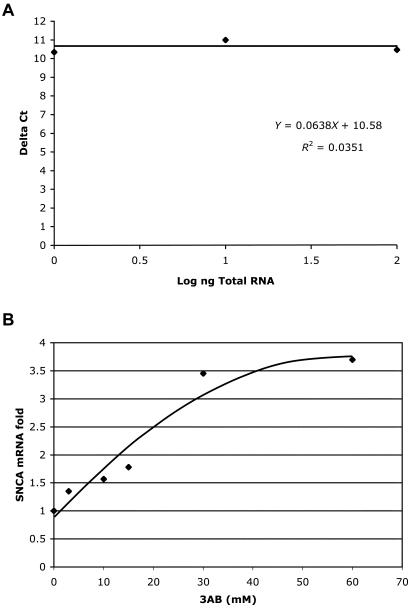
RNA concentration was determined spectrophotometrically at 260 mm and cDNA was synthesized using MultiScribe RT enzyme (Applied Biosystems) and the following conditions: 10 min at 25°C and 120 min at 37°C. Triplicates of each sample were assayed in a relative quantitative PCR for analysis of the level of SNCA message, with GAPDH message as an internal reference, by use of the ABI 7000. Each cDNA (10 ng) was amplified in quadruplicate using TaqMan Universal PCR master mix reagent (Applied Biosystems) under the following conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The target SNCA cDNA was amplified using ABI MGB probe and primer set assay ID Hs00240907_m1, normalized to a GAPDH RNA control (ABI MGB probe and primer set assay ID Hs00999905_m1) (Applied Biosystems). Data were analyzed with a threshold set in the linear range of amplification. The cycle number at which any particular sample crossed that threshold (Ct) was then used to determine fold difference. Fold difference was calculated using the 2−ΔΔCt method; Δ_Ct_=[Ct(SNCA_-Ct(GAPD)]; ΔΔ_Ct_=[Δ_Ct(sample 3_ABtreated_)]-[Δ_Ct_(referenceno 3_AB_)]. Standards curves for each assay using SH-SY5Y RNA samples indicated that the assays were valid while the slope of the relative efficiency plot for SNCA and GAPDH validated the ΔΔ method (fig. 4_A_).
Figure 4.
Real-time analysis of SNCA mRNA levels in SH-SY5Y cell after 3-AB treatment. A, Validation curve of the ΔΔ real-time assay for relative quantization of SNCA mRNA relatively to GAPDH mRNA. A relative efficiency plot of SNCA and GAPDH was formed by plotting the log input amount (ng of total RNA) versus the ΔCt [Ct(SNCA)−Ct(GAPDH)]. The slop is 0.0638<0.1, which indicates the validation of the ΔΔCt calculation in the range of 1–100 ng RNA. B, Cells were treated with 3AB in different concentrations (0, 3, 10, 15, 30, and 60 mM), and the relative levels of SNCA mRNA to GAPDH mRNA were assessed by real-time PCR and were analyzed by the ΔΔ method. The 3-AB concentration (mM) is indicated on the _X_-axis, and the fold increase of SNCA mRNA is indicated on the _Y_-axis. Each data point represents the average of three repeats.
Luciferase Reporter Constructs and Transfection
The construction of the pASP plasmid—carrying the 10.7-kb upstream SNCA gene, including the NACP-Rep1 site—and of the pASP-ΔNACP, in which the NACP-Rep1 site was deleted, has been described elsewhere (Chiba-Falek and Nussbaum 2001). 1 × 106 SH-SY5Y cells were plated onto each well of a six-well dish coated with poly-L-Lysine the day before transfection. For each experiment involving cotransfection into SH-SY5Y cells, 2 μg of pASP or pASP-ΔNACP or 660 ng of pGL 3-Basic were mixed with 10 ng of the reference plasmid, pRL-SV40 (harboring the SV40 early enhancer/promoter region upstream of Renilla luciferase), and cotransfected using the FuGENE 6 Transfection Reagent (Roche), in accordance with the manufacturer’s instructions. Seven hours after transfection, the medium was replaced with DMEM/F-12, containing either 30 mM 3AB/1% DMSO or 1% DMSO alone. Cells were incubated at 37°C for 48 h before harvesting.
For each experiment, three wells were independently transfected, in parallel with three individually prepared aliquots of transfection reaction, and the results from all three replicates were averaged. Each triplicate experiment was repeated twice on separate days.
Luciferase Assay
SH-SY5Y cells were washed and lysed in 200 μl of Passive Lysis Buffer (Promega). Firefly luciferase and Renilla luciferase activities were measured with 10 μl and 20 μl of SH-SY5Y cell lysate, by use of the Dual-Luciferase Reporter assay system (Promega) in a luminometer (EG&G Wallac). “Relative activity” was defined as the ratio of firefly luciferase activity to Renilla luciferase activity and was calculated by dividing luminescence intensity obtained in the assay for firefly luciferase by that obtained for Renilla luciferase. “Fold expression” is defined as the ratio of the relative activity seen with each 3AB treated or untreated test plasmids (pASP, pASP-ΔNACP) to the basal relative activity, and it was calculated by dividing the average value of relative activity of each construct to the relative activity of the pGL 3-Basic plasmid without any insert.
