Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates (original) (raw)
Abstract
Insertion of transposable elements is a major cause of genomic expansion in eukaryotes. Less is understood, however, about mechanisms underlying contraction of genomes. In this study, we show that retroelements can, in rare cases, be precisely deleted from primate genomes, most likely via recombination between 10- to 20-bp target site duplications (TSDs) flanking the retroelement. The deleted loci are indistinguishable from pre-integration sites, effectively reversing the insertion. Through human-chimpanzee-Rhesus monkey genomic comparisons, we estimate that 0.5%-1% of apparent retroelement “insertions” distinguishing humans and chimpanzees actually represent deletions. Furthermore, we demonstrate that 19% of genomic deletions of 200-500 bp that have occurred since the human-chimpanzee divergence are associated with flanking identical repeats of at least 10 bp. A large number of deletions internal to Alu elements were also found flanked by homologies. These results suggest that illegitimate recombination between short direct repeats has played a significant role in human genome evolution. Moreover, this study lends perspective to the view that insertions of retroelements represent unidirectional genetic events.
Current genome size in mammals and other eukaryotes has been greatly affected by massive amplifications of transposable elements (TEs) or retroelements throughout evolution (Brosius 1999; Kidwell 2002; Liu et al. 2003). In mammals, close to 50% of the genome is recognizably TE-derived (Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2004), and in some plant species, the figure is nearly 80% (SanMiguel et al. 1998; Li et al. 2004). The various classes of TEs and their distributions in genomes have been widely studied in many species (C. elegans Sequencing Consortium 1998; Baillie et al. 2004; Adams et al. 2000; Lander et al. 2001; Aparicio et al. 2002; Kidwell 2002; Waterston et al. 2002; Yu et al. 2002; Kirkness et al. 2003; Gibbs et al. 2004; Ma and Bennetzen 2004). In contrast, much less is known about mechanisms that attenuate genome size. Studies in plants have shown that retroelement-driven genome expansion is counteracted by deletions within retroelements, likely mediated by illegitimate recombination between short flanking segments of identity (Devos et al. 2002). Comparison of related rice genomes has also revealed that illegitimate recombination has deleted both retroelement-derived sequences and unique nuclear DNA (Ma and Bennetzen 2004).
A number of studies have documented the prevalence of small deletions and insertions (indels) in primate genomes (Britten et al. 2003; Liu et al. 2003; Watanabe et al. 2004), but there has been no genome-wide analysis to determine the molecular mechanisms that generate these events. Recent availability of the chimpanzee draft sequence has afforded the opportunity to analyze the spectrum of genomic deletions that have occurred in the last 5-6 million years of primate evolution. Moreover, a large-scale comparison of the human and chimpanzee genomes allows examination of the genomic stability of retroelement insertions, which are generally considered to be irreversible with no known mechanism for precise excision from the genome (Hamdi et al. 1999; Roy-Engel et al. 2001; Batzer and Deininger 2002; Salem et al. 2003a,b). Because of this “unidirectional” property, retroelements, particularly Alu elements, are widely viewed as ideal markers for human population genetic studies (Carroll et al. 2001; Roy-Engel et al. 2001; Batzer and Deininger 2002; Salem et al. 2003a) and elucidation of primate phylogenetic relationships (Hamdi et al. 1999; Salem et al. 2003b; Gibbons et al. 2004). In primates, Alu sequences are the most abundant family of retroelements, comprising >10% of the human genome (Lander et al. 2001; Batzer and Deininger 2002). While most of the 1 million Alu elements retrotransposed >40 million years ago, several thousand have integrated into the human genome since divergence from the great apes, and close to a thousand of the “youngest” _Alu_s are polymorphic (Carroll et al. 2001; Roy-Engel et al. 2001; Batzer and Deininger 2002; Salem et al. 2003a; Bennett et al. 2004). Most are associated with flanking direct repeats or target site duplications (TSDs) of 10-20 bp (Jurka 1997). In this study, we have obtained evidence that Alu elements can be precisely deleted from the genome via recombination between these flanking repeats. Similarly, a significant fraction of 200- to 500-bp deletions of nonrepetitive sequence have likely taken place due to recombination between short regions of identical sequence flanking the deleted fragment. We demonstrate that this fraction is much greater than expected if blunt-end joining were responsible for generating all these deletions. Our results are in agreement with a model of genomic deletion occurring both by nonhomologous and error-prone homology-driven mechanisms of DNA double-strand break repair (Helleday 2003).
