A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility (original) (raw)
Abstract
A major goal of cancer research has been to identify genes that contribute to cancer formation. The similar pathology between zebrafish and human tumors, as well as the past success of large-scale genetic screens in uncovering human disease genes, makes zebrafish an ideal system in which to find such new genes. Here, we show that a zebrafish forward genetic screen uncovered multiple cell proliferation mutants including one mutant, crash&burn (crb), that represents a loss-of-function mutation in bmyb, a transcriptional regulator and member of a putative proto-oncogene family. crb mutant embryos have defects in mitotic progression and spindle formation, and exhibit genome instability. Regulation of cyclin B levels by bmyb appears to be the mechanism of mitotic accumulation in crb. Carcinogenesis studies reveal increased cancer susceptibility in adult crb heterozygotes. Gene-expression signatures associated with loss of bmyb in zebrafish are also correlated with conserved signatures in human tumor samples, and down-regulation of the B-myb signature genes is associated with retention of p53 function. Our findings show that zebrafish screens can uncover cancer pathways, and demonstrate that loss of function of bmyb is associated with cancer.
Keywords: cell cycle
Zebrafish is an ideal system for cancer gene discovery owing to its combination of forward genetics and long-standing use as a carcinogenesis model (1, 2). Zebrafish exposed to carcinogens develop a wide variety of tumors in virtually all organs with a histology closely resembling that of human tumors. A genetic link to cancer in fish was established very early on when the fish species Xiphophorus was shown to have a hereditary susceptibility to melanomas (3, 4). More recent studies in zebrafish have established that transgenic expression of human oncogenes can produce strong tumor phenotypes (5, 6). Previous work establishes that zebrafish holds great potential as both a tool to model human disease and as a genetic device that can be used to dissect pathways involved in cancer.
Although no random mutagenesis screens had been previously designed to dissect tumor pathways, previous work in zebrafish has laid the groundwork for the genetic analysis of the cell cycle and cancer formation. Large-scale forward-genetic screens for embryonic lethal mutations have led to the identification of zebrafish lines with increased tumor rates (7). These screens have led to the development of more specific screens designed to dissect particular pathways. Here, we report a screen designed to identify cell-cycle and cancer-susceptibility mutants in the zebrafish. By taking advantage of the established close relationship between tumorigenesis in zebrafish and humans, we sought to identify novel or known genes important for tumor formation.
One gene identified in our screen, whose role in tumorigenesis is not yet fully understood, is B-myb. B-myb is part of a small family of transcription factors with characteristic regions of homology that includes the c-myb proto-oncogene (8). Mammalian studies have shown that B-myb plays a role in cell-cycle regulation, particularly at the G1/S transition. The E2F family of transcription factors directs transcription of B-myb to this phase (9), and B-myb, in cooperation with cyclins, accelerates the G1/S transition (8). In contrast to vertebrates, a single myb gene has been found in Drosophila (10). The dm-myb gene regulates the G2/M transition, maintains normal ploidy and spindle formation, and plays a role in DNA synthesis (11–15). Interestingly, recent evidence has shown that knockdown of human B-myb leads to a reduction of cyclin B and cdc2 expression, suggesting that mammalian B-myb may be involved in G2/M regulation as well (16). Our identification of zebrafish bmyb from a cell proliferation screen has demonstrated that vertebrate B-myb is involved in the regulation of the G2/M transition and maintenance of normal ploidy through potential transcriptional regulation of cyclin B. We also demonstrate that haploinsufficient mutation of zebrafish bmyb leads to an increased rate of tumors after carcinogenesis. Finally, we establish that the gene signature associated with zebrafish bmyb loss is conserved in human tumors, suggesting that loss of human B-myb may also contribute to human cancer.
Materials and Methods
The material and methods used for genetic mapping/positional cloning, embryo injection experiments, immunohistochemistry/in situ hybridization, crb restriction fragment length polymorphism genotyping, DNA content analysis, cytogenetics, and microarray analysis can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Cell Proliferation Screen. Adult male zebrafish from the wik strain were mutagenized by three 1-hr exposures to 3.0 mM ethylnitrosourea as described in ref. 17. The specific-locus frequency of mutation was 1 × 10-3 as indicated by the frequency of non-mosaic mutant progeny for the albino mutation. Beginning 3 weeks after mutagenesis, the males were outcrossed to wild-type wik females to generate F1 heterozygotes. At ≈4–6 months of age, eggs were expressed from anesthetized F1 females and fertilized in vitro with UV-inactivated sperm to generate haploid embryos as described in ref. 18. Thirty-six-hpf (hours postfertilization) haploid embryos were used for whole-mount immunohistochemistry with anti-phosphohistone H3 (pH3). Females giving rise to at least 40% embryos with an abnormal pH3 pattern were outcrossed, and F2 diploid intercrosses were performed to recover the mutation.