Results
Evidence of Protein Binding to the NACP-Rep1 Element by Electrophoretic Mobility Shift Assay (EMSA)
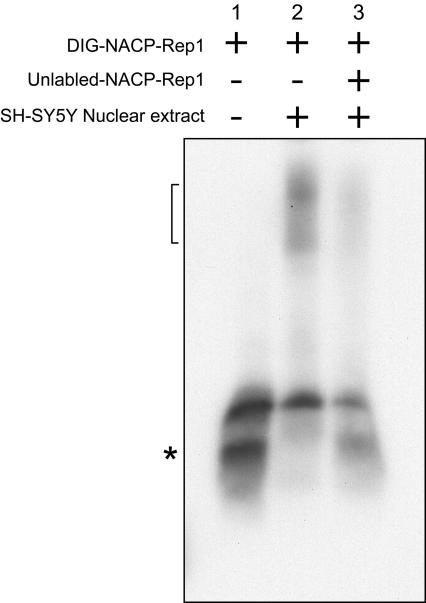
Nuclear extract was prepared from the neuroblastoma cell line, SH-SY5Y. When a digitonin-labeled, double-stranded 80-mer corresponding to the NACP-Rep1 repeat was incubated with the nuclear extract, broad bands forming a smear were seen (fig. 1, lane 2) that were shifted upward relative to what we observed in the absence of nuclear extract (fig. 1, lane 1). The upward shift was resistant to the nonspecific competitors, an excess of unlabeled homopolymers poly[d(CI)] and [d(AT)] (fig. 1, lane 2). This result indicated the existence of SH-SY5Y nuclear proteins with binding activity towards the NACP-Rep1 DNA element. After the addition to the reaction of a 125-fold excess, relative to the DIG-NACP-Rep1, of unlabeled NACP-Rep1, the upwardly shifted band was markedly reduced (fig. 1, lane 3). This result suggested that the protein complexes had specificity for the NACP-Rep1 sequence. The appearance of multiple bands in an apparent smear suggests that a heterogeneous collection of complete or partial protein complexes were binding to the NACP-Rep1 element.
Figure 1.
NACP-Rep1 binds protein/s from nuclear SH-SY5Y extracts. EMSAs were performed with DIG-labeled double-stranded 80 bp oligo including the entire NACP-Rep1 sequence. Lane 1 shows the DIG-NACP-Rep1 only. SH-SY5Y nuclear extract was used in the binding assay (lanes 2 and 3). Unlabeled ds-NACP-Rep1 oligo was used as the specific competitor (lane 3). Unlabeled poly[dCI] and poly[d(AT)] were used in all lanes as a nonspecific competitor to avoid nonspecific DNA-binding. The plus (+) sign indicates what is included in each reaction while a minus sign (−) indicates what is excluded from the reaction. The number in the upper panel indicates the lane number. A bracket indicates the proteins-NACP-Rep1 complex. The lower smear, designated by an asterisk (*), indicates the DIG-NACP-Rep1.
Isolation and Identification of Proteins that Binds to NACP-Rep1
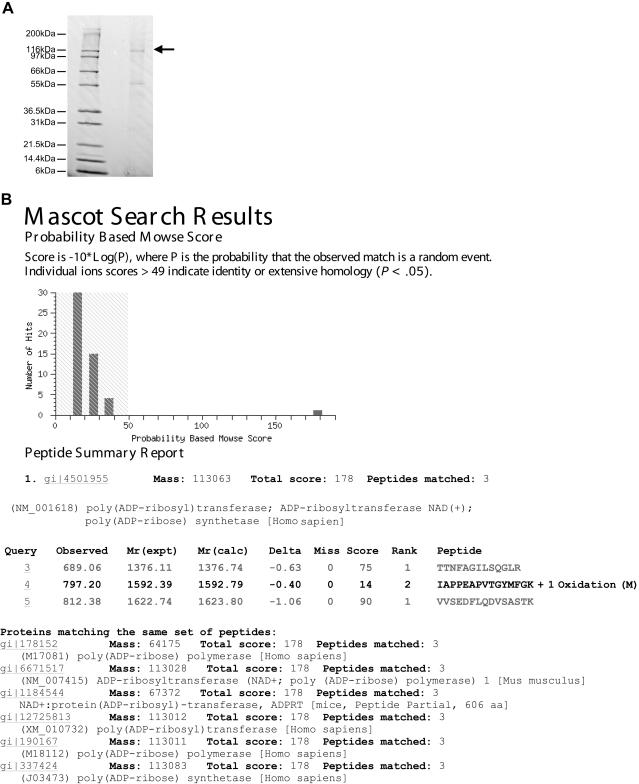
Next, we aimed to identify the NACP-Rep1–binding proteins. NACP-Rep1 ds-oligo was coupled to magnetic particles and used to purify proteins that bind to the NACP-Rep1 sequence from the SH-SY5Y nuclear extract. The purified proteins were subjected to SDS-PAGE and unique bands were visualized by Commassie staining, were cut out of the gel, and were identified by mass spectroscopy (fig. 2_A_). One of the identified proteins, which migrated at ∼120 kDa in the gel, was poly (ADP-ribose) transferase/polymerase-1 (PARP-1) (fig. 2B). Its identification was based on three peptides with the highest score (178) among the other identified proteins (data not shown).
Figure 2.
Identification of PARP-1 as NACP-Rep1 binding protein. A, Commassie stained gel of the NACP-Rep1 binding proteins that were purified from SH-SY5Y nuclear extract. The arrow indicates the 120-kDa protein’s band that was used for mass spectrometry identification. B, Mascot results of the mass spectrometry identification of the 120-kDa band. PARP-1 was identified by three peptides with the highest score value (178).