Results and Discussion
Direct assessment of retroelement deletion frequency
During an analysis to identify TE insertions that occurred after divergence of human and chimpanzee, we detected some apparent insertional differences involving Alu elements of older subfamilies. The _Alu_Y subfamily is the only family known to have been active in the last few million years of human evolution (Batzer and Deininger 2002). However, we identified 187 Alu elements from older families such as _Alu_S and _Alu_J (98 in human and 89 in chimpanzee) that appeared to be insertional differences. This finding raised the possibility that at least some of these cases represent deletions in one species rather than new insertions in the other. To explore this possibility, scripted BLAST searches of the Rhesus macaque whole-genome shotgun trace archive were used to assess the ancestral state of apparent retroelement insertional differences in humans and chimpanzees (see Methods). It should be noted that our requirement that 75% of the totally 100-bp flanking sequence be free of known repeats resulted in only 8389 of 14,765 retroelement loci being tested, and therefore, we expect that our findings represent an underestimate of the overall level of precise deletion of retroelements.
Of 7120 human-chimp indel sites with accepted Rhesus trace matches, 7010 were identified as insertions by our criteria (see Methods). That is, the retroelement was absent in Rhesus. The other 110 sites were examined more closely. Fifty-two of these cases appeared to be rearrangements or multicopy regions in the Rhesus genome due to the existence of multiple Rhesus traces covering the region, some with and some without the retroelement. Three further cases with partial poor trace alignments were likely genomic rearrangements. The remaining 55 cases were subjected to more detailed analysis to confirm that the indel was a case of deletion in human or chimpanzee and not an insertion or other rearrangement.
Multiple sequence alignments of the human, chimpanzee, and Rhesus sequences were done in each of the 55 cases (reproduced in Supplemental data). Only one (no. 23) resulted from poor sequence quality in the chimpanzee assembly. Another (no. 51) was a tandemly duplicated L2 element. Four other cases (nos. 5, 13, 18, and 31) showed evidence of independent insertions in the same site or in sites only several base pairs apart. Independent insertions at the same site have been reported before (Conley et al. 2005).
The remaining 49 cases appeared to be retroelement deletions. Twelve cases, six in humans and six in chimpanzees, were imprecise deletions, removing sequence from older retroelements such as L2 and MIR. A similar case of imprecise Alu deletion has been previously reported (Edwards and Gibbs 1992). In each case, our 12 imprecise deletions had little or no similarity at the deletion breakpoints, suggesting a nonhomologous deletion mechanism as an explanation for these events.
Thirty-seven cases represented apparent precise deletion of previously retrotransposed sequence, and all cases but one were Alu elements. The one anomaly (case no. 6) was a polyadenylated sequence flanked by apparent TSDs. This is a fragment of a ∼340-bp sequence with ∼20 copies mutually ∼6%-10% divergent in the human genome, suggesting possible earlier mobilization as a retrotransposable element.
We found 36 cases of apparent precise deletions of Alu elements. The loss of the Alu was also associated with loss of one copy of the TSD, leaving behind the original, pre-integration site only. This observation raised the possibility that these deletions were mediated by recombination between the flanking identical regions. A possible example of precise Alu deletion on human chromosome 21 has been reported recently by Hedges et al. (2004), but the investigators considered Alu excision to be a remote possibility and instead favored other explanations. Unfortunately, there is no coverage of this region in the chimpanzee scaffolds. Furthermore, recent PCR analysis of human-chimpanzee indels on chimpanzee chromosome 22 revealed two precisely deleted Alu elements; however, sequences and positions of these events were not given. These deletions resulted in loss of the Alu and deletion of one of the TSD copies, leading the investigators to speculate that a homology-dependent recombination mechanism might be responsible for these deletions (Watanabe et al. 2004).
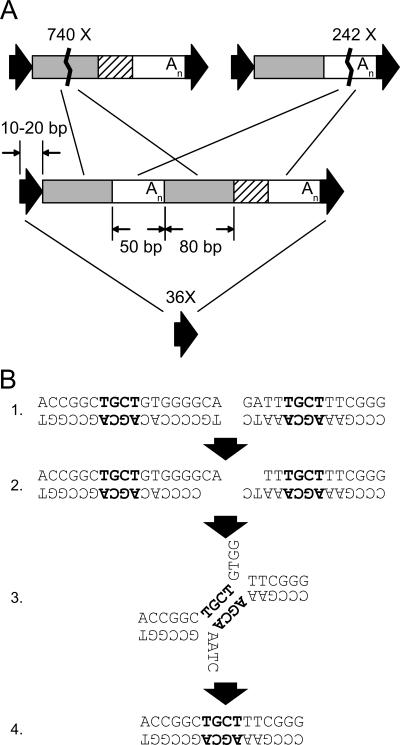
We reasoned that under a null hypothesis of deletions mediated by nonhomologous mechanisms, very few should be flanked by short identical segments. Instead, the majority of the 49 deletions (37 with flanking identical segments and 12 without) had identical regions of ≥10 bp. Compared with the null hypothesis, this association between deletion and flanking identical DNA was highly significant (P < 1e - 100; χ2 test). The skeptical reader could argue that we were only looking at deletions with breakpoints near retroelements, and therefore, we would be more likely to find breakpoints located within TSDs, even with a nonhomologous deletion mechanism. However, the likelihood of locating the breakpoints precisely at the same location within the TSD in the vast majority of the cases by random chance alone remains extremely small. Our findings strongly suggest that short, nonadjacent identical segments recombine, likely during double-strand break repair, to mediate deletion of these sequences. Consistent with this notion is the fact that at least 20-fold more deletions that involve _Alu_s are actually internal to Alu elements and have occurred between the ∼80-bp and 50-bp homologous regions internal to intact Alu elements (Fig. 1A; see Methods). These findings suggest that double-strand DNA breaks internal to _Alu_s are repaired by using the internal Alu homologies, obviating use of the flanking TSDs as repair templates and thus retaining remnants of the Alu element. The proposed mechanism of double-strand break repair is illustrated in Figure 1B, which shows a specific non-Alu small deletion in chimpanzee.