Carcinogenesis. Twenty-eight-day-old fry from backcrosses of mutant heterozygotes were exposed to 1.5 ppm _N_-methyl-_N_′-nitro-_N_-nitrosoguanidine (MNNG) for 24 hr (19). Fish were killed at 3, 6, or 12 months posttreatment, three H&E-stained sagittal step sections from each fish was examined, and each animal was genotyped with an restriction fragment length polymorphism specific to the mutation. A two-tailed Fisher exact test was used to assess whether tumor susceptibility differed between crb heterozygotes and wild-type zebrafish. All time points were combined for this calculation.
Microarray Analysis. Detailed methods are available as part of the Supporting Text, and additional data and information about the signature analysis is available at www.broad.mit.edu/cancer/pub/bmyb.
Results and Discussion
To identify embryonic cell proliferation mutants, we screened the haploid progeny of 750 F1 females obtained from the mating of ethylnitrosourea-mutagenized parents. Clutches were screened for proliferation defects at 36 hpf by using an antibody against serine-10 pH3, a marker of mitotic cells. Eight independent homozygous lethal recessive mutants (death occurs at day 4–5) were identified in the screen, including mutants with increased numbers of mitotic cells (crash&burn, self-destruct, and lollygag), decreased numbers of mitotic cells (standstill, dillydally, sleepyhead, and shortstop), and changes in the size of the mitotic cells (cease&desist) (one example of each class shown in Figs. 1_A_ and 5, which is published as supporting information on the PNAS web site). Our data demonstrate that cell-cycle screens in zebrafish are capable of identifying mutants with a wide range of proliferation phenotypes.
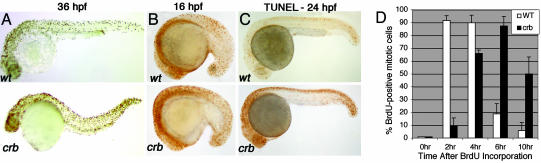
Fig. 1.
A forward genetic screen in zebrafish identified a proliferation mutant, crash&burn.(A) Depiction of the cell-proliferation phenotype (by pH3 staining) of one of the eight mutants isolated in the screen, crash&burn (crb). Embryos are 36-hpf diploids. (B) pH3 staining of crb mutant diploid embryos at 16 hpf. (C) crb has increased numbers of apoptotic cells at 24 hpf as shown by TUNEL staining. (D) Time course of crb and wild type progression from S phase into G2/Mas shown by BrdUrd incorporation followed by pH3 analysis at varying time points after incorporation.
One mutant, crash&burn (crb), has increased pH3 staining observable as early as 16 hpf (Fig. 1_B_) and a 4-fold increase in mitotic cell number by 36 hpf. Increased numbers of apoptotic cells were also found at 24 hpf, as was a decreased number of cells in S phase as assessed by 5-bromo-2-deoxyuridine (BrdUrd) incorporation at 28 hpf (Figs. 1_C_ and 5). The finding of increased pH3 and decreased cell proliferation (BrdUrd incorporation) suggests that cells in crb are delayed or arrested in mitosis.
To examine the cell-cycle kinetics of crb cells from S to M phase and determine whether crb cells are arrested in mitosis, we performed a BrdUrd pulse–chase experiment and double-stained the embryos with anti-BrdUrd antibody and pH3 antibody. Examining the timing of when BrdUrd-positive cells become pH3-positive allows calculation of the length of time between S phase and mitosis in crb as compared with wild types (Fig. 1_D_). Cells in wild-type embryos begin to enter G2/M at 2 hr after BrdUrd labeling and exit from mitosis at 6 hr. Cells in crb embryos do not enter M phase until 4 hr after BrdUrd labeling, demonstrating that the crb cells enter G2/M more slowly than wild-type cells. Fifty percent of crb cells remain in mitosis 10 hr after incorporation. These data suggest that the aberrant accumulation of pH3-positive cells in crb at 16 hpf is a result of the inability of these cells to exit G2/M.