PARP-1 Binds Specifically to NACP-Rep1 DNA Sequence
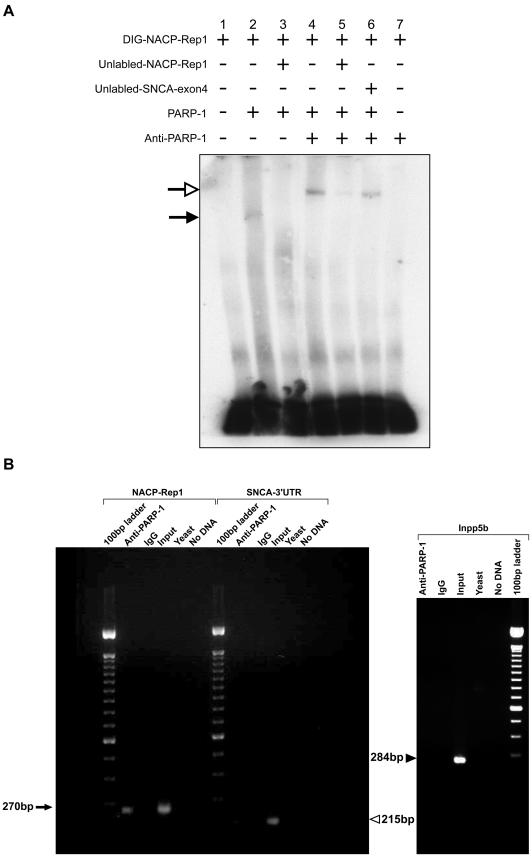
To confirm the binding of PARP-1 to NACP-Rep1, two approaches were taken: EMSA with purified PARP-1 protein (fig. 3A) and chromatin immunoprecipitation (ChIP) (fig. 3B). The in vitro EMSA analysis confirmed both the binding of PARP-1 to NACP-Rep1 and its specificity. In the first lane, the reaction includes the DIG-NACP-Rep1 only. Upon incubation with purified PARP-1, a shifted band appeared (fig. 3_A_, lane 2) that was competed off by unlabeled NACP-Rep1 ds-oligo (fig. 3_A_, lane 3). Incubation with anti-PARP antibody, prior to its addition to the DNA-protein–binding reaction, resulted in the appearance of a supershifted band (fig. 3_A_, lane 4). The supershifted band was competed off by unlabeled NACP-Rep1 oligo (fig. 3_A_, lane 5) but not by a nonspecific DNA fragment (a sequence from exon 4 of SNCA gene) ds-oligo (fig. 3_A_, lane 6). A control reaction of DIG-NACP-Rep1 with the anti-PARP antibody alone led to no shifted bands (fig. 3_A_, lane 7). We would like to note that, to further confirm the specificity of PARP-1 binding to the NACP-Rep1 sequence, we also duplicated the set of EMSA reaction presented above, except for reversing the DIG-labeled oligo and the nonspecific competitor (SNCA exon 4) sequences. Thus, the DIG-labeled ds-oligo, as well as the specific competitor, was the control 80-mer sequence of SNCA exon 4, whereas the nonspecific competitor was the NACP-Rep1 ds-oligo. Upon incubation with purified PARP-1 or PARP-1 preincubated with anti-PARP, neither the shifted nor supershifted bands was detected (data not shown). The above results demonstrate that PARP-1 binds to the NACP-Rep1 sequence in a specific manner under in vitro conditions.
Figure 3.
PARP-1 binds to the NACP-Rep1 sequence. A, EMSAs were performed with DIG-labeled double-stranded 80-bp oligo including the entire NACP-Rep1 sequence. Lane 1 shows the DIG-NACP-Rep1 only. Purified human PARP-1 was used in the binding assay. Anti-PARP-1 antibody was used for the super shift reaction. Unlabeled ds-NACP-Rep1 oligo was used as the specific competitor, and unlabeled ds-SNCA-exon 4 oligo was used as the unspecific competitor. The plus sign (+) indicates what is included in each reaction, whereas a minus sign (−) indicates what is excluded from the reaction. The number in the upper panel is the lane number. Close arrow indicates the PARP-1-NACP-Rep1 complex (shifted band). Open arrow indicates the PARP-1-antibody-NACP-Rep1 complex (super-shift band). B, ChIPs were carried out using anti-PARP-1 antibody and IgG antibody. The input control represents the whole DNA of SH-SY5Y cells. Following the ChIP reactions, a PCR analysis was performed using three different sets of primers. Amplification around the NACP-Rep1 region, control amplifications of the SNCA 3′ UTR and the Inpp5b gene. The yeast DNA and no DNA control reactions for background failed to amplify. The arrow indicates the PCR product of the NACP-Rep1 region (270 bp). The unblackened arrowhead indicates the PCR product of the SNCA 3′ UTR (215 bp), and the blackened arrowhead indicates the PCR product of Inpp5b (284 bp).
ChIP allowed a further in vivo confirmation of the specific binding of PARP-1 to NACP-Rep1 (fig. 3_B_). In the ChIP assay, the SH-SY5Y cells were fixed, thus crosslinking the DNA-binding proteins to the DNA. Subsequent chromatin extraction and immunoprecipitation (IP) with PARP-1 antibody, followed by reversal of the cross linking and DNA purification, gave a DNA fraction enriched for DNA sequences that bind PARP-1. We assayed this fraction for enrichment for the NACP-Rep1 DNA sequence by PCR, with primers flanking the NACP-Rep1 sequence. PCR amplification with NACP-Rep1 specific primers revealed an amplified band when using as template the DNA sample of the ChIP reaction in which anti-PARP antibody was used for IP (fig. 3_B_). PCR of the region flanking the SNCA 3′ UTR was performed on this same ChIP fraction (fig. 3_B_) and resulted in no amplification (fig. 3_B_). Primers flanking the 3′ UTR of a completely unrelated gene, INPP5B, were also used as a comparative control and showed no PCR product with this DNA sample (fig. 3_B_). There was no amplification of NACP-Rep1 in control ChIP reactions in which no antibody was used (data not shown), or an anti-IgG was used for IP (fig. 3_B_), or Saccharomyces cerevisiae DNA (fig. 3_B_) was used as the DNA blocking agent. These control samples also failed to show amplification of SNCA 3′ UTR and Inpp5b, as expected (fig. 3_B_). As a positive control for the PCR itself, we used the supernatant left after a mock ChIP reaction in which no antibody was used, which we expected would represent the total DNA of the SH-SY5Y cells. This sample of input DNA as template led to amplification of NACP-Rep1 region and validated the PCR assay of the NACP-Rep1 region (fig. 3_B_). These results indicate that the NACP-Rep1 DNA region was specifically enriched in the DNA sample derived from the PARP-antibody ChIP reaction, implying that PARP-1 binds to NACP-Rep1 element in vivo and that the binding of PARP-1 to NACP-Rep1 sequence is specific.