Figure 1.
Deletions due to DNA double-strand break repair. (A) Whole and partial Alu element deletions. A full-length Alu is shown in the middle, and black arrows represent target site duplications. Shaded and white internal regions represent internal ∼70% identical homologies. Deletions involving the 84-bp internal Alu homologies (shaded regions) were found 740 times in the human-chimpanzee alignments (top left). Alu internal deletions occurring between the other homologies (white regions) were found 242 times (top right). Precise deletion of entire Alu elements, likely involving the target site duplication (black arrows), was found in 36 cases (bottom) in relatively repeat-free regions since human-chimpanzee divergence. (B) A non-Alu deletion in chimpanzee at human chr1:1448280-1448311. Precise deletions of Alu elements, internal deletions within _Alu_s, and other deletions are explained by an error-prone homology-dependent repair mechanism, involving (1) a double-strand DNA break, (2) resection of DNA and exposure of 3′ tails, (3) homology search, and (4) ligation. In this case, a 4-bp homology mediated a 16-bp deletion.
Several of the apparent deletions from chimpanzee corresponded in human to human-specific Alu families, such as _Alu_Ya5 and _Alu_Yb8. However, in each case the corresponding element in Rhesus monkey shared identical TSDs and was also an _Alu_Y. Two explanations can account for this observation: multiple independent insertions at the identical site, or recent gene conversion in human which converted an existing older _Alu_Y insertion into an apparent human-specific family. Although we cannot rule out independent insertions as an explanation in these cases, we believe gene conversion, reported previously to occur between Alu elements (Salem et al. 2003a), is more likely. It should be noted that both deletion in the chimpanzee lineage and gene conversion in the human lineage, rather than controverting one another, are dual lines of evidence suggesting elevated recombinational or double-strand DNA break repair activity in these loci in recent evolutionary time.
We further noticed a relative paucity of precise deletions in human versus chimpanzee (only nine of 37 occurred in the human lineage). Without further study, it is unclear what this might mean. However, further BLAT alignments confirmed that, with the exception of two events (case no. 25, deleted in human, and case no. 43, deleted in chimpanzee), these events have all occurred in single-copy regions of the human and chimpanzee genomes. Furthermore, we used discontiguous megablast against the chimpanzee sequence trace database at National Center for Biotechnology Information (NCBI) to check for the possibility that some of the putative deletions in chimpanzee were a result of anomalous assembly, in which an _Alu_-containing trace at a locus was overruled by traces not containing the Alu. No such cases were found. By comparison, the numbers of random deletions between 200 and 500 bp long, discussed below, were more similar between human and chimpanzee (1011 and 916, respectively).
Analysis of random genomic deletion by illegitimate recombination
To further investigate the genomic prevalence of deletions that might be mediated by short repeats during the last few million years of primate evolution, we examined all length differences of 200-500 bp (thus approximating the 300-bp size of Alu elements) between human and chimpanzee, and looked for flanking repeats at the breakpoints. After eliminating cases of tandem duplications, insertions (including sequence having additional copies elsewhere in the human genome), indels within TEs, and deletions between homologous TEs (see Methods), 1927 indels remained, and we termed these random deletions. It should be noted that our method did not exclude genomic deletions having one or both breakpoints within repetitive sequence, as long as the repetitive sequence at the endpoints did not belong to homologous repeats. We found that the endpoints of 367, or 19.0%, of 200- to 500-bp random deletions in the human and chimpanzee lineages, are associated with flanking identical repeats of at least 10 bp.
