Genome-Wide Identification of Potential Plant E2F Target Genes (original) (raw)
Abstract
Entry into the S phase of the cell cycle is controlled by E2F transcription factors that induce the transcription of genes required for cell cycle progression and DNA replication. Although the E2F pathway is highly conserved in higher eukaryotes, only a few E2F target genes have been experimentally validated in plants. We have combined microarray analysis and bioinformatics tools to identify plant E2F-responsive genes. Promoter regions of genes that were induced at the transcriptional level in Arabidopsis (Arabidopsis thaliana) seedlings ectopically expressing genes for the E2Fa and DPa transcription factors were searched for the presence of E2F-binding sites, resulting in the identification of 181 putative E2F target genes. In most cases, the E2F-binding element was located close to the transcription start site, but occasionally could also be localized in the 5′ untranslated region. Comparison of our results with available microarray data sets from synchronized cell suspensions revealed that the E2F target genes were expressed almost exclusively during G1 and S phases and activated upon reentry of quiescent cells into the cell cycle. To test the robustness of the data for the Arabidopsis E2F target genes, we also searched for the presence of E2F-cis-acting elements in the promoters of the putative orthologous rice (Oryza sativa) genes. Using this approach, we identified 70 potential conserved plant E2F target genes. These genes encode proteins involved in cell cycle regulation, DNA replication, and chromatin dynamics. In addition, we identified several genes for potentially novel S phase regulatory proteins.
The heterodimeric E2F-DP transcription factors control the cell cycle by regulating transcription of genes required for DNA replication and cell cycle (Helin, 1998; Lavia and Jansen-Dürr, 1999). In mammals, eight E2Fs have been cloned and characterized (Trimarchi and Lees, 2002; de Bruin et al., 2003; Di Stefano et al., 2003; Maiti et al., 2005). E2F1, E2F2, and E2F3 function as potent transcriptional activators of E2F-responsive genes, and the overproduction of one of them is sufficient to drive serum-starved cells into the cell cycle. In contrast, E2F4 and E2F5 are mainly found in quiescent cells and are believed to control cell cycle exit and the onset of terminal differentiation. The physiological role of the E2F6, E2F7, and E2F8 proteins is less well understood, but the lack of a clear trans-activation domain suggests that they may function as repressors of E2F-dependent transcription (Müller and Helin, 2000; Trimarchi and Lees, 2002; de Bruin et al., 2003; Di Stefano et al., 2003).
The E2F pathway is conserved in mammals and plants (De Veylder et al., 2003; Dewitte and Murray, 2003; Inzé, 2005). In the genome of Arabidopsis (Arabidopsis thaliana), three E2F (E2Fa, E2Fb, and E2Fc) and two DP (DPa and DPb) genes have been identified (Vandepoele et al., 2002). Both E2Fa and E2Fb are potent transcriptional activators as demonstrated by their ability to trans-activate reporter genes harboring the E2F consensus cis-acting element (Mariconti et al., 2002; Stevens et al., 2002). Moreover, transient overexpression of E2Fa and DPa induces nondividing mesophyll cells to reenter S phase (Rossignol et al., 2002), whereas their constitutive overexpression induces plant cells to undergo either ectopic cell division or enhanced DNA endoreduplication (De Veylder et al., 2002; Kosugi and Ohashi, 2003). In contrast, E2Fc, which lacks a strong activating domain, functions as a negative regulator of the E2F-responsive genes because its ectopic expression inhibits cell division (del Pozo et al., 2002).
Mammalian E2F target genes have been identified using microarray analysis, chromatin immunoprecipitation assays, or computer-assisted prediction (Ishida et al., 2001; Kel et al., 2001; Müller et al., 2001; Weinmann et al., 2001; Ma et al., 2002; Ren et al., 2002; Stanelle et al., 2002). Only a small number of plant E2F targets are currently known and include mostly homologs of typical mammalian E2F target genes, such as MCM3, PCNA, CDC6, RNR, and CDKB1;1 (Chabouté et al., 2000, 2002; de Jager et al., 2001; Egelkrout et al., 2001, 2002; Kosugi and Ohashi, 2002; Stevens et al., 2002; Boudolf et al., 2004). The characterization of these genes revealed that the E2F DNA-binding site has been conserved during evolution. This observation was exploited to search the Arabidopsis genome for genes that contain the TTTCCCGCC cis-acting element in their promoter region. This in silico search identified 183 potential E2F target genes, including genes that are involved in DNA replication, cell cycle regulation, transcription, defense responses, and signaling (Ramirez-Parra et al., 2003). Available experimental data showed, however, that in addition to the TTTCCCGCC element, other closely related sequences are recognized by the plant E2F transcription factors (Chabouté et al., 2000; Stevens et al., 2002; Vlieghe et al., 2003). Moreover, data from mammalian cell cultures revealed that promoter activation by E2Fs does not depend exclusively on an _E2F_-binding site, but appears to require other regulatory sequences as well (Schlisio et al., 2002; Giangrande et al., 2003, 2004). Thus, it is possible that not all genes with the TTTCCCGCC site in their promoter region are controlled by E2F activity.
