Polymorphisms in the Tyrosine Kinase 2 and Interferon Regulatory Factor 5 Genes Are Associated with Systemic Lupus Erythematosus (original) (raw)
Abstract
Systemic lupus erythematosus (SLE) is a complex systemic autoimmune disease caused by both genetic and environmental factors. Genome scans in families with SLE point to multiple potential chromosomal regions that harbor SLE susceptibility genes, and association studies in different populations have suggested several susceptibility alleles for SLE. Increased production of type I interferon (IFN) and expression of IFN-inducible genes is commonly observed in SLE and may be pivotal in the molecular pathogenesis of the disease. We analyzed 44 single-nucleotide polymorphisms (SNPs) in 13 genes from the type I IFN pathway in 679 Swedish, Finnish, and Icelandic patients with SLE, in 798 unaffected family members, and in 438 unrelated control individuals for joint linkage and association with SLE. In two of the genes—the tyrosine kinase 2 (TYK2) and IFN regulatory factor 5 (IRF5) genes—we identified SNPs that displayed strong signals in joint analysis of linkage and association (unadjusted P<10-7) with SLE. TYK2 binds to the type I IFN receptor complex and IRF5 is a regulator of type I IFN gene expression. Thus, our results support a disease mechanism in SLE that involves key components of the type I IFN system.
Systemic lupus erythematosus (SLE [MIM 152700]) is regarded as the prototypic systemic autoimmune disease; it has a prevalence of 0.06% in white populations. SLE is more common in women than in men and has a strong heritable component, as shown by twin studies (Hochberg 1987; Deapen et al. 1992). Typically, in SLE, immune complexes are formed that consist of autoantibodies against nucleic acids, ribonucleoproteins, and histones. The immune complexes are considered to be the principal cause of inflammation in SLE and give rise to arthritis, skin rashes, nephritis, and vasculitis, among other symptoms (Wallace and Hannahs 2002). Both genetic and environmental factors are thought to contribute to the continuous autoimmune process. In accordance with the apparent multigenic character of SLE, genomewide scans point to multiple chromosomal regions that potentially harbor SLE-susceptibility genes (Cantor et al. 2004), and association studies have suggested several susceptibility genes for SLE. For a review, see Tsao (2004). Genes encoding haplotypes of the human leukocyte antigen (HLA) complex, early components of the complement system, and low-affinity receptors for the Fc region of IgG have shown association with SLE in multiple populations. A polymorphism in the programmed cell death 1 (PDCD1) gene appears to be associated with SLE in Icelandic, Swedish, and Mexican patients (Prokunina et al. 2002). The proteins encoded by all these genes may, in various ways, promote production of autoantibodies and subsequent formation, tissue deposition, and effects of immune complexes. The molecular pathogenesis of SLE, however, is still obscure.
The type I interferon (IFN) system has been proposed as having a pivotal role in the development and maintenance of the disease process in SLE (Rönnblom and Alm 2003). The type I IFNs comprise a multigene family with 13 IFN-α subtype genes and single genes encoding IFN-β, -ω, -ɛ, and -κ (Levy et al. 2003). Patients with SLE have increased serum IFN-α levels that correlate with disease activity and severity, as well as with markers of immune activation, such as levels of antibodies against double-stranded DNA (dsDNA) and complement activation (Bengtsson et al. 2000). The type I IFNs act on a common receptor, the IFN α receptor (IFNAR), resulting in activation of the Janus kinases JAK1 and TYK2 and several signal transducers and activators of transcription (STAT1–6) (David 2002). This results in increased expression of a spectrum of type I IFN–inducible genes (de Veer et al. 2001). Such an “IFN signature” of gene expression was observed in peripheral blood mononuclear cells in patients with SLE and appears to be correlated with the activity and severity of the disease (Baechler et al. 2003; Crow and Wohlgemuth 2003). The reason for the ongoing IFN-α production in SLE is probably activation of the plasmacytoid dendritic cells (PDC), also termed “natural IFN-α–producing cells” (NIPC), by immune complexes consisting of autoantibodies and autoantigens that contain DNA or RNA (Båve et al. 2000; Lövgren et al. 2004). Such autoantigens are formed via the increased apoptosis and impaired scavenging of apoptotic cells in SLE (Kaplan 2004). Induction of IFN-α gene expression by the immune complexes requires both the Fc region of IgG that interacts with the Fc γ receptor IIa (FCGR2A) on NIPC/PDC (Båve et al. 2003) and RNA or DNA that interacts with membrane-bound or intracellular receptors (Akira and Takeda 2004; Yoneyama et al. 2004). Activation of transcription factors, including IFN regulatory factors 3, 5, and 7 (IRF3, IRF5, and IRF7, respectively) is crucial for the expression of the type I IFN genes (Barnes et al. 2002). The IFN-α produced by activated NIPC/PDC can promote autoimmunity by stimulating key cells in the immune system, including lymphocytes and antigen-presenting dendritic cells (Stewart 2003). A causative role of IFN-α in SLE is more directly indicated by the development of SLE and other autoimmune diseases in tumor and in patients with hepatitis during treatment with IFN-α (Rönnblom et al. 1991; Ioannou and Isenberg 2000). The disease that certain patients develop during IFN-α treatment is indistinguishable from spontaneously occurring SLE. Furthermore, deletion of the Ifnar gene in lupus-prone mice markedly reduces SLE disease and mortality (Santiago-Raber et al. 2003).