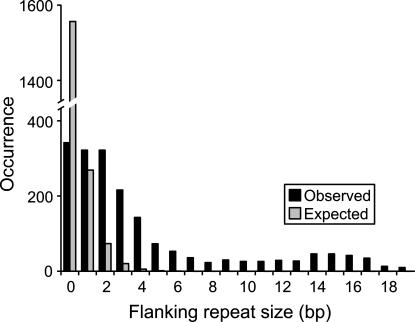
To put this observation in the context of nonrandom sequence composition in primate genomes, we attempted to measure the “background” density of nonadjacent homologies 200-500 bp apart occurring in nonrepetitive human genome sequence. Therefore, repetitive sequence recognized by RepeatMasker (A.F.A. Smit and P. Green, unpubl., http://www.repeatmasker.org) and tandem repeats found by Tandem Repeats Finder 3.21 (Benson 1999) were excised from the genome. This left 1.58 Gbp, or 55.6%, of the human genome. A C++ program was constructed that computed alignments between all genomic positions 200-500 bp apart. From the banded alignments, the program directly calculated the length distribution of randomly-occurring identical segments flanking sequence tracts 200-500 bp long. We then extrapolated the observed homology counts to compute the expected random homology occurrence in a complete genome. This method projected that 1.62 million random homologies of ≥10 bp would exist 200-500 bp apart in the full-size 2.84-Gbp human genome. The 376 random deletions that we observe with ≥10-bp flanking repeats therefore account for 0.0226% of all such homologies available in the genome. This observation again fits well within the paradigm of deletion-prone homology-driven DNA double-strand break repair, known as single-strand annealing (Karran 2000; Helleday 2003). In that model, DNA breakage results in binding of complexes that initiate peeling back of DNA, followed by a stochastic homology search in regions adjacent to the broken ends. In this type of DNA repair, many local homologies may be bypassed before fortuitous matching occurs. Exonucleases break down loose DNA ends, followed by ligation of the broken ends (Fig. 1B). This mechanism accounts for deletion sizes over several orders of magnitude (data not shown), and for varying flanking repeat sizes (Fig. 2).
Figure 2.
Prevalence of direct repeats at deletion boundaries; 1927 random deletions 200-500 bp in length were observed in the UCSC chimpanzee scaffold alignments to the July 2003 human genome. Observed flanking repeat occurrence (black bars) and expected occurrence if these deletions occurred by nonhomologous end joining alone (gray bars) are displayed. Flanking repeats ≥7 bp in size are expected to occur in less than one in 1927 cases.
As observed with Alu deletions, the observed association of random deletions with ≥10-bp flanking repeats appeared much greater than would occur if homology played no role. Indeed, the suggestion that nonadjacent homologies play a role in genomic deletions has also been made based on studies in plants, although no statistical analysis has been done (Devos et al. 2002; Ma and Bennetzen 2004). To statistically confirm a strong association between flanking repeats and deletion, our results were compared to what would be expected in a process of purely random breakage followed by blunt-end rejoining (Fig. 2). We reasoned that, under the hypothesis of no association between homology at breakpoints and deletion occurrence, homology occurrence at breakpoints of 200- to 500-bp deletions should mirror that observed 200-500 bp apart in the nonrepetitive genome. When using the data described above without extrapolation, 0.903 million randomly-occurring homologies occur in the nonrepetitive genome, wherein there exist 300 times as many, or 0.474 trillion, position combinations 200-500 bp apart. Thus ≥10-bp homologies occur randomly at a frequency of 1.9 × 10-6 of any two positions 200-500 bp apart. Therefore, if homology plays no role in these deletions, we would expect much less than one occurrence of ≥10-bp homology in our set of 1927 deletions (1927 × 1.9 × 10-6 = 0.0036 occurrences, precisely), compared with the observed 367 occurrences (P ≪ 1 × 10-100; χ2 test). Furthermore, by plotting the observed number of deletions associated with different lengths of flanking identity, we found that flanking repeats as short as 2 bp were over-represented in the data set (Fig. 2). This strong association of short flanking identities with deletion further confirms that illegitimate recombination between such short sequences has played a highly significant role in sequence deletion during primate evolution.
Direct confirmation of Alu element deletions
Finally, to confirm our findings, we chose nine cases of _Alu_S elements present in human but absent in the draft chimpanzee sequence to examine in more detail. These loci were chosen within and at varying distances from genes. To avoid regions of poor or anomalous alignments, we only investigated cases where the percentage identity between human and chimpanzee sequence surrounding the Alu is very high (>98%) and the Alu is a complete element with recognizable TSDs. Five of the cases (nos. 14, 33, 42, 43, and 52; see Supplemental information) were predicted to be deletions in chimpanzee, and as a control, we selected four cases expected to be insertions in human (nos. C1-C4). The presence or absence of each of these _Alu_s in a range of primate species was then determined by using genomic PCR and the results summarized in Table 1.
Table 1.