To uncover the molecular origin of the crb phenotype, the crb gene was mapped and localized to a ≈5-cM interval on linkage group 11 (Fig. 2_A_). A chromosomal walk was initiated from a closer zebrafish EST marker, fc17h09, and this led to the identification of a gene, bmyb, on one of the BAC clones in the critical interval. Sequence analysis of the crb mutant locus identified a T-to-G mutation in the invariant GT of the splice donor site after exon 13 in the bmyb gene (Fig. 2_B_). This mutation leads to aberrant splicing and the use of a cryptic splice donor site 56 nt downstream in the intron causing a frameshift and the replacement of the last 42 aa of zebrafish Bmyb with a novel 38-aa sequence (Figs. 2_B_ and 6, which is published as supporting information on the PNAS web site). To verify that a defect in the zebrafish bmyb gene causes the crb phenotype, we injected wild-type zebrafish bmyb RNA into crb mutant embryos. After injection, genotypic mutants showed full or partial rescue of the increased mitotic index seen in crb mutants [n = 20/70 (13 full and 7 partial), P < 0.0001] (Fig. 2_C_). Targeted gene knockdowns of bmyb in wild-type embryos by using antisense morpholino-modified oligonucleotides (morpholinos) (20) (targeted to the exon 13 splice donor or the ATG) caused the increased pH3 phenotype seen in the crb mutants as well as the characteristic small head and increased apoptosis (Fig. 2_C_). These data establish that loss of bmyb is responsible for the crb phenotype and suggest that zebrafish B-myb plays a role in G2/M progression.
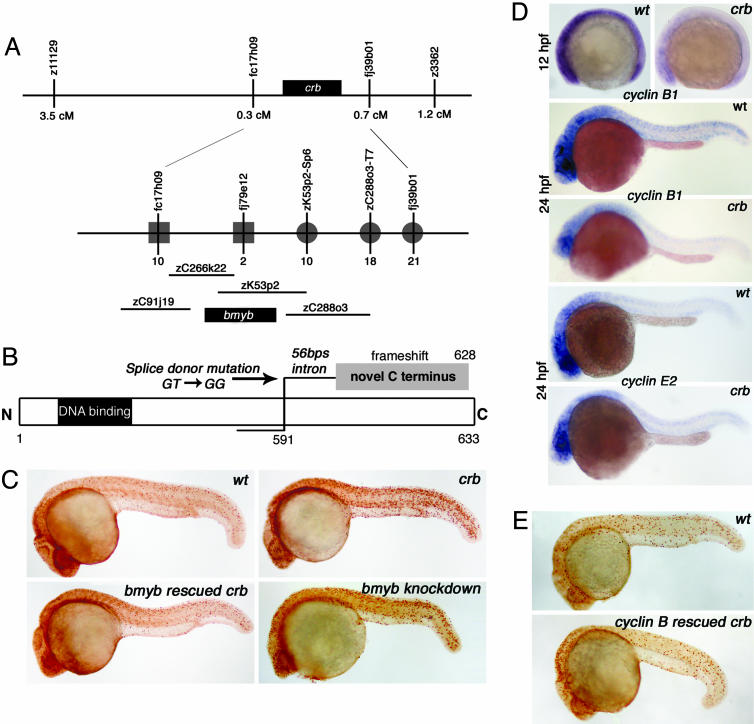
Fig. 2.
bmyb is the crb gene and may act through regulation of cyclin B. (A) crb positional cloning schematic with an enlarged view of the crb locus that depicts the BAC genomic clones identified in a chromosomal walk in analysis of 3,068 meioses from diploid mutants. The number of recombination events identified from the distal side (squares) and proximal side (circles) are indicated. (B) Schematic representation of the mutation in bmyb found in the crb mutants. Splice donor mutation (GT to GG) occurred between exon 13 and exon 14 (just after amino acid 591) causing a frameshift in the protein. (C) Phenotypic rescue (assessed by pH3 staining) of 24-hpf crb embryos by injection of bmyb mRNA and phenocopy of crb by injection of bmyb morpholinos. (D) Whole-mount embryo mRNA in situ hybridization analysis for cyclin B1 and cyclin E2. (E) Phenotypic rescue of 24 hpf crb embryos by injection of cyclin B1 mRNA.
Because bmyb is a transcription factor, the cell-cycle defect in crb may be due to abnormal gene expression of critical regulators of the G2/M transition. Levels of cyclin B1 mRNA in crb embryos are greatly reduced at 12 hpf, before the crb pH3 phenotype is apparent, and at 24 hpf (Fig. 2_D_). Levels of cyclin E2 are unchanged, strongly suggesting that cyclin B1 is a specific target of bmyb (Fig. 2_D_). To support a major role for cyclin B, injection of cyclin B1 mRNA into embryos caused a full or partial rescue of the pH3 phenotype of crb mutant 24 hpf embryos (n = 12/64) (Fig. 2_E_). This finding establishes a model in which reduction of cyclin B1 levels leads to at least some of the crb defects. This finding supports evidence in both plants and flies that Myb genes play a role in cyclin B expression (21–23). In fact, recent evidence shows that knockdown of B-myb in human cells leads to a reduction of cyclin B and cdc2 expression demonstrating that B-myb plays a role in G2/M regulation in mammals (16). Our evidence shows that reduced cyclin B levels cause the accumulation of crb cells in G2/M and a delay in mitotic progression.