In a previous study, we compared the transcriptome of Arabidopsis plants ectopically expressing E2Fa-DPa with that of wild-type plants and discovered a previously unrecognized genetic network between DNA replication and nitrogen assimilation (Vlieghe et al., 2003). The number of potential E2F target genes that could be identified in this study was small, however, because the array represented only 4,571 cDNAs. Therefore, we performed a new transcriptome analysis of plants ectopically expressing E2Fa-DPa using the Affymetrix ATH1 GeneChip microarrays that represent nearly all genes in the Arabidopsis genome. By combining microarray analysis and bioinformatics tools, we were able to identify 181 putative E2Fa-DPa target genes. Most of the genes encode proteins that function in DNA replication, chromatin dynamics, and cell cycle regulation. In addition, comparison of the promoter regions of the Arabidopsis target genes with the promoters of orthologous genes in the rice (Oryza sativa) genome revealed a group of evolutionarily conserved plant E2F target genes, of which several encode proteins with unknown functions.
RESULTS
Genome-Wide Transcriptome Analysis of Transgenic Arabidopsis Plants Ectopically Expressing E2Fa-DPa Genes
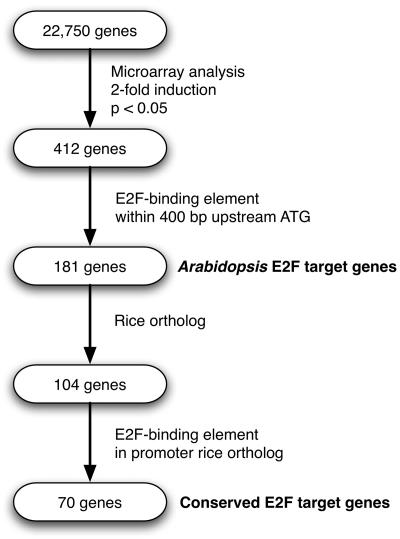
In order to identify E2F target genes of Arabidopsis on a genome-wide scale, we compared the transcriptomes of wild-type plants and plants ectopically expressing the E2Fa-DPa genes (E2Fa-DPaOE) using the Affymetrix ATH1 microarray that contains 22,810 probe sets, representing 22,750 annotated genes of the Arabidopsis genome (Fig. 1). In four independent experiments, E2Fa-DPaOE plants were grown side-to-side with wild-type (Columbia-0) plants. RNA was extracted from 6-d-old seedlings. Each biological sample was harvested and processed independently and finally all probed individually to a microarray, resulting in eight hybridization signals for each probe set. Statistical analysis indicated that 2,069 genes had a significant change in expression levels, of which 412 and 220 were more than 2-fold up- or down-regulated, respectively (see Supplemental Tables I and II online).
Figure 1.
Flow chart representation of the experimental strategy. For details, see text.