Inhibition of the Enzymatic Activity of PARP-1 Elevates SNCA Levels
3-Aminobenzamide (3-AB) is known for its ability to inhibit the PARP-1 catalytic domain. 3-AB (3–60 mM) was used to inhibit PARP-1 activity in SH-SY5Y cells, to study the effect on endogenous steady-state SNCA mRNA levels. Cells were treated with six different concentrations of 3-AB (0, 3, 10, 15, 30, and 60 mM) for 48 h, after which RNA was extracted and levels of endogenous SNCA mRNA were analyzed by real-time PCR (see assay validation in fig. 4_A_). In general, PARP-1 inhibition resulted in an increase in the levels of SNCA mRNA in a dose-responsive manner (fig. 4_B_). A slight (1.4-fold) increase in SNCA mRNA was detected on treatment with 3-AB at a concentration as low as 3 mM (fig. 4_B_). Doses of 3-AB from 0 to 30 mM led to a significant increase in the levels of SNCA mRNA, up to 3.5-fold, which appeared to be linear (fig. 4_B_). However, further increase in 3-AB concentration from 30 to 60 mM produced only a slight additional increase in SNCA mRNA, from 3.5 to 3.7 fold, suggesting the response had reached a plateau (fig. 4_B_). These results indicate that inhibition of the PARP-1 catalytic domain was associated with in an increase in steady-state SNCA mRNA level of ∼3.7 fold over untreated cells.
Inhibition of the Enzymatic Activity of PARP-1 Increases the Activity of SNCA Promoter/Enhancer Region
Our previous studies showed the importance of the region containing the human NACP-Rep1 repeat in the transcriptional regulation of human SNCA gene expression (Chiba-Falek and Nussbaum 2001; Touchman et al. 2001). To investigate whether alterations in transcriptional activity of the SNCA gene contributed to the increase in steady-state SNCA mRNA levels induced by 3-AB treatment, we examined the effect of 3-AB on the activity of luciferase driven by the 10.7-kb region upstream of SNCA translational start (Touchman et al. 2001) using constructs that either contained the NACP-Rep1 sequence (pASP) or were deleted for the sequence (pASP-ΔNACP) (Chiba-Falek and Nussbaum 2001). These constructs were transfected into the neuroblastoma cell line SH-SY5Y and treated with 3-AB or left untreated. Each plasmid was cotransfected with pRL-SV40. Eight hours after transfection, half of the wells were exposed to 30 mM of 3-AB, and half remained untreated for 48 h. Subsequently, cells were harvested, protein extracts were prepared, and the levels of firefly and Renilla luciferase expression were measured. As a control for luciferase basal expression, we also cotransfected the commercial promoter-less plasmid, pGL 3-Basic, with the pRL-SV40 plasmid. For each cotransfection experiment, the relative activity of luciferase to the Renilla enzyme was calculated, to eliminate the effect of transfection efficiency and cell number (see the “Material and Methods” section).
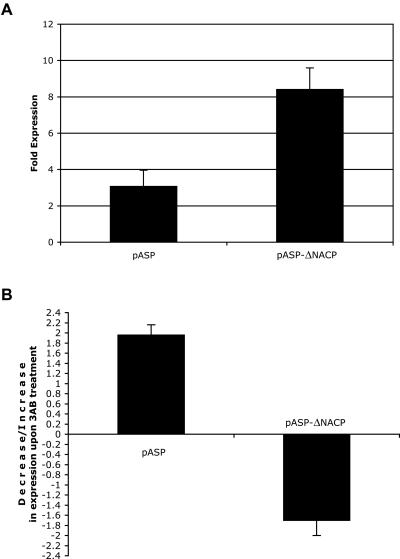
The full-length plasmid, pASP demonstrated a ∼2.7-fold lower luciferase activity in SH-SY5Y as compared with the deleted plasmid, pASP-ΔNACP; 3.1-fold expression compared with 8.4-fold expression, respectively (fig. 5_A_). This result confirmed our previous observations that the NACP-Rep1 DNA sequence acts as a transcription-repressor element (Chiba-Falek and Nussbaum 2001). Upon treatment with 3-AB, the full-length construct, pASP, gave a twofold increase in the expression of the luciferase, relative to the untreated cells (fig. 5_B_). This result indicates that PARP-1 inhibition led to higher promoter activity in SH-SY5Y cells transfected with the full-length construct (pASP). Thus, PARP-1 may be one element of the transcriptional machinery that acts on the NACP-Rep1 element to down regulate SNCA transcription. In contrast, treatment with 3-AB of SH-SY5Y cells transfected with the deleted NACP-Rep1 construct showed a decrease (1.7 fold) in luciferase activity (fig. 5_B_). Thus, PARP-1 inhibition resulted in lower promoter activity for the deleted construct (pASP-ΔNACP). Thus, inhibition of PARP-1 catalytic domain by 3-AB treatment had the opposite effect on luciferase expression driven by pASP and by pASP-ΔNACP, with pASP showing an overall ∼4-fold higher elevated expression, compared with pASP-ΔNACP (fig. 5_B_). These results illustrate that active PARP-1 could have an overall repressing effect on SNCA transcription via its interaction with NACP-Rep1 but other elements may reside within the SNCA promoter/enhancer region, through which PARP-1 acts to increase SNCA transcription, either directly, through DNA binding, or indirectly, through an effect on other transcription factors that can affect SNCA expression.