_Alu_S indels assayed in primates by PCR and BLAST
| Case no. | Fam. | Positiona | TSD (bp) | H | C | G | O | Gi | B | R | Location/nearest genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | Sx | 20:11512274 | 13 | Y | N | N | N | N | N | N | ∼354 kb 5′ of BTBD3 (BTB/POZ domain containing-3) |
| C2 | Sg | 15:83819720 | 16 | Y | N | N | N | N | N | N | In intron of AKAP13 (A-kinase anchor protein) |
| C3 | Sg | 7:104197804 | 19 | Y | N | N | N | I | N | N | ∼17 kb 5′ of MLL5 (Myeloid/lymphoid leukemia 5) |
| C4 | Sg | 20:18254452 | 15 | Y | N | N | N | N | N | N | ∼9.7 kb 5′ of ZNF133 (Kruppel Zn-finger protein) |
| 14 | Sg | 3:127318836 | 17 | Y | N | Y | Y | ? | ? | Y | ∼63 kb 3′ of KLF15 (Kruppel-like factor 15) |
| 33 | Sx | 12:48585272 | 16 | Y | N | Y | Y | Y | Y | Y | ∼1.3 kb 5′ of FAIM2 (Fas apoptotic inhibitory molecule 2) |
| 42 | Sq | 16:69279114 | 16 | Y | N | Y | Y | Y | Y | Y | ∼5.2 kb 3′ of CYB5-M (cytochrome b5) |
| 43 | Sx | 16:74232245 | 17 | Y | Y/Nb | Y | Y | Y | ? | Y | In intron of LOC348174 (secretory protein) |
| 52 | Sq | 22:45658137 | 15 | Y | N | Y | ? | Y | ? | Y | In intron of C22orf4 (putative GTPase activator) |
As expected, our four controls demonstrate _Alu_S presence only in human and no other primate, consistent with insertion in the human lineage after divergence from chimpanzee (Table 1; Fig. 3A). In accord with this finding is a study suggesting that some _Alu_Sx elements may still be active (Johanning et al. 2003). Therefore, some of the non-_Alu_Y differences between human and chimpanzee may reflect recent low levels of retrotranspositional activity of _Alu_S elements. An alternative explanation is that “young” _Alu_Y elements inserted in these locations, followed by gene conversion templated by older _Alu_S elements. We therefore more carefully examined these Alu sequences to look for nucleotide positions diagnostic of young _Alu_Y subfamilies (Batzer and Deininger 2002). Although we found no convincing evidence for partial gene conversion, this mechanism cannot be ruled out. Interestingly, in control no. 3, gibbon has an independent _Alu_Y insertion at this locus, offset by 4 bp (NCBI accession no. AY953324). Independent “parallel” retroelement insertions at or near the same genomic site have been previously noted (Salem et al. 2003a; Conley et al. 2005).
Figure 3.
PCR and sequence evidence for precise Alu element deletion. (_A_-F) Cases C4, 14, 33, 42, 52, and 43 from Table 1; lanes are human (H), chimpanzee (C), gorilla (G), orangutan (O), gibbon (Gi), baboon (Ba), and no-template control (-). (G) Case 43; genomic PCR in six additional chimpanzees, labeled _1_-6.
In the remaining five cases, PCR evidence confirms deletion in chimpanzee rather than lineage-specific insertion in human (Table 1). In four cases (nos. 14, 33, 42, and 52), the Alu element was found to be uniformly present in 10 of 10 humans and absent in 10 of 10 chimpanzee DNA samples (data not shown). These four regions are apparently unique in the human genome with no evidence of segmental duplication. Insertion of these Alu elements could be verified by PCR in orangutan, which diverged from the higher apes 12-15 million years ago (Glazko and Nei 2003), or in even more distantly related primates (Fig. 3B-E). (For case no. 14 in gibbon, the PCR product was of unexpected size [Fig. 3B] suggesting rearrangement or other insertions in the region.) Given these long periods of time, it is unlikely that these loci reflect lineage sorting of ancestral polymorphisms, proposed previously to explain unexpected Alu presence/absence relationships in the great apes (Salem et al. 2003b; Hedges et al. 2004). Rather, these results suggest that pre-existing fixed Alu elements have been deleted in the chimpanzee lineage. To verify that the loci in other primates contain the same Alu insertion, we sequenced the region in gorilla for case nos. 33 and 52 (NCBI accession nos. AY953323 and AY953322) and compared with the human, chimpanzee, and Rhesus macaque genomic sequences from the databases (Fig. 4A,B). In both cases, the gorilla and Rhesus loci are occupied by the same ancestral Alu as in human with the same TSD. Moreover, the sequence in chimpanzee has the expected structure of the pre-integration locus, with only one copy of the TSD generated upon Alu insertion.
Figure 4.
Sequence evidence for precise Alu element deletion. (A, B) Cases 33 and 52, sequenced in gorilla and compared with the database sequences of human, chimpanzee, and Rhesus macaque. (C) Case 43, showing available human, chimpanzee, and Rhesus loci. Target site duplications are boxed.