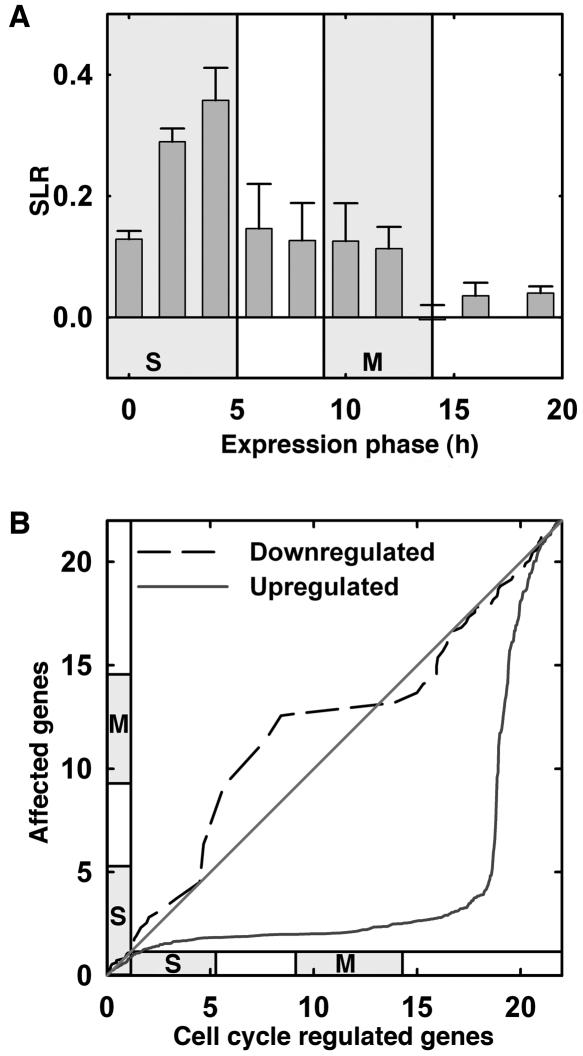
Previously, 9,910 genes were identified for their differential expression during the cell cycle (Menges et al., 2003). These genes were sorted into 10 bins according to their timing of maximal expression during the cell cycle, where the bins represent the 10 time points measured by Menges and colleagues. For all genes in every bin, the signal log ratios (SLRs) between expression signals in wild-type and E2Fa-DPaOE plants were averaged. This analysis revealed transcriptional induction of most proliferation-associated genes, corroborating with the previous observation that cells in E2Fa-DPaOE plants undergo extra rounds of cell division and endoreduplication (Fig. 2A; De Veylder et al., 2002). Genes expressed during S were clearly more strongly induced that those expressed during the G2 and M phases, whereas genes specifically transcribed during G1 were not up-regulated or only slightly up-regulated. Quantile-quantile (q-q) plots represent a graphical means to compare the distribution between different data sets. We analyzed the distribution of expression maxima during the cell cycle and compared this distribution for the 412 and 220 genes that were more than 2-fold up- or down-regulated in E2Fa-DPaOE seedlings to the distribution for all genes expressed in a cell cycle phase-specific manner in synchronized Arabidopsis cells (Fig. 2B). The q-q plots show that genes specifically expressed during mid-S phase were clearly enriched in the set of genes up-regulated in E2Fa-DPaOE seedlings (Fig. 2B). In contrast, genes that were down-regulated in E2Fa-DPaOE seedlings were not enriched for any of the specific classes of genes that show cell-cycle-specific gene expression patterns, as seen by the near-diagonal line (Fig. 2B). Because down-regulated genes had no cell-cycle-specific expression pattern and because E2Fa-DPa operates as a transcriptional activator (Mariconti et al., 2002; Rossignol et al., 2002), we focused on the up-regulated genes only in the downstream analysis to identify E2F target genes (Fig. 1).
Figure 2.
Increased expression of cell-cycle-phase-specific genes in E2Fa-DPaOE plants. Expression peaks for 9,910 genes during the 22-h cell cycle of Arabidopsis (Menges et al., 2003) were used. In this experiment, transcript levels were measured at 10 time points. The first time point (0 h) represents cells blocked in early S phase. Synchronized cells completed S phase at 5 h and went through mitosis from 9–14 h after release from the block. Identification of 9,910 genes that were differentially expressed during the cell cycle and determination of their expression peaks were described before (Menges et al., 2003). A, The 9,910 genes identified by Menges et al. (2003) were sorted into 10 bins, which represent the 10 time points of measurements, based on their maximal expression during the cell cycle. Shown are the mean ± se of the SLRs (logarithms to base 2) between expression signals in wild-type and E2Fa-DPaOE plants for all genes in a bin. Note that the 8th bin, left to the time point of 15 h, holds only a small number of genes. Shaded areas represent periods of S phase and mitosis, respectively. B, The q-q plots for genes significantly modified in E2Fa-DPaOE seedlings versus all cell-cycle-regulated genes. Slopes close to 0 indicate an overrepresentation of the number of E2Fa-DPa-affected genes among the number of cell-cycle-regulated genes in the time window indicated on the y axis.