To examine crb for mitotic abnormalities, whole-mount embryos were stained with antibodies against α-tubulin and γ-tubulin. Spindle abnormalities were seen in 64% of cells in crb, 44% in the bmyb splice knockdown, and 77% in the bmyb ATG knockdown (Fig. 7, which is published as supporting information on the PNAS web site). These abnormalities include monopolar spindles originating from a single centrosome or two unseparated centrosomes (Fig. 3_A_). In addition to monopolar spindles, crb embryos also have cells with multipolar or disorganized spindles (Fig. 3_A_). Our studies indicate that the loss of bmyb in zebrafish has dramatic effects on spindle formation that will likely lead to catastrophic mitoses.
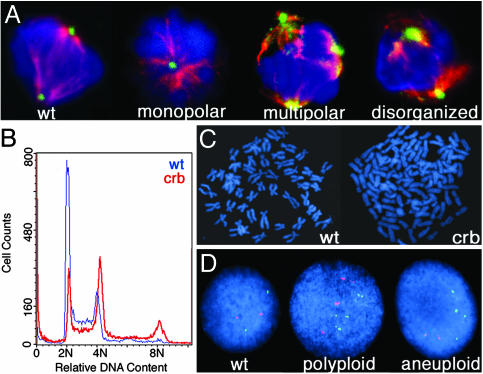
Fig. 3.
crb embryos exhibit abnormal mitoses and genomic instability. (A) Mitotic cells shown stained with DAPI (blue), α-tubulin (red), and γ-tubulin (green). (B) DNA content of crb (red) and wild-type (blue) embryos as measured by flow cytometry. (C) Metaphase chromosome spreads from a diploid cell containing 50 chromosomes (Left) and a crb hypertetraploid cell containing 108 chromosomes (Right). (D) FISH analysis on crb mutant embryos with probes for linkage group 2 (red) and linkage group 16 (green) and DAPI counterstaining.
Defects in the mitotic spindle and centrosome number have been associated with genomic instability and aneuploidy in mammalian cells and are often observed in cancer cells (24). Whole-embryo DNA-content FACS analysis indicated that as compared with wild-type siblings, mutant crb embryos have a higher fraction of 4N cells and have a proportion of cells with 8N DNA content (Fig. 3_B_). Metaphase spreads demonstrate that crb mutants contain polyploid and hypertetraploid cells (Fig. 3_C_). Further evidence of aneuploidy was obtained from interphase fluorescent in situ hybridization (FISH), which showed that crb embryos contain 30% polyploid and 23% aneuploid cells versus 0% and 4%, respectively, in wild types (n = 100 cells) (Fig. 3_D_). Because aneuploidy is thought to contribute to malignant transformation and tumor progression, these data provide a mechanism for the role of bmyb loss-of-function in cancer formation (24).
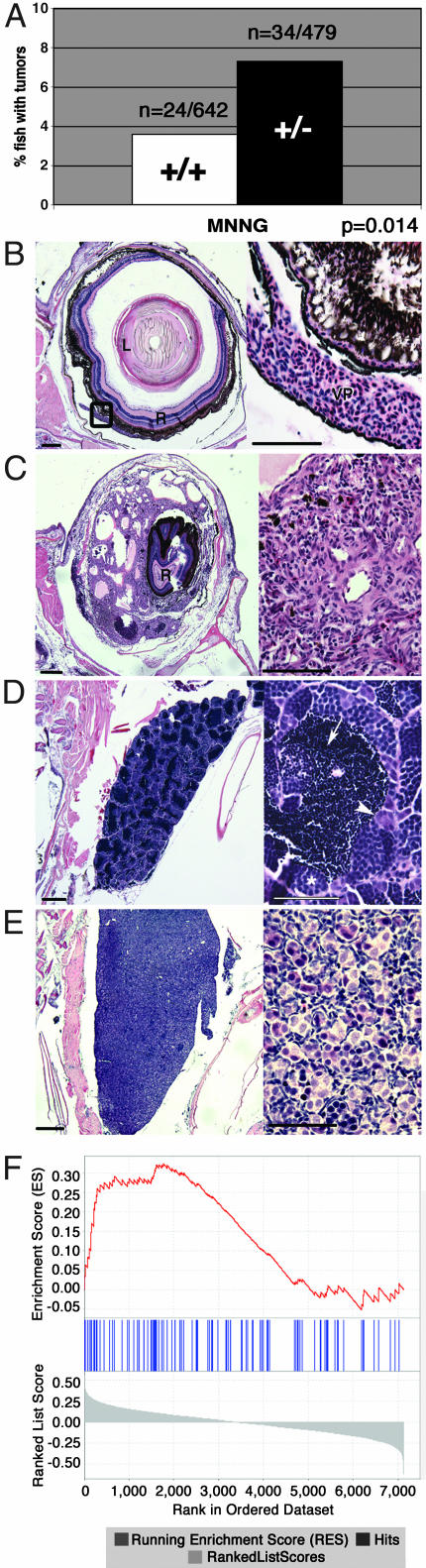
We next asked whether the bmyb mutation affected cancer susceptibility in adults. Because there was no increase in spontaneous tumor rate in the crb heterozygotes at 1 year of age, 28-day-old zebrafish (heterozygotes and wild-type siblings) were treated with the carcinogen MNNG, which is known to accelerate the development of tumors in fish (19). At 3, 6, or 12 months after exposure, the 1,121 treated fish were killed, and serial step sections of these fish were examined for the presence of tumors. crb heterozygotes have an ≈2-fold increase (3.7–7.1%) in cancer susceptibility relative to wild-type siblings after MNNG exposure (Fig. 4_A_; P = 0.014). Vascular tumors and testicular germ cell tumors are the most common tumor types induced by MNNG (Fig. 4 B_–_E), although various tumor types are seen (Fig. 8, which is published as supporting information on the PNAS web site). The tumor spectra of crb heterozygotes and wild types were not significantly different (Fig. 9, which is published as supporting information on the PNAS web site). We performed quantitative real-time PCR analysis to determine whether the tumors arose from a loss of the wild-type bmyb allele (25, 26). In analysis of DNA from 17 tumors from MNNG-treated crb heterozygotes, no loss of heterozygosity was detected (Fig. 10, which is published as supporting information on the PNAS web site). These data suggest that bmyb acts as a haploinsufficient tumor suppressor in zebrafish.