Figure 5.
Fold expression of luciferase activity derived by full-length and NACP-Rep1–deleted pASP constructs in SH-SY5Y cells before and after 3-AB treatment. A, Cells were transfected with each construct or pGL 3-Basic. The relative activity for each pASP or pGL 3-Basic plasmid was calculated by dividing the luminescence intensity of the firefly luciferase by that of the cotransfected Renilla luciferase in each independent aliquot of cells and then averaging the three relative luciferase activities seen. The fold expression for pASP and pASP-ΔNACP was then determined by dividing the average relative activity of each construct to that of the average obtained with pGL 3-Basic. B, The ratio of fold expression of luciferase activity derived by the full-length pASP and pASP-ΔNACP in SH-SY5Y cells upon 3-AB treatment. Cells were cotransfected with each of the two constructs (full-length and NACP-Rep1 deletion) or pGL 3-Basic and pRL. In each experiment for each construct, three well were untreated and three were treated with 30 mM 3-AB for 48 h. For each construct, two to three independent experiments were performed. The relative activity for each pASP, with and without 3-AB treatment or pGL 3-Basic plasmid, was calculated by dividing the luminescence intensity of the firefly luciferase by that of the cotransfected Renilla luciferase in each independent aliquot of cells and then averaging the three relative luciferase activities seen. The fold expression for each pASP with and without 3-AB treatment was then determined by dividing the average relative activity of each construct to that of the average obtained with pGL 3-Basic. Then, the ratio of the fold expression for each of the 3-AB treated cells transfected with each construct (pASP and pASP-ΔNACP) relative to the untreated cells transfected with the same construct was determined. The average of the ratios of two to three independent experiments performed on separate days was calculated. A bar in the positive scale of the _Y_-axis indicates an increase in luciferase activity upon 3-AB treatment, and a bar in the negative scale of the _Y_-axis indicates a decrease in luciferase activity upon 3-AB treatment.
Discussion
One of the more important outstanding questions for research into the molecular pathogenesis of PD is the role of α-synuclein in sporadic PD. α-Synuclein may be involved in PD through altered regulation of α-synuclein expression, among other possible mechanisms. Although efforts to identify α-synuclein mutations in sporadic PD have failed, there is accumulating evidence in both in vitro systems (Hashimoto et al. 1998) and in vivo models (Masliah et al. 2000) suggesting that higher expression levels of this protein are sufficient to cause the disease. Several studies imply that overexpression of wild-type α-synuclein can be toxic to cells (Ostrerova et al. 1999) and that dopaminergic neurons might be particularly vulnerable to toxicity from α-synuclein overexpression. More recently, genomic duplication or triplication of the region containing SNCA has been documented in affected individuals from families with autosomal dominant PD (Singleton et al. 2003; Chartier-Harlin et al. 2004; Ibanez et al. 2004). In the case of the triplication, four fully functional copies of α-synuclein were associated with twofold overexpression of α-synuclein mRNA and protein (Farrer et al. 2004; Miller et al. 2004). This finding emphasizes the importance of α-synuclein levels in PD, showing that elevated levels of wild-type α-synuclein are sufficient to cause familial early-onset PD. Thus, the regulation of α-synuclein expression might play an important role in the susceptibility to develop PD more generally.
Several association studies have shown that certain alleles of the NACP-Rep1 locus may confer an increased risk of sporadic PD (Kruger et al. 1999; Tan et al. 2000, 2003, 2004; Farrer et al. 2001; Mizuta et al. 2002). Our previous study suggested a model in which the polymorphic NACP-Rep1 element is involved in the regulation of SNCA expression, but exactly how the repeat can modulate the expression level is still unknown. As a first step towards understanding the role of NACP-Rep1 in the control of SNCA expression, we identified a protein, PARP-1, that binds NACP-Rep1 and found it to be involved in the regulation of SNCA expression
Poly (ADP-ribose) polymerase-1 (PARP-1) is a 116-kDa nuclear protein widely known for its DNA-binding properties (Soldatenkov et al. 2002) and for a unique enzymatic activity, ADP-ribosylation. PARP-1 can bind with high affinity in a cooperative manner to different DNA structures (e.g., cruciform, curved, supercoiled, crossover, stem-loop, and base unpairing region [BUR], as well as some specific double-stranded sequences) (Soldatenkov et al. 2002; Kraus and Lis 2003). Its enzymatic activity is to catalyze the covalent attachment of ADP-ribose units from its substrate β-NAD+ to several nuclear-acceptor proteins, including itself (D’Amours et al. 1999). Initial studies implicated this enzyme in many crucial biological functions, including DNA repair, replication, recombination, apoptosis, and cancer (D’Amours et al. 1999; Virag and Szabo 2002). A role for PARP-1 in transcriptional regulation of specific genes has also been demonstrated using a variety of experimental approaches (Kraus and Lis 2003). PARP-1 was identified as a constituent of the positive cofactor-1 complex, which is essential for the activity of several transcription factors (Guermah et al. 1998). The role of PARP-1 in transcription is well established by the demonstration by several groups of its potent effect on some transcription factors (Whitacre et al. 1995; Nie et al. 1998; Butler and Ordahl 1999; Kannan et al. 1999; Oliver et al. 1999; Anderson et al. 2000; Cervellera and Sala 2000; Hassa and Hottiger 2002; Virag and Szabo 2002). Gene-expression studies using microarrays indicated the effect of PARP-1 on the expression of >90 genes (Simbulan-Rosenthal et al. 2000).