The final case (no. 43) is more complex in that chimpanzee appears to have both occupied and unoccupied alleles or loci (Fig. 3F). This pattern was seen in DNA from six of six additional chimpanzees tested (Fig. 3G), suggesting that it does not reflect allelic polymorphism. Indeed, database analysis revealed that this locus is part of complex segmental duplications that resulted in three copies in the human genome, all of which have the Alu insertion. The draft chimpanzee sequence has two copies, one of which lacks the Alu insertion. We cannot determine if a third copy exists in chimpanzee because of gaps and poor sequence coverage in these regions. An alignment of the three human and two chimpanzee sequences, as well as one Rhesus sequence is depicted in Figure 4C and shows that the chimpanzee locus without the Alu has the expected structure of a pre-integration allele. We confirmed the database entries by sequencing the two loci in chimpanzee (NCBI accession nos. AY953325 and AY953326). The most probable explanation for this finding is that the Alu integrated prior to duplication of the region followed by loss of the Alu in one chimpanzee copy.
Conclusions
In summary, our analysis strongly suggests an important role for short nonadjacent segments of DNA identity in genomic deletions. In rare cases, even retroelement insertions deeply fixed in the primate lineage can apparently be precisely excised from the genome in a manner involving the flanking TSDs, leaving behind no footprint of their insertion. We believe that illegitimate recombination between short identical stretches of DNA, likely involving a DNA double-strand break repair mechanism, is the most likely and simplest molecular mechanism to explain the findings reported here. This conclusion is supported by the fact that a large fraction of non-TE-associated deletions distinguishing human and chimpanzee have short repeats at the breakpoints. Furthermore, this study provides new insights into genomic attenuation and contradicts a rigid view that all insertions of retroelements represent unidirectional events. On the other hand, this study demonstrates that, for Alu elements in particular, homoplasy freedom is a mostly valid assumption and implicates internal homologous regions as preventing wholesale deletion of _Alu_s.
Finally, an aspect of Alu biology that has provoked interest is the slight preferential localization of younger elements in AT-rich regions but higher density of older elements in more GC-rich DNA (Lander et al. 2001). Several theories have been proposed to explain the differences in Alu distributions with element age (Schmid 1998; Brookfield 2001; Pavlicek et al. 2001; Medstrand et al. 2002; Jurka 2004;). While our findings indicate that precise deletion of Alu elements makes reversal of retroelement insertions possible, the phenomenon is nevertheless quite rare (∼0.5% of length polymorphisms) and is likely insufficient to explain the shifts in Alu distribution. However, ectopic illegitimate recombination not involving TSDs may help to explain overall Alu sequence loss and distribution patterns.
Methods
Direct assessment of retroelement deletion rate
Putative retroelement insertions were obtained from the chimpanzee scaffold alignments to the University of California at Santa Cruz (UCSC) July 2003 human genome (Kent et al. 2002) using RepeatMasker (A.F.A. Smit and P. Green, unpubl.), MaskerAid (Bedell et al. 2000), and libraries from the RepBase Update (Jurka 2000). Pseudogenes were detected using BLAT (Kent 2002) and the human RefSeq mRNA records. Insertions were defined as having a single retroelement (including pseudogenes) filling all but up to 90 bp of the indel and not extending beyond the indel by >10 bp on either side. Search queries were then constructed of the 50-bp sequences upstream and downstream of each putative retroelement insertion location. Scripted discontiguous Mega-BLAST searches of the relevant NCBI trace archive were then carried out by using perl scripts and the QBLAST application programming interface (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi; http://www.ncbi.nlm.nih.gov/BLAST/Doc/urlapi.html) (Altschul et al. 1990; McGinnis and Madden 2004). Our BLAST queries used a noncoding template of size 21 and required only one seed hit per high-scoring segment pair. To minimize false positives and ensure nonredundant hits, we required that 75% of the query be free of known human repeats. Further, the accepted hits were required to match the query at least 30 bp on either side of the putative breakpoint. All traces not fulfilling these requirements were ignored. Deletions in human relative to chimpanzee or vice versa were diagnosed by the presence in Rhesus of an insertion at least 80% of the size expected and no traces with less than this amount of sequence. A site was considered an insertion if one or more Rhesus traces matched the empty site and no traces had extra sequence. Putative deletions in human or chimpanzee were further individually aligned with their Rhesus counterpart by using ClustalW version 1.82 (Higgins et al. 1996), and the alignments were edited using Jalview (Clamp et al. 2004) to check for the presence of the same element in the expected position in the Rhesus trace. The alignments are provided in Supplemental information.
Detection of deletions internal to Alu elements
All indel loci in the chimpanzee scaffold alignments to the UCSC July 2003 human genome, masked as described above, were reanalyzed. Deletions occurring entirely within Alu elements were analyzed for involvement of the ∼80-bp and 50-bp internal homologies. Putative deletions occurring between the 80-bp homologous regions were detected by having one deletion endpoint occurring within positions 1-84 of the consensus and the other endpoint within positions 136 to 219. Similarly deletions between positions 85-135 and 219 to the end of the consensus were considered as occurring between the 50-bp homologies.