Identification of E2F DNA Consensus Motifs in Arabidopsis Promoter Regions
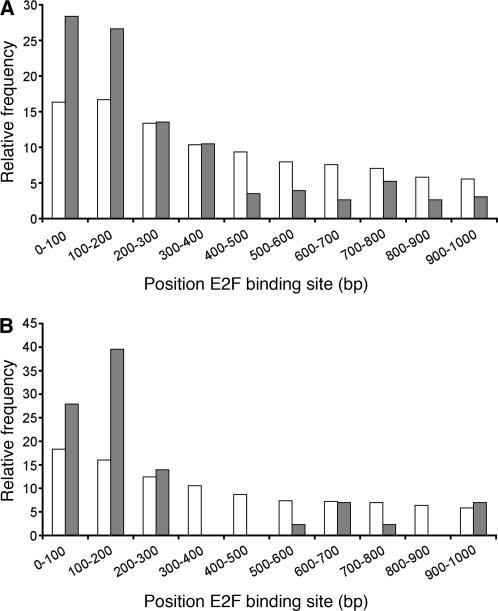
The 1-kb promoter DNA sequence upstream of the ATG start codon of each of the 412 genes that were significantly up-regulated >2-fold was extracted from the Arabidopsis genome sequence and scanned for the presence of the E2F-cis-acting element with the sequence WTTSSCSS (where W = A or T, and S = C or G), which represents the consensus DNA sequence of all different E2F-DP-binding motifs that were experimentally verified in plants (Chabouté et al., 2000, 2002; de Jager et al., 2001; Egelkrout et al., 2001, 2002; Kosugi and Ohashi, 2002; Stevens et al., 2002). We searched the full 1-kb sequence upstream of the ATG rather than only the sequence upstream of the transcription start site because functional E2F elements can reside in the 5′ untranslated region (UTR; Chabouté et al., 2002; Ren et al., 2002). Using this approach, we found that 229 out of the 408 nuclear-encoded E2Fa-DPa-responsive genes possess at least one potential E2F-binding site. We then plotted the frequency of occurrence of an E2F consensus motif in 100-bp intervals against the distance from the ATG and compared the frequencies between the _E2Fa-DPaOE_-responsive genes and all genes represented on the ATH1 microarray. Figure 3A shows that the frequency of the E2F consensus motif was relatively constant along promoter regions on a genome-wide scale. By contrast, among the genes that were 2-fold up-regulated in E2Fa-DPaOE seedlings, the E2F consensus motif was clearly more abundant inside the 200-bp region immediately upstream of the start codon. Further analysis, however, indicated that applying a 200-bp promoter cutoff selection criterion resulted in a high number of false negatives in our strategy to screen for putative E2F target genes. Therefore, a 400-bp criterion was used because up to this limit the E2F consensus motif was more abundant in the group of E2Fa-DPa up-regulated genes than randomly expected. Beyond the 400-bp limit, the number of E2F-binding sites was clearly underrepresented in the data set of E2Fa-DPa-induced genes (Fig. 3A).
Figure 3.
E2F element distribution in promoters of Arabidopsis and rice. A, Frequency of occurrence of the E2F motif in Arabidopsis promoter sequences. White bars represent the relative frequency of the consensus E2F motif in 100-bp intervals upstream from the ATG for promoter sequences on a genome-wide scale. Gray bars represent the relative frequency of the consensus E2F motif in promoters of genes up-regulated in E2Fa-DPaOE plants. B, Frequency of occurrence of the E2F motif in rice promoter sequences. White bars represent the relative frequency of the consensus E2F motif for promoter sequences on a genome-wide scale. Gray bars represent the relative frequency of the consensus E2F motif in promoter sequences for rice orthologs of the Arabidopsis E2F target genes.
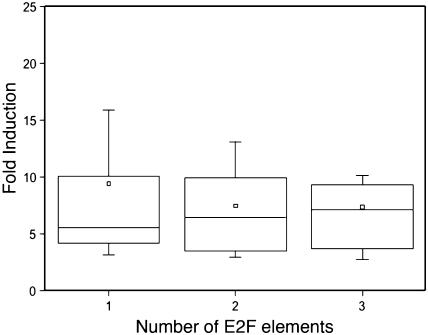
In total, 181 genes (or 44.4% of all 408 nuclear-encoded up-regulated genes) harbored an E2F consensus motif within the 400 bp upstream of the ATG (Fig. 1). This percentage is significantly (_P_-value Fisher's exact test < 0.0001) higher than the frequency of the E2F consensus motif to occur within the first 400 bp for all Arabidopsis promoters in the genome, which is 14.9%. The genes identified include all previously reported E2F target genes represented on the ATH1 array (PCNA, RNR large subunit, CDC6, and MCM3), thus validating our combined analysis of large-scale RNA expression data and genome sequences. In addition, we identified other genes that encode proteins involved in DNA replication (e.g. replication factors, DNA polymerases, and DNA primase), initiation of DNA replication (e.g. ORC1, CDC45, and CDC6), chromatin dynamics (e.g. MSI3, CMT3, trihorax-like protein gene, and FAS1), cell cycle (E2Fb, E2Fc, RBR1, DEL3, and CYCA3;2), and DNA repair (e.g. RAD17, mismatch repair protein MSH6-1, and UVR3; see Supplemental Table III online). In 28 of the identified genes, the E2F consensus motif was located in the 5′ UTR. Two-thirds of the 181 genes contained only one E2F consensus motif in their promoter, whereas the remaining genes had two or more E2F consensus motifs. Interestingly, the promoter of the gene encoding the G/T DNA mismatch repair enzyme has five E2F consensus motifs within the first 200 bp proximal to the ATG. No direct correlation between the number of E2F consensus motifs present in a promoter and the fold transcriptional induction of the corresponding gene was observed (Fig. 4), suggesting that some of the identified motifs might represent low-affinity E2F-binding sites that are rarely occupied in planta. Alternatively, other factors might be required in conjunction with E2F transcription factors to induce gene expression.