Two mechanisms, at least, have been suggested for how PARP-1 might be involved in transcriptional regulation: (1) modifying histones to alter chromatin structure and (2) participating in forming promoter/enhancer binding complexes, in conjunction with other DNA-binding factors and coactivators (Kraus and Lis 2003). There is evidence that PARP-1 can act as a transcription activator (Anderson et al. 2000; Cervellera and Sala 2000; Hassa and Hottiger 2002), but other data show that PARP-1 may repress transcription (Butler and Ordahl 1999; Miyamoto et al. 1999; Soldatenkov et al. 2002). Recently, PARP-1 was shown to have a distinctly dualistic role, with opposing effects in AP-2α-mediated transcriptional regulation. It was demonstrated that separate regions of PARP-1 interact with AP-2α and independently control its transcriptional activation; while PARP-1’s middle region enhances transcription, its catalytic domain functions to repress transcription (Li et al. 2004).
In this study, we found, using both in vitro and in vivo approaches, that PARP-1 interacts with the NACP-Rep1 site and contributes to the regulation of α-synuclein expression. NACP-Rep1 does not contain a known PARP-1 recognition sequence, and thus the basis of the recognition of NACP-Rep1 by PARP-1 is not clear. One can speculate that this repeat sequence might tend to form certain DNA conformations such as a BUR, a structure that is recognized by PARP-1. However, we cannot exclude the possibility that NACP-Rep1 contains a new sequence-specific recognition site of PARP-1. It is important to note that, when we used a whole nuclear extract in the EMSA, the labeled DNA was shifted upward to form a smear, rather than a discrete band as was seen when purified PARP-1 was used. This result implies that PARP-1 might be a part of a protein complex that interacts with NACP-Rep1 and that PARP-1 is likely to be one of the proteins in the complex that binds NACP-Rep1 directly.
Our previous and current findings indicate that NACP-Rep1 functions as a repressor element. In light of these data, one can speculate that PARP-1 acts as a transcription repressor of the SNCA gene through its interaction with NACP-Rep1. Inhibition of PARP-1’s enzymatic activity with 3-aminobenzamide abrogated this repressive effect and led to elevated levels of endogenous SNCA mRNA in SH-SY5Y cells. The repressive effect of PARP-1 on SNCA expression and its reversal by 3-aminobenzamide appears to be transcriptional, at least in part, as shown by our luciferase reporter assays. A number of hypothetical models could be envisioned for how inhibition of PARP-1’s enzymatic activity might either directly or indirectly up-regulate SNCA expression. (1) Enzymatically active PARP-1 might alter chromatin structure at the NACP-Rep1 region to a transcriptionally inaccessible form of DNA, while inhibition of the PARP-1 catalytic domain results in an open DNA conformation and transcriptional up-regulation. (2) Enzymatically active PARP-1 might recruit transcriptional repressors and/or prevent the binding and interaction of transcriptional activators while inhibition of PARP-1 would have the opposite effect (i.e., to reduce the binding of the repressors or allow transcriptional activators to bind), leading to up-regulation of expression. Each of these models can act either via (a) PARP-1's catalytic property (i.e., ADP-ribosylation) or (b) its DNA-binding property, if enzymatically active and inactive PARP-1 bind the NACP-Rep1 segment differently. It should be noted that the effect on SNCA promoter/enhancer function of inhibiting PARP-1’s enzymatic activity depends on the NACP-Rep1 sequence being present: inhibition of PARP-1 in cells transfected with the NACP-Rep1 deleted construct resulted in an opposite effect, namely a decrease in SNCA promoter/enhancer transcriptional activity. This result may reflect a nonspecific effect of PARP-1 inhibition or may indicate that other recognition site(s) of PARP-1 may reside along the SNCA promoter/enhancer region and allow PARP-1 to play a dualistic role in SNCA transcriptional regulation. If this were the case, the actual effect of PARP-1 on SNCA transcription could be the result of a balance between the effect of its interaction with NACP-Rep1 and its interaction with other sites upstream of the start of SNCA transcription.
The mechanism of how PARP-1 might be involved in transcriptional regulation of SNCA is unclear. Previously, we demonstrated that the two segments that flank the NACP-Rep1 appeared responsible, in an additive manner, for expression, whereas the NACP-Rep1 itself was shown to have a negative effect on expression (Chiba-Falek and Nussbaum 2001). These results suggest a model of two regulatory regions, one located upstream of the NACP-Rep1 and the other downstream of the repeat. The transcription factors binding in these two regions may interact to function in an additive manner, while different alleles of NACP-Rep1 modulate this interaction to different degrees. PARP-1 binding to the repeat might contribute to the modulation of this interaction. Exactly how PARP-1 functions in this modulation is unknown, but it could be altering how effectively or efficiently the transcription factors that bind upstream and downstream of the repeat can interact, or it could be modulating the recruitment, binding, and/or activation of these transcription factors. In the future, it will be of interest to analyze the variation among the different NACP-Rep1 alleles, with respect to PARP-1 binding affinity and its effect on the transcriptional regulation of SNCA.
Our current and previous experiments lend support to the hypothesis that the NACP-Rep1 microsatellite in cooperation with its binding protein, PARP-1, have a physiological function in the regulation of SNCA gene expression. Thus, one can speculate that polymorphism at this microsatellite might be involved in sporadic PD via its effect on the expression of the SNCA gene, thus contributing, among other factors, to the risk for disease.