Fig. 4.
crb adult heterozygotes have an increased susceptibility to cancer. (A) Percentage of wild-type or crb heterozygous fish that developed tumors after MNNG treatment. (B_–_E) The most common tumors seen in crb heterozygotes after MNNG treatment (C and E) and the corresponding wild-type structures (B and D). (Scale bars: Left, 200 μm; Right,50 μm.) (B) Sagittal section through normal zebrafish eye. Lens (L), retina (R), and normal choroidal vascular plexus (VP) are shown. (C) Vascular neoplasm arising in the retinal vascular plexus and pushing the retina to the center of the eye. The tumor is composed of plump, atypical endothelial cells forming vascular channels. (D) Sagittal section through normal zebrafish testis containing spermatogonia located at the tubule periphery (white arrowhead), spermatocytes with more condensed nuclei (white asterisk), and fully differentiated spermatozoa (white arrow). (E) Testicular germ cell tumor where the lobular architecture of the testis is completely effaced by a massive proliferation of undifferentiated spermatogonia. Spermatocytes and spermatozoa have been replaced by the tumor. (F) Enrichment plot of the B-myb signature genes in the human lung adenocarcinoma data set. The plot shows the location of the B-myb signature genes (blue bars) in the gene list ranked by the p53 phenotype. The running enrichment score as a function of position in the gene list is shown in red. The signal-to-noise scores of all 7,129 genes in the gene list are shown in gray, with positive scores representing markers for the p53 mutant class and negative scores representing markers for the p53 wild-type class. The signature genes are clearly overrepresented at the left side of the gene list, representing their enrichment in the p53 mutant class.
Our data demonstrating that B-myb acts as a tumor suppressor in a vertebrate system led us to explore a potential role for B-myb in human tumors using gene-expression profiling. Microarray profiles were generated from wild-type and crb mutant 24-hpf embryo pools (n = 3 and 3, respectively), and the signal-to-noise metric was used to rank genes by their differential expression in wild-type and crb mutant classes. To extract a signature of B-myb loss, the 500 unique genes whose expression was most decreased in the crb mutant were selected. Confirming the uniqueness of the B-myb signature, microarray analysis of other zebrafish cell-cycle mutants reveals signatures that are distinct from crb (data not shown). Mapping of genes between zebrafish and humans yielded a set of 198 human homologues that was used as the B-myb loss-of-function signature (Table 1, which is published as supporting information on the PNAS web site). No human homologues for the remaining 302 zebrafish genes could be conclusively identified. This list of 198 genes will allow us to determine whether a similar signature exists in human cancers, indicating conservation of this pathway.