Figure 4.
Lack of correlation between the number of E2F binding sites and transcriptional activation in E2Fa-DPaOE plants. The fold induction level of the putative E2F target genes was plotted against the number of E2F elements found in their promoter. Only populations with more than five genes were plotted. The horizontal lines in the box denote the 25th, 50th, and 75th percentile values. The error bars indicate the 5th and 95th percentile values. The square symbol in the box corresponds to the mean of the column of data.
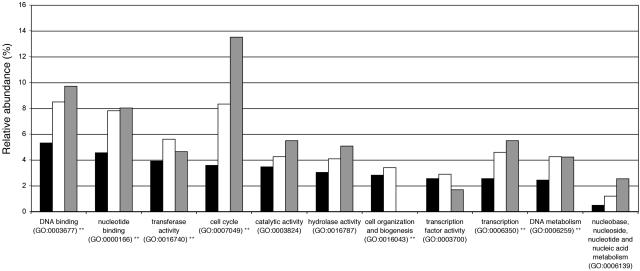
The 181 putative E2F target genes we identified belong to different functional classes according to the gene ontology (GO) biological process classification system. Figure 5 shows that E2F target genes were among others significantly overrepresented in the categories “cell cycle” and “DNA metabolism.”
Figure 5.
Functional distribution of E2F target genes using the GO slim biological process classification system. Black bars indicate the relative frequency of GO classes in the set of 408 up-regulated Arabidopsis genes, white bars indicate relative frequency of GO classes in the set of up-regulated Arabidopsis genes with an E2F binding site, and gray bars indicate relative frequency of GO classes for the set of E2F target genes conserved between Arabidopsis and rice. Significantly overrepresented classes for the up-regulated genes containing an E2F binding site (white bars) are indicated (**, _P_-value < 0.01). Note that GO classes covering <2.5% of the genes were omitted for clarity and that cellular component GO classes and unknown classes are not shown.
To identify a preferred E2F-binding site, we compared the relative abundance of all possible motifs of the WTTSSCSS E2F consensus motif between the set of genes up-regulated in the E2Fa-DPaOE seedlings and all genes present on the ATH1 microarray. Table I shows that three motifs were significantly enriched (_P_-value Fisher's exact test < 0.01). These motifs are found more frequently in the set of E2Fa-DPa-induced genes than randomly expected on the basis of the number of E2F elements found among all genes on the ATH1 array. The most abundant motif was TTTCCCGC, representing 25.4% of all motifs found in the 400-bp promoter regions upstream of the ATG. The other two motifs, ATTCCCGC and TTTGGCGC, were represented by 5.1% and 9.9%, respectively. Although the TTTGGCGG motif was relatively highly abundant as well (representing 9.2% of all motifs found), it was not significantly overrepresented in the data set.
Table I.
Frequency of E2F elements found among the E2F target genes
| E2F Motifa | E2F Elements in the Set of Up-Regulatedb Genes | Relative Frequency (%) in the Set of Up-Regulated Genes | E2F Elements in Genomebc | Relative Frequency (%) in Genomec |
|---|---|---|---|---|
| ATTCCCCC | 2 (2) | 0.7 | 88 (88) | 2.3 |
| ATTCCCCG | 6 (6) | 2.2 | 74 (74) | 1.9 |
| ATTCCCGC | 14 (14) | 5.1 | 78 (78) | 2.0 |
| ATTCCCGG | 4 (4) | 1.5 | 94 (91) | 2.4 |
| ATTCGCCC | 0 (0) | 0.0 | 30 (29) | 0.8 |
| ATTCGCCG | 4 (4) | 1.5 | 159 (150) | 4.1 |
| ATTCGCGC | 3 (3) | 1.1 | 43 (43) | 1.1 |
| ATTCGCGG | 5 (5) | 1.8 | 55 (52) | 1.4 |
| ATTGCCCC | 2 (2) | 0.7 | 62 (62) | 1.6 |
| ATTGCCCG | 0 (0) | 0.0 | 27 (27) | 0.7 |
| ATTGCCGC | 5 (5) | 1.8 | 51 (51) | 1.3 |
| ATTGCCGG | 1 (1) | 0.4 | 64 (64) | 1.6 |
| ATTGGCCC | 4 (4) | 1.5 | 187 (182) | 4.8 |
| ATTGGCCG | 1 (1) | 0.4 | 69 (69) | 1.8 |
| ATTGGCGC | 9 (8) | 3.3 | 62 (58) | 1.6 |
| ATTGGCGG | 11 (11) | 4.0 | 97 (96) | 2.5 |
| TTTCCCCC | 9 (9) | 3.3 | 266 (265) | 6.8 |
| TTTCCCCG | 3 (3) | 1.1 | 137 (137) | 3.5 |
| TTTCCCGC | 69 (67) | 25.4 | 257 (244) | 6.6 |
| TTTCCCGG | 13 (12) | 4.8 | 318 (285) | 8.1 |
| TTTCGCCC | 2 (2) | 0.7 | 52 (52) | 1.3 |
| TTTCGCCG | 9 (9) | 3.3 | 269 (259) | 6.9 |
| TTTCGCGC | 5 (5) | 1.8 | 69 (67) | 1.8 |
| TTTCGCGG | 6 (6) | 2.2 | 92 (91) | 2.4 |
| TTTGCCCC | 3 (3) | 1.1 | 163 (162) | 4.2 |
| TTTGCCCG | 4 (4) | 1.5 | 65 (63) | 1.7 |
| TTTGCCGC | 7 (7) | 2.6 | 111 (110) | 2.8 |