Acknowledgments
This work was supported in part by grants from the National Cancer Institute (PO1CA-74175 and the U.S. Air Force Office of Scientific Research (AF-FA9550-04-1-0395) (to M.E.S.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PD and SNCA)
References
- Anderson MG, Scoggin KE, Simbulan-Rosenthal CM, Steadman JA (2000) Identification of poly(ADP-ribose) polymerase as a transcriptional coactivator of the human T-cell leukemia virus type 1 Tax protein. J Virol 74:2169–2177 10.1128/JVI.74.5.2169-2177.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AJ, Ordahl CP (1999) Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol 19:296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervellera MN, Sala A (2000) Poly(ADP-ribose) polymerase is a B-MYB coactivator. J Biol Chem 275:10692–10696 10.1074/jbc.275.14.10692 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A (2004) Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364:1167–1169 10.1016/S0140-6736(04)17103-1 [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Nussbaum RL (2001) Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet 10:3101–3109 10.1093/hmg/10.26.3101 [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Touchman JW, Nussbaum RL (2003) Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum Genet 113:426–431 10.1007/s00439-003-1002-9 [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342:249–268 10.1042/0264-6021:3420249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, Hulihan M, Maraganore D, Gwinn-Hardy K, Wszolek Z, Dickson D, Langston JW (2004) Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol 55:174–179 10.1002/ana.10846 [DOI] [PubMed] [Google Scholar]
- Farrer M, Maraganore DM, Lockhart P, Singleton A, Lesnick TG, de Andrade M, West A, de Silva R, Hardy J, Hernandez D (2001) α-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum Mol Genet 10:1847–1851 10.1093/hmg/10.17.1847 [DOI] [PubMed] [Google Scholar]
- Feany MB, Bender WW (2000) A Drosophila model of Parkinson’s disease. Nature 404:394–398 10.1038/35006074 [DOI] [PubMed] [Google Scholar]
- Golbe LI (1999) Alpha-synuclein and Parkinson’s disease. Mov Disord 14:6–9 [DOI] [PubMed] [Google Scholar]
- Guermah M, Malik S, Roeder RG (1998) Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol Cell Biol 18:3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Hsu LJ, Sisk A, Xia Y, Takeda A, Sundsmo M, Masliah E (1998) Human recombinant NACP/α-synuclein is aggregated and fibrillated in vitro: relevance for Lewy body disease. Brain Res 799:301–306 10.1016/S0006-8993(98)00514-9 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO (2002) The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cell Mol Life Sci 59:1534–1553 10.1007/s00018-002-8527-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A (2004) Causal relation between α-synuclein gene duplication and familial Parkinson’s disease. Lancet 364:1169–1171 10.1016/S0140-6736(04)17104-3 [DOI] [PubMed] [Google Scholar]
- Izumi Y, Morino H, Oda M, Maruyama H, Udaka F, Kameyama M, Nakamura S, Kawakami H (2001) Genetic studies in Parkinson’s disease with an α-synuclein/NACP gene polymorphism in Japan. Neurosci Lett 300:125–127 10.1016/S0304-3940(01)01557-9 [DOI] [PubMed] [Google Scholar]
- Johnson J, Hague SM, Hanson M, Gibson A, Wilson KE, Evans EW, Singleton AA, McInerney-Leo A, Nussbaum RL, Hernandez DG, Gallardo M, McKeith IG, Burn DJ, Ryu M, Hellstrom O, Ravina B, Eerola J, Perry RH, Jaros E, Tienari P, Weiser R, Gwinn-Hardy K, Morris CM, Hardy J, Singleton AB (2004) SNCA multiplication is not a common cause of Parkinson disease or dementia with Lewy bodies. Neurology 63:554–556 [DOI] [PubMed] [Google Scholar]
- Kannan P, Yu Y, Wankhade S, Tainsky MA (1999) PolyADP-ribose polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Res 27:866–874 10.1093/nar/27.3.866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Graham E, Dixon P, Morris C, Mander A, Clayton D, Vaughan J, Quinn N, Lees A, Daniel S, Wood N, de Silva R (2001) Parkinson’s disease is not associated with the combined alpha-synuclein/apolipoprotein E susceptibility genotype. Ann Neurol 49:665–668 10.1002/ana.1027.abs [DOI] [PubMed] [Google Scholar]
- Kraus WL, Lis JT (2003) PARP goes transcription. Cell 113:677–683 10.1016/S0092-8674(03)00433-1 [DOI] [PubMed] [Google Scholar]
- Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O (1998) Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat Genet 18:106–108 10.1038/ng0298-106 [DOI] [PubMed] [Google Scholar]
- Kruger R, Vieira-Saecker AM, Kuhn W, Berg D, Muller T, Kuhnl N, Fuchs GA, Storch A, Hungs M, Woitalla D, Przuntek H, Epplen JT, Schols L, Riess O (1999) Increased susceptibility to sporadic Parkinson’s disease by a certain combined α-synuclein/apolipoprotein E genotype. Ann Neurol 45:611–617 [DOI] [PubMed] [Google Scholar]
- Li M, Naidu P, Yu Y, Berger NA, Kannan P (2004) Dual regulation of AP-2α transcriptional activation by poly(ADP-ribose) polymerase-1. Biochem J 382:323–329 10.1042/BJ20040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K (2003) The role of α-synuclein in Parkinson’s disease: insights from animal models. Nat Rev Neurosci 4:727–738 10.1038/nrn1199 [DOI] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L (2000) Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science 287:1265–1269 10.1126/science.287.5456.1265 [DOI] [PubMed] [Google Scholar]