Given this proposed crucial role of the type I IFN system in SLE, we dissected the function of the type I IFN system in SLE by joint linkage and association analysis of a panel of 11 genes encoding important type I IFN signaling molecules in Swedish and Finnish patients with SLE, their family members, and unrelated control individuals. The analyzed genes encode the two IFNAR subunits (IFNAR1 and IFNAR2), the two IFNAR-associated protein tyrosine kinases (JAK1 and TYK2), two signal transducers and activators of transcription (STAT1 and STAT3), an IFN regulatory factor (IRF5), an IFN inducible protein (IFI1), and type I IFNs (IFNA21, IFNA6, and IFNB1). Because of their connection to the type I IFN system, the genes encoding FCGR2A and PDCD1 were also included in the analysis. FCGR2A is required for IFN-α production (Båve et al. 2003), and the ligand of PDCD1, called “PDL1,” is upregulated by IFN-α (Eppihimer et al. 2002).
The study subjects comprised a Swedish set of 499 patients with SLE, 442 unaffected family members, and 256 unrelated controls, and a Finnish set of 150 patients with SLE, 237 unaffected pedigree members, and 182 unrelated controls (table 1). The Swedish patients with SLE and their family members were recruited from Lund, Uppsala, and Umeå University hospitals. A group of 256 volunteer blood donors, including 60 individuals age- and sex-matched to patients from Uppsala and a set of 182 anonymous Finnish population samples, served as unrelated control individuals. The Finnish families with SLE were recruited from the major hospitals in Finland (Koskenmies et al. 2004). A set of 11 Icelandic families with multicase SLE (Kristjansdottir et al. 2000) from the Center for Rheumatology Research, in Reykjavik, were analyzed to confirm our findings in the Swedish and Finnish samples. The affected individuals fulfilled the criteria for SLE defined by the American College of Rheumatology (ACR) (Tan et al. 1982). Informed consent was provided by each individual included in the study. The samples were collected according to the Helsinki Declaration. Ethical approvals for the study were obtained from the ethics committees of each center.
Table 1.
Sample Overview
| No. of Families | No. of Samples | ||||
|---|---|---|---|---|---|
| Sample andLocation | Multicase | Single Case | Affecteda | Unaffected | Total |
| Family: | |||||
| Uppsala, Sweden | 3 | 38 | 45 | 66 | 111 |
| Lund, Sweden | 3 | 128 | 146 | 376 | 522 |
| Finland | 37 | 72 | 150 | 237 | 387 |
| Iceland | 11 | … | 30 | 119 | 149 |
| Unrelated: | |||||
| Uppsala, Sweden | … | … | 33 | 256 | 289 |
| Lund, Sweden | … | … | 15 | … | 15 |
| Umeå, Sweden | … | … | 260 | … | 260 |
| Finland | … | … | … | 182 | 182 |
| Total | 54 | 238 | 679 | 1,236 | 1,915 |
First, we genotyped a panel of 33 SNPs in the candidate genes from the type I IFN pathway, by use of DNA extracted from blood samples of the study subjects. The SNPs were randomly selected from databases; their assay performance had been validated in a previous study (Lindroos et al. 2002). The SNP rs1800593 (R/H131) in FCGR2A (Karassa et al. 2002) and the SNP rs11568821 (PD-1.3) in PDCD1 (Prokunina et al. 2002) have been associated elsewhere to SLE in Swedish patients (Prokunina et al. 2002; Magnusson et al. 2004). For genotyping the SNPs, we used fluorescent minisequencing with an “in-house” developed multiplex tag-array system (Lindroos et al. 2002), the 12-plex SNPStream system (Beckman Coulter) (Bell et al. 2002), or the homogeneous template directed-dye terminator assay with fluorescence polarization detection (FP-TDI [Perkin Elmer]) for individual SNPs (Hsu et al. 2001). We assessed the quality of the genotype data by testing for Hardy-Weinberg equilibrium in the control samples, using Fisher’s exact test (_P_>.05) and using the program Mendel (Lange 2001) to identify possible non-Mendelian inheritance of alleles in the family samples. No significant deviation from Hardy-Weinberg equilibrium and no Mendelian inheritance errors were observed for any of the SNPs included in the study. The overall genotype call rate was 92%, and the accuracy was >99%, according to duplicate genotyping of all samples and by repeated genotyping of 10 SNPs in the two most interesting genes in all samples, with another of the three methods used. Table 2 provides information about the SNPs and their minor-allele frequencies. The allele frequencies for the affected individuals given in table 2 are based on all affected singleton individuals and one randomly selected individual from each pedigree with SLE. We did not observe any significant differences in allele frequencies for any of the analyzed SNPs between affected individuals from Umeå (northern Sweden), Uppsala (central Sweden), and Lund (southern Sweden), nor between unaffected pedigree members from Uppsala and Lund (data not shown).
Table 2.