We used an analytical procedure, Kolmogorov–Smirnov scanning (KSS) (27), that can detect subtle correlations to search for conservation of the signature in humans. Because it is possible that only a subset of human cancers display B-myb activity, this analysis was performed in a large database of diverse human cancers (190 samples and 14 types). Using the Global Cancer Map (GCM) to perform KSS (28), we found that the B-myb signature is significantly correlated with the expression of both B-myb (MYBL2) and cyclin B1 (CCNB1) in human cancers. To determine this, the expression of a gene across all 190 samples in the GCM was used to rank order all other genes in the data set, producing 16,063 ordered gene lists. Using the Kolmogorov–Smirnov (KS) nonparametric statistic to quantify the position of the B-myb gene set within each ordered gene list, we found that the KS score for B_-myb_ ranked very highly: only 94 genes from the 16,063 profiles tested scored better. Cyclin B (CCNB1) was the 86th-ranked gene, thereby demonstrating that its activity was correlated both with B_-myb_ as well as the B-myb signature gene set (Table 2, which is published as supporting information on the PNAS web site). To estimate the statistical significance of the observed KS score for B_-myb_, we randomly permuted the expression profile of the index gene, while keeping the other profiles in the data set as well as the signature gene set fixed. Applying this procedure by permuting the B_-myb_ expression profile 10,000 times, we obtained a P value of 0.033. We conclude that our zebrafish model indeed recapitulates B-myb activity seen in human cancers.
To examine a clinical outcome association, the B-myb signature was used to probe a collection of 14 microarray data sets of human cancer by using the Gene Set Enrichment Analysis (GSEA) method (29). GSEA is a method for determining whether a rank-ordered list of genes for a particular comparison of interest (e.g., tumor vs. normal) is enriched in genes derived from an independently generated gene set (e.g., B-myb model-derived marker genes). A statistically significant enrichment of the B-myb signature genes (i.e., up-regulation of the signature genes that are down-regulated in crb mutants) was found in p53 mutant lung adenocarcinomas, the only data set among the 14 for which p53 functional information was available (nominal P = 0.007, family-wise error rate P = 0.015) (Table 3, which is published as supporting information on the PNAS web site, and Fig. 4_F_). To confirm the B-_myb_-p53 association, we examined the gene expression profiles of the NCI-60 panel of human cancer cell lines for which p53 mutational status is also known (database from Novartis, http://dtp.nci.nih.gov/mtargets/mt_index.html). Again, we observed enrichment of the B-myb signature in p53 mutant lines (P = 0.019), consistent with the notion that B-myb loss of function (inferred by absence of the B-myb signature) is preferentially observed in p53 wild-type cells. To confirm that the enrichment of B-myb signature genes in p53 mutant cells is not merely a result of common gene expression changes, we identified the signature of zebrafish p53 mutants (30) and demonstrated that this gene list was significantly different from the B-myb signature (Fig. 11, which is published as supporting information on the PNAS web site). Although initial sequencing of cDNAs from 52 human tumor cell lines revealed no mutations in the human B-myb gene (data not shown), the strong correlation seen in the gene-expression profiling suggests a model in which decreased levels of B-myb could contribute to tumor formation in the context of wild-type p53. This involvement of B-myb in tumorigenesis could occur by various mechanisms. The strong effect of a p53 mutation may mask the contribution of B-myb deficiency to tumorigenesis. Alternatively, gene-expression changes caused by p53 mutation during the genesis of a tumor may lead to loss of the B-myb signature (i.e., upregulation of the signature genes). The absence of gene-expression-profile overlap between the B-myb and p53 mutants suggests that there may not be a direct regulatory interaction between these two pathways. Elucidation of this novel B-myb/p53 correlation would provide further insight into the pathways involved in tumor formation.
Our studies have shown that a screen for cell-proliferation mutants in zebrafish can identify embryonic cell-cycle mutants with varying phenotypes including a predisposition to cancer as adults. Although overexpression data from human tumors suggest that B-myb acts as an oncogene (31), our work in zebrafish suggests that genes that play a role in mitotic progression may affect cancer in both zebrafish and humans. Studies in humans and mice have shown that inactivation of the MAD2 tumor suppressor can cause similar defects as seen in our crb embryos, including mitotic defects and chromosomal instability (32, 33). Recent work has shown that overexpression of Mad2 in the context of Rb mutation also causes mitotic defects and aneuploidy (34). These findings suggest that genes controlling the mitotic checkpoint may affect tumor formation through either inactivation or overexpression. It is also interesting to note that E2F1, which is a transcriptional regulator of B-myb, has also been shown to contribute to tumorigenesis through either inactivation or overexpression (35, 36). Recent findings have also shown that proteins associated with Drosophila Myb participate in the Rb pathway, suggesting that exploring connections between the B-myb and Rb pathways in human cancer will be fruitful (37). The genetic tractability of zebrafish and their physiological similarity to humans make them an ideal system to fully explore the complex pathways that underlie cancer.