Assessment of deletion frequency due to illegitimate recombination
Human chromosomal sequence files with human repeats pre-masked to lower case by RepeatMasker were used. These files were further masked by using Tandem Repeats Finder 3.21 (Benson 1999). All repetitive sequence was excised, including human repeats and tandem repeats. We then constructed a C++ program that used an alignment method to find all nonredundant nonadjacent identical segments up to 20 bp long between 200 and 500 bp apart in the nonrepetitive genome. These were tallied by homologous length, giving the expected distribution of potential sites for illegitimate recombination in this distance range.
We then analyzed all fully-sequenced insertions and deletions (indels) 200-500 bp long present in the alignments of the UCSC July 2003 human sequence to the chimpanzee scaffolds (Kent et al. 2002) for the presence of flanking identical segments beginning at the deletion breakpoints. Specifically, indels with 50-bp flanking sequence on each were analyzed, and those containing putative new retroelement insertions, tandem duplications, or flanking homologous retroelements corresponding to the indel breakpoints were removed from consideration, as were all indels occurring inside TEs. The remaining indels were classified as deletions.
Retroelement insertions were detected as described above. Other insertions were diagnosed if the indel internal sequence was found by BLAT elsewhere in the human genome. Tandem duplications were diagnosed by running Tandem Repeats Finder (Benson 1999) on a sequence including the indel extra sequence and equivalent lengths of sequence flanking the indel upstream and downstream. An indel was considered to be a case of tandem duplication if a tandem duplication was found covering one of the breakpoints and extending to within 1 bp of the other. Manual checks confirmed the validity of this criterion. After disqualifying putative insertions, tandem duplications, and indels with flanking homologous repeats, the remaining 1927 indels were termed random deletions and were analyzed for the distribution of flanking repeat sizes. Flanking repeats were considered to begin at the breakpoint positions and consist of a tract of identical sequence. No mismatches in the flanking repeat or off-sets of the identical segment from the indel breakpoints were allowed.
Genomic PCR and sequencing
Primate genomic DNA was isolated from various cell lines as described previously (Goodchild et al. 1993). Additional chimpanzee DNA samples were kindly provided by Dr. Peter Parham (Stanford University, Stanford, CA). One hundred fifty nanograms of human or primate genomic DNA was amplified in a 50-μL reaction with 200 μM each dNTP, 200 nM each primer (see Supplemental information), 1.5 mM MgCl2, and 1 U of Platinum Taq (Invitrogen) in 1× PCR buffer (Invitrogen). The conditions for the PCR were 94°C for 1 min followed by 30 cycles of the amplification step (30 sec at 94°C, 30 sec at 48°C-60°C, and 30 sec to 1 min at 72°C). The annealing temperature and extension time varied for different primer combinations. Sequencing was performed directly on PCR products by using the BigDye Terminator v3.1 Cycle Sequencing Kit (ABI) in an ABI PRISM 3730XL DNA Analyzer system at the McGill University sequencing facility.
Supplementary Material
[Supplemental Research Data]
[ [The Chimpanzee Genome]]
Acknowledgments
We thank Peter Parham for providing samples of chimpanzee DNA. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to D.L.M. L.N.L. is supported by a studentship from CIHR.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. of gorilla, gibbon, and chimpanzee sequences: AY953322, AY953323, AY953324, AY953325, and AY953326. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: P. Parham, A.F.A. Smit, and P. Green.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3910705.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287**:** 2185-2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215**:** 403-410. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297**:** 1301-1310. [DOI] [PubMed] [Google Scholar]
- Baillie, G.J., van de Lagemaat, L.N., Baust, C., and Mager, D.L. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78**:** 5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3**:** 370-379. [DOI] [PubMed] [Google Scholar]
- Bedell, J.A., Korf, I., and Gish, W. 2000. MaskerAid: A performance enhancement to RepeatMasker. Bioinformatics 16**:** 1040-1041. [DOI] [PubMed] [Google Scholar]
- Bennett, E.A., Coleman, L.E., Tsui, C., Pittard, W.S., and Devine, S.E. 2004. Natural genetic variation caused by transposable elements in humans. Genetics 168**:** 933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27**:** 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R.J., Rowen, L., Williams, J., and Cameron, R.A. 2003. Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. 100**:** 4661-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J.F. 2001. Selection on Alu sequences? Curr. Biol. 11**:** R900-R901. [DOI] [PubMed] [Google Scholar]
- Brosius, J. 1999. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107**:** 209-238. [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. The C. elegans Sequencing Consortium. Science 282**:** 2012-2018. [DOI] [PubMed] [Google Scholar]
- Carroll, M.L., Roy-Engel, A.M., Nguyen, S.V., Salem, A.H., Vogel, E., Vincent, B., Myers, J., Ahmad, Z., Nguyen, L., Sammarco, M., et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311**:** 17-40. [DOI] [PubMed] [Google Scholar]