| TTTGCCGG | 6 (6) | 2.2 | 133 (130) | 3.4 |
| TTTGGCCC | 11 (11) | 4.0 | 246 (239) | 6.3 |
| TTTGGCCG | 2 (2) | 0.7 | 119 (119) | 3.0 |
| TTTGGCGC | 27 (24) | 9.9 | 159 (152) | 4.1 |
| TTTGGCGG | 25 (25) | 9.2 | 214 (199) | 5.5 |
| TOTAL | 272 (181) | 100.0 | 3910 (3788) | 100.0 |
Cell-Cycle-Regulated Gene Expression of E2F Target Genes
Using the Arabidopsis MM2d cell culture, Menges et al. (2003) reported changes in gene expression during the synchronous cell cycle reentry of starved cells following Suc addition. The analysis revealed two major classes of gene expression. The first cluster included genes that had low expression levels during the starvation period but were up-regulated after Suc addition (up-regulated genes). The second cluster included genes with a peak of expression immediately following the initiation of the starvation procedure but that were down-regulated after Suc supplementation (down-regulated genes). Both clusters represent about the same amount of genes (2,101 and 2,132 for the cluster of up-regulated and down-regulated genes, respectively). Of the 181 putative E2F target genes we identified in our analysis, 110 genes fall into one of these two categories. Remarkably, among those, 98.2% belong to the class of genes that are up-regulated during cell cycle reentry. The remaining two genes belong to the class of down-regulated genes (see Supplemental Table IV online).
In a second experiment reported by Menges et al. (2003), Arabidopsis MM2d cells were released from an aphidicolin arrest and followed by gene expression profiling as they moved synchronously from S phase through G2 and M into G1. After statistic analysis, a total of 1,016 genes with a clear cell-cycle-periodic expression profile were defined. Among these, 65.8% of the oscillating genes were expressed in S phase, 2.0% in G2, 19.5% in M, and 12.7% in G1. Of all predicted 181 Arabidopsis E2F target genes, 43 displayed a cell-cycle-phase-dependent gene expression pattern as defined by Menges et al. (2003). This low number can be explained by the fact that aphidicolin arrests cells in early S phase. As such, the G1-to-S transition, where E2Fa-DPa is supposed to be active, is missed. Of the 43 E2F target genes with a clear cell-cycle-periodic expression profile, 42 genes displayed a peak of expression during G1 or S, and only one gene (CDT1a) was expressed during M phase (see Supplemental Table V online).
Conservation of E2F-cis-Acting Elements between Arabidopsis and Rice Promoters
E2F activity was recently demonstrated in rice (Kosugi and Ohashi, 2002). Because the E2F pathway is highly conserved and the E2F DNA consensus motif is conserved between plants and mammals, we reasoned that a large number of E2F target genes should also be conserved between rice and Arabidopsis. To generate a robust set of E2F target genes, we therefore searched the rice genome for genes orthologous to the 181 Arabidopsis E2Fa-DPa target genes. We based our analysis on the 44,498 rice gene sequences annotated by The Institute for Genome Research (TIGR release 3.0, excluding transposable elements; Yuan et al., 2005). Because it is difficult to define orthology based on DNA or amino acid sequence similarity only, we selected only those Arabidopsis genes with one or two rice homologs. After removing rice genes with only partial homology to their Arabidopsis counterparts (see “Materials and Methods”), 104 Arabidopsis and 128 rice genes were found to be significantly homologous, representing putative orthologous gene pairs (Fig. 1). For eight Arabidopsis genes, a one-to-two relation with homologous rice genes was found (e.g. RBR1, RNR large subunit, and CDC45), whereas for some paralogous Arabidopsis genes, only a single rice copy was detected (e.g. PCNA and CDC6). For approximately 50% of the Arabidopsis genes, the homology relation was one-to-one, suggesting orthology between Arabidopsis and rice. The remaining genes either had no homologs in rice or belonged to large gene families, implying complex many-to-many relationships.