SNP Allele Frequencies, P Values from Joint Linkage and Association Analysis and P Values from Association Analysis in Swedish and Finnish Samples[Note]
| Minor Allele Frequency | P Values for | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sweden | Finland | Joint Linkage and Associationb | Associationc | |||||||||
| Gene anddbSNP Number | SNP Alleles | Positiona | Affected(_n_=480) | Controls(_n_=256) | Affected(_n_=109) | Controls(_n_=121) | Sweden | Finland | Combinedd | Sweden | Finland | Combinedd |
| FCGR2A: | ||||||||||||
| rs1801274 | T/C | Exon (H/R) 4481 | .45 | .45 | .47 | .52 | .42 | .29 | .38 | .67 | .10 | .24 |
| IFI27: | ||||||||||||
| rs2799 | G/C | Exon (5′UTR) 4897 | .03 | .04 | .07 | .06 | .52 | .88 | .82 | .84 | .67 | .89 |
| IFNA21: | ||||||||||||
| rs2939 | C/T | Exon (5′UTR) 607 | .19 | .16 | .18 | .16 | .19 | .96 | .50 | 1.00 | 1.00 | 1.00 |
| rs1053887 | T/C | Exon (A/A) 488 | <.01 | <.01 | .01 | <.01 | .35 | .05 | .09 | .21 | .12 | .12 |
| IFNA6: | ||||||||||||
| rs614541 | T/C | Promoter −1976 | .20 | .15 | .16 | .14 | .12 | .033 | .07 | .81 | 1.00 | .98 |
| rs2383187 | G/A | Promoter −2093 | <.01 | <.01 | .01 | <.01 | .88 | .08 | .26 | .57 | 1.00 | .89 |
| IFNAR1: | ||||||||||||
| rs2257167 | G/C | Exon (V/L) 18338 | .10 | .15 | .12 | .08 | .3 | .89 | .62 | .02 | .20 | .03 |
| rs914142 | G/A | Intron 28446 | .25 | .26 | .25 | .35 | .48 | .52 | .60 | .82 | .12 | .34 |
| rs1041868 | A/G | Intron 29969 | .14 | .17 | .16 | .10 | .41 | .94 | .75 | .09 | .23 | .10 |
| IFNAR2: | ||||||||||||
| rs2073362 | G/A | Intron 6573 | .10 | .07 | .05 | .07 | .03 | .23 | .04 | .02 | .84 | .08 |
| rs3153 | T/C | Intron −4723 | .26 | .29 | .33 | .35 | .55 | .69 | .75 | .45 | .60 | .63 |
| IFNB1: | ||||||||||||
| rs1424855 | C/T | Exon (Y/Y) −947 | .35 | .34 | .35 | .36 | .85 | .84 | .96 | .68 | 1.00 | .94 |
| rs1424856 | A/G | Promoter −872 | <.01 | <.01 | <.01 | <.01 | .38 | .53 | .53 | 1.00 | 1.00 | 1.00 |
| rs1051922 | C/G | Promoter 152 | .39 | .34 | .37 | .44 | .29 | .09 | .12 | .17 | .46 | .28 |
| IRF5e: | ||||||||||||
| rs729302 | −13176 | .25 | .3 | .26 | .42 | .027 | 6.30×10-3 | 1.70×10-3 | .076 | .011 | 6.70×10-3 | |
| rs2004640 | C/A | Intron −3835 | .38 | .48 | .37 | .52 | 8.50×10-5 | 1.50×10-4 | 2.40×10-7 | 2.50×10-4 | 9.50×10-5 | 4.40×10-7 |
| rs752637 | −2716 | .31 | .34 | .23 | .41 | .49 | 2.20×10-4 | 1.10×10-3 | .54 | 4.40×10-3 | .017 | |
| rs3807306 | −1456 | .42 | .47 | .57 | .47 | .068 | .16 | .06 | .12 | .25 | .13 | |
| rs1874328 | C/T | Intron 2968 | .36 | .41 | .36 | .36 | .59 | .76 | .81 | .1 | .48 | .20 |
| rs2280714e | G/A | 3′ UTR 12589 | .29 | .29 | .22 | .30 | .85 | .10 | .29 | .82 | .23 | .51 |
| JAK1: | ||||||||||||
| rs2991269 | C/T | Exon (P/P) 38639 | .29 | .28 | .74 | .72 | .11 | .86 | .32 | .23 | .91 | .53 |
| rs310229 | A/G | I/E boundary 27740 | .24 | .24 | .23 | .25 | .013 | .82 | .061 | .32 | .83 | .62 |
| rs310227 | A/G | Intron 25986 | .23 | .24 | .76 | .77 | .05 | .51 | .12 | .37 | .82 | .66 |
| PDCD1: | ||||||||||||
| rs11568821 | T/C | Intron 7146 | .09 | .09 | .03 | .06 | .67 | .46 | .67 | .69 | .29 | .52 |
| STAT1: | ||||||||||||
| rs1400657 | C/A | 3′ UTR 41189 | .12 | .03 | .07 | .06 | .04 | .43 | .08 | .87 | .68 | .02 |
| rs1547550 | C/G | Intron 29004 | .33 | .35 | .26 | .28 | .78 | .12 | .31 | .79 | .91 | .96 |
| rs1914408 | A/G | Intron 34753 | .21 | .26 | .11 | .09 | 6.60×10-3 | .61 | .026 | .035 | .22 | .04 |
| rs2066798 | A/G | I/E boundary −24 | <.01 | .01 | <.01 | <.01 | .35 | .58 | .52 | .25 | 1.00 | .60 |
| rs2066802 | C/T | Exon (L/L) 62 | .08 | .06 | .05 | .03 | .57 | .42 | .58 | .73 | .35 | .60 |
| rs908598 | T/C | Intron 3442 | .02 | .03 | .04 | .02 | .43 | .04 | .09 | .65 | .50 | .70 |
| rs952798 | A/G | Intron 7882 | .01 | <.01 | .01 | <.01 | .27 | .25 | .25 | 1.00 | 1.00 | 1.00 |
| STAT3: | ||||||||||||
| rs744166 | C/T | Intron −13667 | .43 | .40 | .42 | .39 | .29 | .45 | .40 | .73 | 1.00 | .96 |
| rs1026916 | A/G | Intron −29301 | .34 | .35 | .39 | .36 | .12 | .76 | .32 | .66 | .68 | .81 |
| TYK2e | ||||||||||||
| rs280501 | A/G | Promoter −2240 | .09 | .11 | .07 | .08 | .76 | .72 | .88 | .45 | .26 | .37 |
| rs2304258e | G/T | Exon (5′UTR) −2104 | .01 | .01 | .03 | .03 | .84 | .08 | .24 | 1.00 | .76 | .97 |
| rs280500 | A/G | Exon (5′UTR) −1320 | .18 | .19 | .16 | .17 | 1.00 | .48 | .83 | 1.00 | .44 | .80 |
| rs280523 | A/G | Exon (T/T) 11876 | .09 | .07 | .11 | .11 | .32 | .44 | .42 | .82 | .88 | .96 |