Supplementary Material
Supporting Information
Acknowledgments
We thank Ryan Murphey and Christian Straub for feedback and immunohistochemistry assistance; Cassandra Belair for laser-capture microscopy; Amanda Smith for technical assistance with cytogenetic analysis; Adam Amsterdam for providing cyclin B embryos; Daphne Bell and Daniel Haber for supplying the human tumor cell lines cDNAs; and William Kaelin, Nicholas Dyson, and Caroline Erter Burns for critical review of the manuscript. This work was supported by National Institutes of Health Grants 1R01 DK55381 (to L.I.Z.), 1R01 HD044930 (to L.I.Z.), K08 HL04082 (to J.F.A.), and 5K08 DK061849 (to H.M.S.), and by the Albert J. Ryan Fellowship (to J.L.S. and K.L.P.). L.I.Z. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: J.L.S., J.F.A., and L.I.Z. designed research; J.L.S., J.F.A., H.M.S., D.F., J.Z., K.R.F., K.L.P., C.H., B.B., and M.F. performed research; J.L.S. and J.S. contributed new reagents/analytic tools; J.L.S., J.F.A., H.M.S., A.S., Y.Z., C.L., D.N., G.W., T.R.G., J.N.G., J.L.K., and J.C.A. analyzed data; and J.L.S. and A.S. wrote the paper.
Abbreviations: hpf, hours postfertilization; MNNG, _N_-methyl-_N_′-nitro-_N_-nitrosoguanidine; pH3, phosphohistone H3.
References
- 1.Amatruda, J. F., Shepard, J. L., Stern, H. M. & Zon, L. I. (2002) Cancer Cell 1**,** 229-231. [DOI] [PubMed] [Google Scholar]
- 2.Stern, H. M. & Zon, L. I. (2003) Nat. Rev. Cancer 3**,** 533-539. [DOI] [PubMed] [Google Scholar]
- 3.Nairn, R. S., Kazianis, S., McEntire, B. B., Della Coletta, L., Walter, R. B. & Morizot, D. C. (1996) Proc. Natl. Acad. Sci. USA 93**,** 13042-13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schartl, M. (1995) Trends Genet. 11**,** 185-189. [DOI] [PubMed] [Google Scholar]
- 5.Patton, E. E., Widlund, H. R., Kutok, J. L., Kopani, K. R., Amatruda, J. F., Murphey, R. D., Berghmans, S., Mayhall, E. A., Traver, D., Fletcher, C. D., et al. (2005) Curr. Biol. 15**,** 249-254. [DOI] [PubMed] [Google Scholar]
- 6.Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., Lin, S., Prochownik, E., Trede, N. S., Zon, L. I. & Look, A. T. (2003) Science 299**,** 887-890. [DOI] [PubMed] [Google Scholar]
- 7.Amsterdam, A., Sadler, K. C., Lai, K., Farrington, S., Bronson, R. T., Lees, J. A. & Hopkins, N. (2004) PLoS Biol. 2**,** E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joaquin, M. & Watson, R. J. (2003) Cell Mol. Life Sci. 60**,** 2389-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam, E. W.-F., Robinson, C. & Watson, R. J. (1992) Oncogene 7**,** 1885-1890. [PubMed] [Google Scholar]
- 10.Katzen, A. L., Kornberg, T. B. & Bishop, J. M. (1985) Cell 41**,** 449-456. [DOI] [PubMed] [Google Scholar]
- 11.Katzen, A. L., Jackson, J., Harmon, B. P., Fung, S. M., Ramsay, G. & Bishop, J. M. (1998) Genes Dev. 12**,** 831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung, S. M., Ramsay, G. & Katzen, A. L. (2002) Development (Cambridge, U.K.) 129**,** 347-359. [DOI] [PubMed] [Google Scholar]
- 13.Beall, E. L., Manak, J. R., Zhou, S., Bell, M., Lipsick, J. S. & Botchan, M. R. (2002) Nature 420**,** 833-837. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick, C. A., Sharkov, N. V., Ramsay, G. & Katzen, A. L. (2002) Development (Cambridge, U.K.) 129**,** 4497-4507. [DOI] [PubMed] [Google Scholar]