- Clamp, M., Cuff, J., Searle, S.M., and Barton, G.J. 2004. The Jalview Java alignment editor. Bioinformatics 20**:** 426-427. [DOI] [PubMed] [Google Scholar]
- Conley, M.E., Partain, J.D., Norland, S.M., Shurtleff, S.A., and Kazazian Jr., H.H. 2005. Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum. Mutat. 25**:** 324-325. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K., and Bennetzen, J.L. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12**:** 1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M.C. and Gibbs, R.A. 1992. A human dimorphism resulting from loss of an Alu. Genomics 14**:** 590-597. [DOI] [PubMed] [Google Scholar]
- Gibbons, R., Dugaiczyk, L.J., Girke, T., Duistermars, B., Zielinski, R., and Dugaiczyk, A. 2004. Distinguishing humans from great apes with _Alu_Yb8 repeats. J. Mol. Biol. 339**:** 721-729. [DOI] [PubMed] [Google Scholar]
- Gibbs, R.A., Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428**:** 493-521. [DOI] [PubMed] [Google Scholar]
- Glazko, G.V. and Nei, M. 2003. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20**:** 424-434. [DOI] [PubMed] [Google Scholar]
- Goodchild, N.L., Wilkinson, D.A., and Mager, D.L. 1993. Recent evolutionary expansion of a subfamily of RTVL-H human endogenous retrovirus-like elements. Virology 196**:** 778-788. [DOI] [PubMed] [Google Scholar]
- Hamdi, H., Nishio, H., Zielinski, R., and Dugaiczyk, A. 1999. Origin and phylogenetic distribution of Alu DNA repeats: Irreversible events in the evolution of primates. J. Mol. Biol. 289**:** 861-871. [DOI] [PubMed] [Google Scholar]
- Hedges, D.J., Callinan, P.A., Cordaux, R., Xing, J., Barnes, E., and Batzer, M.A. 2004. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 14**:** 1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday, T. 2003. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 532**:** 103-115. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266**:** 383-402. [DOI] [PubMed] [Google Scholar]
- Johanning, K., Stevenson, C.A., Oyeniran, O.O., Gozal, Y.M., Roy-Engel, A.M., Jurka, J., and Deininger, P.L. 2003. Potential for retroposition by old Alu subfamilies. J. Mol. Evol. 56**:** 658-664. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 94**:** 1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16**:** 418-420. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Evolutionary impact of human Alu repetitive elements. Curr. Opin. Genet. Dev. 14**:** 603-608. [DOI] [PubMed] [Google Scholar]
- Karran, P. 2000. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10**:** 144-150. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT: The BLAST-like alignment tool. Genome Res. 12**:** 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12**:** 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M.G. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115**:** 49-63. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301**:** 1898-1903. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409**:** 860-921. [DOI] [PubMed] [Google Scholar]
- Li, W., Zhang, P., Fellers, J.P., Friebe, B., and Gill, B.S. 2004. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40**:** 500-511. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhao, S., Bailey, J.A., Sahinalp, S.C., Alkan, C., Tuzun, E., Green, E.D., and Eichler, E.E. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13**:** 358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. and Bennetzen, J.L. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. 101**:** 12404-12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, S. and Madden, T.L. 2004. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32**:** W20-W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand, P., van de Lagemaat, L.N., and Mager, D.L. 2002. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 12**:** 1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek, A., Jabbari, K., Paces, J., Paces, V., Hejnar, J.V., and Bernardi, G. 2001. Similar integration but different stability of _Alu_s and LINEs in the human genome. Gene 276**:** 39-45. [DOI] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., Vogel, E., Garber, R.K., Nguyen, S.V., Salem, A.H., Batzer, M.A., and Deininger, P.L. 2001. Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159**:** 279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, A.H., Kilroy, G.E., Watkins, W.S., Jorde, L.B., and Batzer, M.A. 2003a. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20**:** 1349-1361. [DOI] [PubMed] [Google Scholar]
- Salem, A.H., Ray, D.A., Xing, J., Callinan, P.A., Myers, J.S., Hedges, D.J., Garber, R.K., Witherspoon, D.J., Jorde, L.B., and Batzer, M.A. 2003b. Alu elements and hominid phylogenetics. Proc. Natl. Acad. Sci. 100**:** 12787-12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20**:** 43-45. [DOI] [PubMed] [Google Scholar]
- Schmid, C.W. 1998. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26**:** 4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T.D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R., et al. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429**:** 382-388. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520-562. [DOI] [PubMed] [Google Scholar]
- Yu, J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296**:** 79-92. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Traces/trace.cgi; NCBI Sequence Trace Archive.
- http://www.ncbi.nlm.nih.gov/BLAST/Doc/urlapi.html; NCBI QBLAST Interface.
- http://genome.ucsc.edu; UCSC Genome Browser.
- http://www.repeatmasker.org; RepeatMasker Interface.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Research Data]
[ [The Chimpanzee Genome]]