| rs12720270e | T/C | Intron 13322 | .18 | .17 | .14 | .18 | .25 | .11 | .12 | .51 | .38 | .51 |
| rs2304256 | A/C | Exon (V/F) 13430 | .24 | .32 | .21 | .27 | 1.00×10-6 | .018 | 3.40×10-7 | 9.60×10-5 | .043 | 5.60×10-5 |
| rs2304255e | T/C | Exon (G/S) 13433 | .08 | .09 | .05 | NAf | .74 | .39 | .65 | 1.00 | 1.00 | 1.00 |
| rs280521e | A/G | I/E boundary 15690 | .16 | .16 | .14 | .14 | .45 | .60 | .62 | .54 | .86 | .82 |
| rs280519e | G/A | I/E boundary 16149 | .49 | .49 | .54 | .48 | .16 | .15 | .12 | .57 | .11 | .24 |
| rs12720356 | T/C | Exon (I/S) 19107 | .07 | .13 | .08 | .07 | 5.50×10-4 | .35 | 1.80×10-3 | 1.80×10-5 | .70 | 1.50×10-4 |
| rs280497e | T/C | I/E boundary 24395 | .48 | .45 | .55 | .50 | .19 | .02 | .02 | .53 | .14 | .26 |
We performed joint linkage and association analysis on individual SNPs, analyzing all of our genotype data from pedigrees and singletons together (table 1), using the software package Pseudomarker, which implements penetrance model-based analogs of popular so-called model-free methods for statistical gene localization (Göring and Terwilliger 2000). The key advantage of Pseudomarker is that it extracts both the linkage and association information from a data set. We analyzed the samples from Sweden and Finland separately, to avoid possible problems due to potential genetic differences between these populations. We used Fisher’s procedure for combining P values from different data sets to derive overall P values, using the analytical expression of Jost. Table 2 shows the results from this analysis in a dominant model of inheritance. We obtained strong signals for joint linkage and association for one of the SNPs in TYK2 (TYK2 rs2304256) and one of the SNPs in IRF5 (IRF5 rs2004640) in the Swedish and Finnish populations analyzed separately. Combination of the P values from both populations increased significance further, with P values of 3.4×10-7 for TYK2 rs2304256 and 2.4×10-7 for IRF5 rs2004640. To adjust the P values for multiple testing, we applied a Bonferroni correction with each individual SNP as an independent variable. Despite this conservative adjustment, our results remained highly significant, with adjusted P values of 1.1×10-5 for TYK2 rs2304256 and 7.9×10-6 for IRF5 rs2004640. An additional SNP in TYK2 (TYK2 rs12720356) resulted in a low P value (_P_=5.5×10-4; adjusted _P_=1.7×10-2) in the Swedish population only. We were not able to reproduce the association results for the SNPs in FCGR2A and PDCD1 in the Swedish or Finnish samples. Analysis of linkage alone, without allowance for association, did not reach significance for any of the SNPs in either population, separately or jointly (data not shown).
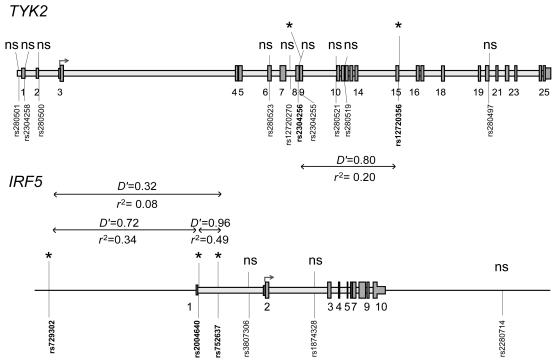
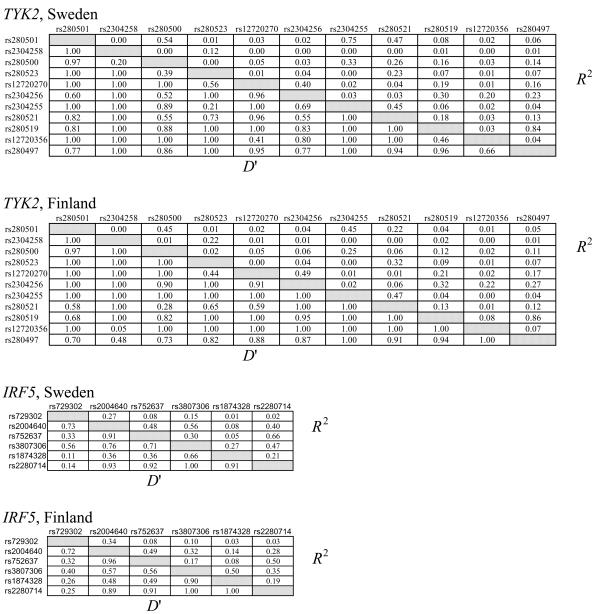
Given these results, we genotyped seven additional SNPs in TYK2 and four additional SNPs in IRF5, and we performed the same analyses as for the previous SNPs. The results are included in table 2 (in which these additional SNPs are footnoted). Two of the added IRF5 SNPs, rs729302 and rs752637, gave a nearly significant signal in the Finnish samples. To verify our results, we also performed case-control association analysis, using Fisher’s exact test, for data from all singleton affected individuals and one affected individual from each of the pedigrees (480 Swedish and 109 Finnish subjects) and unrelated control individuals (256 Swedish and 121 Finnish individuals). These results, also shown in table 2, confirm the results from our joint linkage and association analyses, albeit at slightly lesser significance. The ORs were 1.6 (95% CI 1.3–1.9) for both the TYK2 rs2304256 and for the IRF5 rs2004640 SNP and support our association results obtained by Fisher’s exact test. Figure 1 displays the positions of the 11 analyzed SNPs on TYK2, the 6 analyzed SNPs on IRF5, and the pairwise linkage-disequilibrium (LD) values between the SNPs with low P values in either of the populations. The LD-strength patterns are similar in both populations, as shown in figure A1 (online only), which provides the pairwise LD values for all genotyped TYK2 and IRF5 SNPs in Swedish and Finnish control samples. When we used the Haploview program (Barrett et al. 2005), we did not detect any statistically significant differences in haplotype frequencies between the two populations, and haplotype association analyses revealed no association with SLE with higher statistical significance than analysis of individual SNPs in either of the populations (data not shown).