- 15.Manak, J. R., Mitiku, N. & Lipsick, J. S. (2002) Proc. Natl. Acad. Sci. USA 99**,** 7438-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu, W., Giangrande, P. H. & Nevins, J. R. (2004) EMBO J. 23**,** 4615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solnica-Krezel, L., Schier, A. F. & Driever, W. (1994) Genetics 136**,** 1401-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corley-Smith, G. E., Brandhorst, B. P., Walker, C. & Postlethwait, J. H. (1999) Methods Cell Biol. 59**,** 45-60. [DOI] [PubMed] [Google Scholar]
- 19.Spitsbergen, J. M., Tsai, H. W., Reddy, A., Miller, T., Arbogast, D., Hendricks, J. D. & Bailey, G. S. (2000) Toxicol. Pathol. 28**,** 716-725. [DOI] [PubMed] [Google Scholar]
- 20.Nasevicius, A. & Ekker, S. C. (2000) Nat. Genet. 26**,** 216-220. [DOI] [PubMed] [Google Scholar]
- 21.Ito, M. (2000) Plant Mol. Biol. 43**,** 677-690. [DOI] [PubMed] [Google Scholar]
- 22.Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., Doonan, J. H. & Watanabe, A. (2001) Plant Cell 13**,** 1891-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, M., Akimaru, H., Hou, D. X., Takahashi, T. & Ishii, S. (2002) EMBO J. 21**,** 675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer, A., Neben, K. & Ho, A. D. (2002) Leukemia 16**,** 767-775. [DOI] [PubMed] [Google Scholar]
- 25.Reis, P. P., Rogatto, S. R., Kowalski, L. P., Nishimoto, I. N., Montovani, J. C., Corpus, G., Squire, J. A. & Kamel-Reid, S. (2002) Oncogene 21**,** 6480-6487. [DOI] [PubMed] [Google Scholar]
- 26.Berggren, P., Kumar, R., Sakano, S., Hemminki, L., Wada, T., Steineck, G., Adolfsson, J., Larsson, P., Norming, U., Wijkstrom, H. & Hemminki, K. (2003) Clin. Cancer Res. 9**,** 235-242. [PubMed] [Google Scholar]
- 27.Ramaswamy, S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C. H., Angelo, M., Ladd, C., Reich, M., Latulippe, E., Mesirov, J. P., et al. (2001) Proc. Natl. Acad. Sci. USA 98**,** 15149-15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb, J., Ramaswamy, S., Ford, H. L., Contreras, B., Martinez, R. V., Kittrell, F. S., Zahnow, C. A., Patterson, N., Golub, T. R. & Ewen, M. E. (2003) Cell 114**,** 323-334. [DOI] [PubMed] [Google Scholar]
- 29.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34**,** 267-273. [DOI] [PubMed] [Google Scholar]
- 30.Berghmans, S., Murphey, R. D., Wienholds, E., Neuberg, D., Kutok, J. L., Fletcher, C. D., Morris, J. P., Liu, T. X., Schulte-Merker, S., Kanki, J. P., et al. (2005) Proc. Natl. Acad. Sci. USA 102**,** 407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sala, A. & Watson, R. (1999) J. Cell Physiol. 179**,** 245-250. [DOI] [PubMed] [Google Scholar]
- 32.Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. & Benezra, R. (2001) Nature 409**,** 355-359. [DOI] [PubMed] [Google Scholar]
- 33.Michel, L., Diaz-Rodriguez, E., Narayan, G., Hernando, E., Murty, V. V. & Benezra, R. (2004) Proc. Natl. Acad. Sci. USA 101**,** 4459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernando, E., Nahle, Z., Juan, G., Diaz-Rodriguez, E., Alaminos, M., Hemann, M., Michel, L., Mittal, V., Gerald, W., Benezra, R., et al. (2004) Nature 430**,** 797-802. [DOI] [PubMed] [Google Scholar]
- 35.Yamasaki, L., Bronson, R., Williams, B. O., Dyson, N. J., Harlow, E. & Jacks, T. (1998) Nat. Genet. 18**,** 360-364. [DOI] [PubMed] [Google Scholar]
- 36.Pierce, A. M., Gimenez-Conti, I. B., Schneider-Broussard, R., Martinez, L. A., Conti, C. J. & Johnson, D. G. (1998) Proc. Natl. Acad. Sci. USA 95**,** 8858-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipsick, J. S. (2004) Genes Dev. 18**,** 2837-2844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information