- Miller DW, Hague SM, Clarimon J, Baptista M, Gwinn-Hardy K, Cookson MR, Singleton AB (2004) α-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology 62:1835–1838 [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Kakizawa T, Hashizume K (1999) Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol Cell Biol 19:2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta I, Nishimura M, Mizuta E, Yamasaki S, Ohta M, Kuno S (2002) Meta-analysis of α synuclein/ NACP polymorphism in Parkinson’s disease in Japan. J Neurol Neurosurg Psychiatry 73:350 10.1136/jnnp.73.3.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Sakamoto S, Song D, Qu Z, Ota K, Taniguchi T (1998) Interaction of Oct-1 and automodification domain of poly(ADP-ribose) synthetase. FEBS Lett 424:27–32 10.1016/S0014-5793(98)00131-8 [DOI] [PubMed] [Google Scholar]
- Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G (1999) Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly (ADP-ribose) polymerase-1 deficient mice. Embo J 18:4446–4454 10.1093/emboj/18.16.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrerova N, Petrucelli L, Farrer M, Mehta N, Choi P, Hardy J, Wolozin B (1999) α-Synuclein shares physical and functional homology with 14-3-3 proteins. J Neurosci 19:5782–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A, Racette B, Zhang ZH, Chakraverty S, Rundle M, Goate A, Perlmutter JS (1998) Mutation, sequence analysis, and association studies of α-synuclein in Parkinson’s disease. Neurology 51:1757–1759 [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL (1997) Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047 10.1126/science.276.5321.2045 [DOI] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Ly DH, Rosenthal DS, Konopka G, Luo R, Wang ZQ, Schultz PG, Smulson ME (2000) Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc Natl Acad Sci USA 97:11274–11279 10.1073/pnas.200285797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K (2003) α-Synuclein locus triplication causes Parkinson’s disease. Science 302:841 10.1126/science.1090278 [DOI] [PubMed] [Google Scholar]
- Smulson ME, Pang D, Jung M, Dimtchev A, Chasovskikh S, Spoonde A, Simbulan-Rosenthal C, Rosenthal D, Yakovlev A, Dritschilo A (1998) Irreversible binding of poly(ADP)ribose polymerase cleavage product to DNA ends revealed by atomic force microscopy: possible role in apoptosis. Cancer Res 58:3495–3498 [PubMed] [Google Scholar]
- Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A (2002) Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem 277:665–670 10.1074/jbc.M108551200 [DOI] [PubMed] [Google Scholar]
- Spadafora P, Annesi G, Pasqua AA, Serra P, Ciro Candiano IC, Carrideo S, Tarantino P, Civitelli D, De Marco EV, Nicoletti G, Annesi F, Quattrone A (2003) NACP-REP1 polymorphism is not involved in Parkinson’s disease: a case-control study in a population sample from southern Italy. Neurosci Lett 351:75–78 10.1016/S0304-3940(03)00859-0 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M (1997) α-synuclein in Lewy bodies. Nature 388:839–840 10.1038/42166 [DOI] [PubMed] [Google Scholar]
- Stone KL, Williams KR (1993) Enzymatic digestion of proteins and HPLC peptide isolation. Academic Press, San Diego, CA [Google Scholar]
- Tan EK, Chai A, Teo YY, Zhao Y, Tan C, Shen H, Chandran VR, Teoh ML, Yih Y, Pavanni R, Wong MC, Puvan K, Lo YL, Yap E (2004) Alpha-synuclein haplotypes implicated in risk of Parkinson’s disease. Neurology 62:128–131 10.1159/000080514 [DOI] [PubMed] [Google Scholar]
- Tan EK, Matsuura T, Nagamitsu S, Khajavi M, Jankovic J, Ashizawa T (2000) Polymorphism of NACP-Rep1 in Parkinson’s disease: an etiologic link with essential tremor? Neurology 54:1195–1198 [DOI] [PubMed] [Google Scholar]
- Tan EK, Tan C, Shen H, Chai A, Lum SY, Teoh ML, Yih Y, Wong MC, Zhao Y (2003) Alpha synuclein promoter and risk of Parkinson’s disease: microsatellite and allelic size variability. Neurosci Lett 336:70–72 10.1016/S0304-3940(02)01178-3 [DOI] [PubMed] [Google Scholar]
- Touchman JW, Dehejia A, Chiba-Falek O, Cabin DE, Schwartz JR, Orrison BM, Polymeropoulos MH, Nussbaum RL (2001) Human and mouse alpha-synuclein genes: comparative genomic sequence analysis and identification of a novel gene regulatory element. Genome Res 11:78–86 10.1101/gr.165801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag L, Szabo C (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev 54:375–429 10.1124/pr.54.3.375 [DOI] [PubMed] [Google Scholar]
- Whitacre CM, Hashimoto H, Tsai ML, Chatterjee S, Berger SJ, Berger NA (1995) Involvement of NAD-poly(ADP-ribose) metabolism in p53 regulation and its consequences. Cancer Res 55:3697–3701 [PubMed] [Google Scholar]
- Xia Y, Rohan de Silva HA, Rosi BL, Yamaoka LH, Rimmler JB, Pericak-Vance MA, Roses AD, Chen X, Masliah E, DeTeresa R, Iwai A, Sundsmo M, Thomas RG, Hofstetter CR, Gregory E, Hansen LA, Katzman R, Thal LJ, Saitoh T (1996) Genetic studies in Alzheimer’s disease with an NACP/α-synuclein polymorphism. Ann Neurol 40:207–215 10.1002/ana.410400212 [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez Tortosa E, del Ser T, Munoz DG, de Yebenes JG (2004) The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55:164–173 10.1002/ana.10795 [DOI] [PubMed] [Google Scholar]