Figure 1.
Schematic illustration of the structure of TYK2 and IRF5. The positions of exons are shown as numbered gray boxes, and the translation initiation sites are shown by arrows on both genes. The positions of the 11 TYK2 SNPs and the 6 IRF5 SNPs analyzed in the study are shown by vertical lines and dbSNP rs numbers. SNPs with low P values in joint linkage and association analysis in the Swedish and/or Finnish sample sets are marked with an asterisk (*). The D′ and _r_2 values for pairwise linkage disequilibrium for the TYK2 SNPs were calculated from the genotypes of Swedish unrelated control individuals, and the D′ and _r_2 values for the IRF5 SNPs were calculated from Finnish unrelated control individuals, by use of the Haploview program (Barrett et al. 2005). ns=nonsignificant.
The IRF5 SNP rs2004640 and TYK2 SNP rs2304256 were also analyzed for joint linkage and association in 11 Icelandic families with multicase SLE. Table 3 gives the unadjusted P values obtained in this analysis in the Swedish, Finnish, and Icelandic populations, under dominant and recessive models, with the overall P value derived from the P values from the data sets from the three individual countries. Both models gave similar results. Inclusion of the Icelandic families in the analysis further increased the significance of our findings by one order of magnitude, with P values of 4.2×10-8 for TYK2 rs2304256 and 1.5×10-8 for IRF5 rs2004640 under the dominant model, and with P values of 2.0×10-7 for TYK2 rs2304256 under a recessive model. Inclusion of the Icelandic families did not increase the significance of our findings for IRF5 rs2004640 under the recessive model.
Table 3.
Pseudomarker Analysis of SNPs TYK2 rs2304256 and IRF5 rs2004640 in the Swedish, Finnish, and Icelandic Populations, in Dominant and Recessive Models[Note]
| P Valuesa for | ||||||||
|---|---|---|---|---|---|---|---|---|
| Joint Linkage and Association | Linkage Only | |||||||
| Model anddbSNP rs Number | Sweden | Finland | Icelandb | Combinedc | Sweden | Finland | Iceland | Combinedc |
| Dominant: | ||||||||
| TYK2 rs2304256 | 1.0×10-6 | .018 | 4.1×10-3 | 2.2×10-8 | .23 | .23 | .063 | .077 |
| IRF5 rs2004640 | 8.5×10-5 | 1.5×10-4 | .015 | 5.2×10-8 | .50 | .35 | 5.4×10-3 | .031 |
| Recessive: | ||||||||
| TYK2 rs2304256 | 2.5×10-5 | 6.0×10-4 | .054 | 2.0×10-7 | .50 | .17 | .32 | .30 |
| IRF5 rs2004640 | 8.1×10-4 | 4.1×10-4 | .25 | 1.2×10-5 | .50 | .42 | .3 | .49 |
The frequencies of the rare A allele of TYK2 rs2304256 (A/C) and the rare C allele of IRF5 rs2004640 (C/A) are lower in the Swedish and Finnish patients with SLE than in the corresponding controls (table 2). We used logistic regression analysis to correlate the genotypes of these SNPs in TYK2 and IRF5, with the ACR classification criteria (Tan et al. 1982) fulfilled by the patients with SLE and with the presence of anti-dsDNA antibodies. This analysis revealed no significant correlation between the TYK2 or IRF5 genotypes and ACR criteria, although there was a tendency of association of the rare A allele of TYK2 rs2304256 with reduced occurrence of lupus nephritis (ACR criterion 7) in the Swedish patients (_P_=.026) and of the rare C allele of IRF5 rs2004640 with reduced occurrence of CNS involvement (ACR criterion 8) in Finnish patients (_P_=.023). The data from the logistic regression analysis are provided in table A1 (online only). Obviously, this analysis must be applied to a larger number of patients. It is, however, possible that the TYK2 and IRF5 genes are more generally involved in development of SLE via multiple effects, particularly on the type I IFN system, than with the specific manifestations represented by the ACR criteria. The latter may well be controlled by other genes. The TYK2 gene is located on chromosome 19p13.2, and, interestingly, this locus has been linked to SLE in white pedigrees stratified by the presence of anti-dsDNA antibodies (Namjou et al. 2002). Moreover, the A allele of the intronic SNP PD-1.3 in the PDCD1 gene (Prokunina et al. 2002) was recently found to be associated with a reduced occurrence of antiphospholipid antibodies both in patients with SLE and in the general population (Sanghera et al. 2004). The IRF5 gene is located on chromosome 7q32, a region lacking previously published linkage results.