In order to define objectively the length of rice promoters for our analysis, we again compared the frequency of the E2F DNA consensus motif in 100-bp intervals between all rice genes and the subset of 59 rice genes, which showed a one-to-one homology relation to the Arabidopsis genes that were up-regulated in E2Fa-DPaOE seedlings and that contain the E2F consensus motif. We reasoned that the subset of putative orthologous rice genes should provide a good approximation of the size of promoter sequences that were enriched for the E2F consensus motif and therefore could be E2F target genes in rice. Figure 3B shows that the first 300-bp proximal to the ATG in the promoters of the putative rice E2F target genes were enriched for E2F consensus motifs when compared to all rice promoter sequences. Together, our comparative analysis confirmed that for 70 of the Arabidopsis E2F targets genes (67%), the corresponding E2F DNA sequence motif was conserved in the promoter of the putative homologous rice gene (Fig. 1). This suggests that the group of Arabidopsis genes we identified represents evolutionarily conserved plant E2F target genes (Table II). Most of these genes belonged to the GO functional class of cell cycle genes (Fig. 5). For three out of six E2F target genes of Arabidopsis with two putative rice orthologs, an E2F element was found in both rice genes, whereas three had an E2F-binding site in only one rice ortholog. Similarly, in only 7 out of 12 rice single-copy genes, both co-orthologous Arabidopsis genes harbored an E2F binding site. These results suggest that during evolution a number of genes created by gene duplication might have escaped transcriptional control by E2F, giving rise to novel functions or control of the gene by other regulatory signals (Van de Peer et al., 2001; Prince and Pickett, 2002).
Table II.
Arabidopsis E2F target genes and their putative rice orthologs sorted according to functional category
| Gene Category | Gene Description | Fold Induction | Arabidopsis Locus | Paralog in Rice | Rice Locus | Conserved E2F Motif |
|---|---|---|---|---|---|---|
| Cell cycle | ||||||
| Retinoblastoma-related protein (RBR1) | 11.2 | At3g12280 | 2 | Os08g42600 | Y | |
| Os11g32900 | N | |||||
| DP-E2F-related protein (DEL3) | 8.4 | At3g01330 | 2 | Os02g50630 | Y | |
| Os06g13670 | Y | |||||
| Replication enzymes | ||||||
| Helicase-related protein | 14.2 | At1g20750 | 1 | Os09g37920 | Y | |
| DEAD/DEAH box helicase | 13.7 | At1g35530 | 1 | Os11g07870 | N | |
| DNA helicase | 13.6 | At3g05740 | 1 | Os11g44910 | Y | |
| DNA topoisomerase III α | 13.4 | At5g63920 | 1 | Os03g06900 | Y | |
| Ribonucleoside-diphosphate reductase (RNR) small subunit | 11.7 | At3g27060 | 2 | Os06g03720 | Y | |
| Os06g14620 | Y | |||||
| DNA primase large subunit | 11.6 | At1g67320 | 1 | Os07g22400 | Y | |
| DNA polymerase _α_-subunit B | 11.0 | At1g67630 | 1 | Os12g13950 | Y | |
| DNA polymerase III catalytic subunit | 10.0 | At5g63960 | 1 | Os11g08330 | N | |
| DNA primase small subunit | 10.0 | At5g41880 | 1 | Os05g29010 | N | |
| DNA polymerase α 1 | 9.5 | At5g67100 | 1 | Os01g64820 | N | |
| Ribonucleoside-diphosphate reductase (RNR) large subunit | 9.3 | At2g21790 | 2 | Os02g56100 | Y | |
| Os06g07210 | Y | |||||
| Helicase-related protein | 8.0 | At1g20720 | 1 | Os09g37920 | Y | |