The Janus kinase TYK2 consists of seven JAK-homology (JH) regions. The SNP TYK2 rs2304256 is located in exon 8 of the gene, where the rare A allele of this SNP causes a substitution of Val→Phe at position 362 in the JH4 region of TYK2. This region is part of a larger domain of TYK2 that is crucial not only for the interaction of TYK2 with IFNAR1 and its function (Richter et al. 1998) but also for maintaining expression of IFNAR1 on cell membranes (Ragimbeau et al. 2003). The Val→Phe substitution in TYK2 is characterized as “benign,” according to the PolyPhen (polymorphism phenotyping) and “intolerant” according to the SIFT (sorting intolerant from tolerant) programs, respectively (as described at the University of Washington–Fred Hutchinson Cancer Research Center SeattleSNPs Web site). The TYK2 rs12720356 SNP causes a substitution of Ile→Ser at amino acid position 684 in the pseudokinase region JH2 of TYK2. The Ile→Ser substitution is predicted to be “damaging” and “intolerant” by the PolyPhen and SIFT tools, respectively. The JH2 region of TYK2 is required for binding of type I IFN to IFNAR1, but it does not influence the expression of IFNAR1 (Yeh et al. 2000). Although we originally selected TYK2 for study because of its connection with the type I IFN system, it should be noted that TYK2 also interacts with the receptors for colony stimulating factor 1, angiotensin II, platelet-activating factor, and several interleukins (Kisseleva et al. 2002). Thus, an altered function of TYK2 may well also affect other cytokines, such as IL-10, for which an association to SLE has been reported (Chong et al. 2004). The importance of TYK2 in autoimmunity is also supported by the finding that _Tyk2_-deficient mice are resistant to experimental arthritis (Shaw et al. 2003). Consequently, we speculate that the amino acid substitutions caused by the TYK2 SNPs (rs2304256 and rs12720356) may reduce the function of TYK2, resulting in a decreased susceptibility to SLE.
IRF5 is one of the nine so-far-identified members of the IRF family that, together with IRF3 and IRF7, play an important role in the innate immune response by inducing type I IFN production during viral infections (Barnes et al. 2002). IRF5 is constitutively expressed mainly in cells of the immune system, particularly in NIPC, monocytes, and monocyte-derived dendritic cells, as well as in B cells (Izaguirre et al. 2003). Expression of IRF5 can be enhanced by type I IFN (Barnes et al. 2001). IRF5 is phosphorylated in cells upon viral infections and translocates to the nucleus, which results in activation of a spectrum of IFN-α genes that differ from those activated by IRF3 and IRF7 (Barnes et al. 2004). IRF5 also increases the expression of many genes other than the type I IFN genes, such as genes coding for signaling molecules, proteins involved in cell signaling, apoptosis, cell-cycle regulation, and early immune response. The IRF5 gene may be directly upregulated by p53, which indicates that IRF5 can be a p53-dependent mediator of the control of cell cycle and apoptosis (Mori et al. 2002; Barnes et al. 2003). Thus, polymorphism within the IRF5 gene may affect several cellular functions of importance for the development of an autoimmune disease such as SLE. The IRF5 SNP rs2004640 is the first nucleotide of IRF5 intron 1 and may thus have a functional role by altering the splicing of exon 1 of the IRF5.
In conclusion, by analyzing candidate genes from the type I IFN system, we identified polymorphisms in the TYK2 and IRF5 genes that displayed strong signals of joint linkage and association with SLE. The possible consequences of the identified SNPs on the function or expression levels of TYK2 and IRF5 remain to be elucidated experimentally. Our finding that genes involved in both the production of type I IFN and response to these cytokines are associated with SLE further strengthens the view that the type I IFN system is pivotal in the etiopathogenesis of this autoimmune disease. The results are also remarkable because we analyzed only 11 genes, selected on the basis of their connection with the type I IFN system. In fact, >200 genes may be involved in the activation and regulation of the type I IFN system and as mediators of the effects of IFN. We anticipate that several other genes within the type I IFN system are associated with SLE and possibly also other autoimmune disorders.
Acknowledgments
We thank Raul Figueroa and Per Lundmark (Molecular Medicine, Department of Medical Sciences, Uppsala University) for producing the tag-microarrays and for advice on the Haploview analysis, respectively; Marta Alarcón-Riquelme (Department of Genetic and Pathology, Uppsala University), for sequence information on the PDCD1 gene; and Lars Berglund (Uppsala Clinical Research Center), for performing the logistic regression analysis. Financial support for the study was provided by the Swedish Research Council and by the Knut and Alice Wallenberg Foundation (Wallenberg Consortium North) (to A.-C.S.), the Swedish Research Council, the Swedish Rheumatism Foundation, the Agnes and Mac Rudbergs Foundation, the King Gustaf V 80-Year Foundation, the Nanna Svartz Foundation, the Magnus Bergvall Foundation, the Börje Dahlin Foundation, the Brunnberg Foundation, the Astrid Karlsson Foundation, and the Uppsala University Hospital Research and Development Fund (to L.R. and G.A.)
Appendix A
Figure A1.
Pairwise LD values determined using the Haploview program for 11 SNPs in the tyrosine kinase 2 (TYK2) gene and 6 SNPs in the IFN regulatory factor 5 (IRF5) gene in control individuals from the Swedish population (_n_=256) and the Finnish population (_n_=121). The dbSNP rs numbers for the SNPs are given to the left and above the boxes in the diagrams. D′ values are given in the lower left half and _R_2 values in the upper right half of the diagrams.
Table A1.
Logistic Regression Analysis between TYK2 rs304256 and IRF5 rs402939 SNP Genotype and Disease Manifestations in Patients with SLE
| P Valueb for | ||||||
|---|---|---|---|---|---|---|
| TYK2 rs2304256 | IRF5 rs2004640 | |||||
| ACR Criteriona | Swedish | Finnish | Combined | Swedish | Finnish | Combined |
| Malar rash | .49 | .25 | 1.00 | .98 | .41 | .63 |
| Discoid rash | .91 | .40 | .84 | .14 | .62 | .24 |
| Photosensitivity | .90 | .86 | .85 | .90 | .86 | 1.00 |
| Oral ulcers | .35 | .70 | .29 | .28 | .40 | .78 |
| Arthritis | .26 | .16 | .12 | .85 | .32 | .41 |
| Serositis | .16 | .11 | .69 | .66 | .17 | .76 |
| Nephritis | .026c | .29 | .16 | .58 | .60 | .92 |
| CNS | .96 | .66 | .93 | .42 | .023d | .30 |
| Hematological | .83 | .30 | .60 | .55 | .51 | .49 |
| Immunological | .80 | .23 | .41 | .66 | .56 | .57 |
| Antinuclear antibodies | .59 | .76 | .76 | .47 | .47 | .98 |
| Anti-dsDNA antibodiese | .97 | .32 | .40 | .71 | .57 | .63 |
Electronic-Database Information
The URLs for data presented herein are as follows:
- CHIP Bioinformatics Tools, http://snpper.chip.org/bio/top (for SNPer)
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- Haploview, http://www.broad.mit.edu/mpg/haploview/
- Jost Web site, http://www.loujost.com/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLE) [PubMed]
- Pseudomarker, http://www.helsinki.fi/~tsjuntun/pseudomarker/
- University of Washington–Fred Hutchinson Cancer Research Center SeattleSNPs, http://pga.gs.washington.edu/data/tyk2/ (for TYK2)
References
- Akira S, Takeda K (2004) Toll-like receptor signalling. Nat Rev Immunol 4:499–511 [DOI] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 100:2610–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B, Lubyova B, Pitha PM (2002) On the role of IRF in host defense. J Interferon Cytokine Res 22:59–71 [DOI] [PubMed] [Google Scholar]
- Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM (2003) Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res 63:6424–6431 [PubMed] [Google Scholar]
- Barnes BJ, Moore PA, Pitha PM (2001) Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon α genes. J Biol Chem 276:23382–23390 [DOI] [PubMed] [Google Scholar]
- Barnes BJ, Richards J, Mancl ME, Hanash S, Beretta L, Pitha PM (2004) Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem 279:45194–45207 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Båve U, Alm GV, Rönnblom L (2000) The combination of apoptotic U937 cells and lupus IgG is a potent IFN-α inducer. J Immunol 165:3519–3526 [DOI] [PubMed] [Google Scholar]
- Båve U, Magnusson M, Eloranta ML, Perers A, Alm GV, Rönnblom L (2003) FcγRIIa is expressed on natural IFN-α-producing cells (plasmacytoid dendritic cells) and is required for the IFN-α production induced by apoptotic cells combined with lupus IgG. J Immunol 171:3296–3302 [DOI] [PubMed] [Google Scholar]
- Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, Kopla R, Modica F, Pohl M, Varde S, Zhao R, Zhao X, Boyce-Jacino MT, Yassen A (2002) SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques Suppl 32:70–77 [PubMed] [Google Scholar]
- Bengtsson AA, Sturfelt G, Truedsson L, Blomberg J, Alm G, Vallin H, Rönnblom L (2000) Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus 9:664–671 [DOI] [PubMed] [Google Scholar]
- Cantor RM, Yuan J, Napier S, Kono N, Grossman JM, Hahn BH, Tsao BP (2004) Systemic lupus erythematosus genome scan: support for linkage at 1q23, 2q33, 16q12-13, and 17q21-23 and novel evidence at 3p24, 10q23-24, 13q32, and 18q22-23. Arthritis Rheum 50:3203–3210 [DOI] [PubMed] [Google Scholar]
- Chong WP, Ip WK, Wong WH, Lau CS, Chan TM, Lau YL (2004) Association of interleukin-10 promoter polymorphisms with systemic lupus erythematosus. Genes Immun 5:484–492 [DOI] [PubMed] [Google Scholar]
- Crow MK, Wohlgemuth J (2003) Microarray analysis of gene expression in lupus. Arthritis Res Ther 5:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M (2002) Signal transduction by type I interferons. Biotechniques Suppl 33:58–65 [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR (2001) Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 69:912–920 [PubMed] [Google Scholar]
- Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM (1992) A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum 35:311–318 [DOI] [PubMed] [Google Scholar]
- Eppihimer MJ, Gunn J, Freeman GJ, Greenfield EA, Chernova T, Erickson J, Leonard JP (2002) Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation 9:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg MC (1987) The application of genetic epidemiology to systemic lupus erythematosus. J Rheumatol 14:867–869 [PubMed] [Google Scholar]
- Hsu TM, Chen X, Duan S, Miller RD, Kwok PY (2001) Universal SNP genotyping assay with fluorescence polarization detection. Biotechniques 31:560–570 [DOI] [PubMed] [Google Scholar]
- Ioannou Y, Isenberg DA (2000) Current evidence for the induction of autoimmune rheumatic manifestations by cytokine therapy. Arthritis Rheum 43:1431–1442 [DOI] [PubMed] [Google Scholar]
- Izaguirre A, Barnes BJ, Amrute S, Yeow WS, Megjugorac N, Dai J, Feng D, Chung E, Pitha PM, Fitzgerald-Bocarsly P (2003) Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J Leukoc Biol 74:1125–1138 [DOI] [PubMed] [Google Scholar]
- Kaplan MJ (2004) Apoptosis in systemic lupus erythematosus. Clin Immunol 112:210–218 [DOI] [PubMed] [Google Scholar]
- Karassa FB, Trikalinos TA, Ioannidis JP (2002) Role of the Fcγ receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum 46:1563–1571 [DOI] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW (2002) Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1–24 [DOI] [PubMed] [Google Scholar]
- Koskenmies S, Lahermo P, Julkunen H, Ollikainen V, Kere J, Widen E (2004) Linkage mapping of systemic lupus erythematosus (SLE) in Finnish families multiply affected by SLE. J Med Genet 41:e2–e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjansdottir H, Bjarnadottir K, Hjalmarsdottir IB, Grondal G, Arnason A, Steinsson K (2000) A study of C4AQ0 and MHC haplotypes in Icelandic multicase families with systemic lupus erythematosus. J Rheumatol 27:2590–2596 [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E (2001) Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet Suppl 69:212 [Google Scholar]
- Levy DE, Marie I, Prakash A (2003) Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr Opin Immunol 15:52–58 [DOI] [PubMed] [Google Scholar]
- Lindroos K, Sigurdsson S, Johansson K, Ronnblom L, Syvänen AC (2002) Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res 30:e70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövgren T, Eloranta ML, Båve U, Alm GV, Rönnblom L (2004) Induction of interferon-α production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum 50:1861–1872 [DOI] [PubMed] [Google Scholar]
- Magnusson V, Johanneson B, Lima G, Odeberg J, Alarcon-Segovia D, Alarcon-Riquelme ME (2004) Both risk alleles for FcγRIIA and FcγRIIIA are susceptibility factors for SLE: a unifying hypothesis. Genes Immun 5:130–137 [DOI] [PubMed] [Google Scholar]
- Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H (2002) Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene 21:2914–2918 [DOI] [PubMed] [Google Scholar]
- Namjou B, Nath SK, Kilpatrick J, Kelly JA, Reid J, Reichlin M, James JA, Harley JB (2002) Genome scan stratified by the presence of anti-double-stranded DNA (dsDNA) autoantibody in pedigrees multiplex for systemic lupus erythematosus (SLE) establishes linkages at 19p13.2 (SLED1) and 18q21.1 (SLED2). Genes Immun 3 Suppl 1:S35–S41 [DOI] [PubMed] [Google Scholar]
- Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, Brookes AJ, Tentler D, Kristjansdottir H, Grondal G, Bolstad AI, Svenungsson E, Lundberg I, Sturfelt G, Jonssen A, Truedsson L, Lima G, Alcocer-Varela J, Jonsson R, Gyllensten UB, Harley JB, Alarcon-Segovia D, Steinsson K, Alarcon-Riquelme ME (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 32:666–669 [DOI] [PubMed] [Google Scholar]
- Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S (2003) The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. Embo J 22:537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter MF, Dumenil G, Uze G, Fellous M, Pellegrini S (1998) Specific contribution of TYK2 JH regions to the binding and the expression of the interferon α/β receptor component IFNAR1. J Biol Chem 273:24723–24729 [DOI] [PubMed] [Google Scholar]
- Riva A, Kohane IS (2004) A SNP-centric database for the investigation of the human genome. BMC Bioinformatics 5:33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnblom L, Alm GV (2003) Systemic lupus erythematosus and the type I interferon system. Arthritis Res Ther 5:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnblom LE, Alm GV, Oberg KE (1991) Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann Intern Med 115:178–183 [DOI] [PubMed] [Google Scholar]
- Sanghera DK, Manzi S, Bontempo F, Nestlerode C, Kamboh MI (2004) Role of an intronic polymorphism in the PDCD1 gene with the risk of sporadic systemic lupus erythematosus and the occurrence of antiphospholipid antibodies. Hum Genet 115:393–398 [DOI] [PubMed] [Google Scholar]
- Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN (2003) Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med 197:777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MH, Boyartchuk V, Wong S, Karaghiosoff M, Ragimbeau J, Pellegrini S, Muller M, Dietrich WF, Yap GS (2003) A natural mutation in the TYK2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. Proc Natl Acad Sci USA 100:11594–11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart TA (2003) Neutralizing interferon alpha as a therapeutic approach to autoimmune diseases. Cytokine Growth Factor Rev 14:139–154 [DOI] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25:1271–1277 [DOI] [PubMed] [Google Scholar]
- Tsao BP (2004) Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol 16:513–521 [DOI] [PubMed] [Google Scholar]
- Wallace DJ, Hannahs BH (eds) (2002) Dubois' Lupus Erythematosus. Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Yeh TC, Dondi E, Uze G, Pellegrini S (2000) A dual role for the kinase-like domain of the tyrosine kinase TYK2 in interferon-α signaling. Proc Natl Acad Sci USA 97:8991–8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737 [DOI] [PubMed] [Google Scholar]