| DNA replication protein-related | 7.1 | At5g49010 | 1 | Os05g05150 | N | |
| Deoxyuridine 5′-triphosphate nucleotidohydrolase | 5.7 | At3g46940 | 1 | Os03g46640 | Y | |
| Helicase-related protein | 5.6 | At1g79890 | 1 | Os05g13300 | Y | |
| Replication protein A-like | 5.4 | At2g24490 | 2 | Os02g42230 | Y | |
| Os02g58220 | Y | |||||
| Proliferating cell nuclear antigen 2 (PCNA2) | 5.3 | At2g29570 | 1 | Os02g56130 | Y | |
| Proliferating cell nuclear antigen 1 (PCNA1) | 4.5 | At1g07370 | 1 | Os02g56130 | Y | |
| DNA polymerase δ small subunit-related | 3.5 | At2g42120 | 1 | Os03g03650 | Y | |
| DNA ligase | 3.3 | At1g08130 | 1 | Os10g34750 | N | |
| Replication protein A-like | 3.0 | At3g02920 | 2 | Os02g42230 | Y | |
| Os02g58220 | Y | |||||
| DNA polymerase ζ catalytic subunit | 2.6 | At1g67500 | 1 | Os07g47280 | N | |
| DNA gyrase subunit A | 2.6 | At3g10690 | 1 | Os03g59750 | N | |
| Putative replication factor C | 2.4 | At5g27740 | 1 | Os03g57870 | Y | |
| Origin factors | ||||||
| DNA replication licensing factor CDC6a | 20.5 | At2g29680 | 1 | Os01g63710 | Y | |
| Origin recognition complex subunit 1 (ORC1) | 12.4 | At4g12620 | 1 | Os06g08790 | Y | |
| Origin recognition complex subunit 3 (ORC3) | 10.3 | At5g16690 | 1 | Os10g26280 | Y | |
| Minichromosome maintenance family protein (MCM) | 9.4 | At2g14050 | 1 | Os06g11500 | Y | |
| Origin recognition complex subunit 6 (ORC6) | 9.0 | At1g26840 | 1 | Os07g43540 | Y | |
| Minichromosome maintenance family protein (MCM) | 8.1 | At1g44900 | 1 | Os11g29380 | Y | |
| Minichromosome maintenance family protein (MCM) | 6.6 | At2g16440 | 1 | Os01g36390 | Y | |
| Minichromosome maintenance family protein (MCM) | 6.2 | At3g09660 | 1 | Os05g38850 | N | |
| CDT1a | 4.9 | At2g31270 | 1 | Os04g10650 | Y | |
| DNA replication licensing factor CDC6b | 4.0 | At1g07270 | 1 | Os01g63710 | Y | |
| Cell division control protein 45 (CDC45) | 3.7 | At3g25100 | 2 | Os12g03130 | Y | |
| Os11g03430 | Y | |||||
| Chromatin structure | ||||||
| High mobility group protein (HMGB7) | 22.1 | At5g23420 | 1 | Os08g01100 | Y | |
| Structural maintenance of chromosomes (SMC) family protein | 17.8 | At5g07660 | 1 | Os09g03370 | Y | |
| SET domain-containing protein (SDG27) | 16.5 | At5g24330 | 2 | Os01g73460 | Y | |
| Os02g03030 | Y | |||||
| Arabidopsis homolog-pairing 2 (AHP2) | 16.1 | At1g13330 | 1 | Os03g50220 | N | |
| Structural maintenance of chromosomes (SMC) family protein | 9.7 | At5g61460 | 1 | Os09g03370 | Y | |
| Chromatin assembly factor-1 (FAS1) | 8.9 | At1g65470 | 1 | Os01g67100 | N | |
| DNA (cytosine-5-)-methyltransferase (DMT1) | 8.2 | At5g49160 | 2 | Os03g58400 | N | |
| Os07g08500 | N | |||||
| Histone acetyltransferase (HAG2) | 5.2 | At5g56740 | 1 | Os09g17850 | Y | |
| Sister chromatid cohesion 1 protein (RAD21-2) | 5.0 | At3g59550 | 1 | Os08g16610 | Y | |
| Trithorax-like protein 1 (SDG27) | 4.9 | At2g31650 | 1 | Os09g04890 | Y | |
| Antisilencing factor 1-related protein (SGA1) | 4.3 | At5g38110 | 1 | Os05g48030 | Y | |
| DNA-dependent ATPase (CHR10) | 4.2 | At2g44980 | 1 | Os03g01200 | N | |
| SMC2-like condensing (CPE1) | 4.2 | At5g62410 | 1 | Os01g67740 | Y | |
| Chromomethylase 3 (DMT6) | 3.7 | At1g69770 | 1 | Os10g01570 | N | |
| DNA-dependent ATPase (CHR17) | 3.4 | At5g18620 | 2 | Os01g27040 | Y | |
| Os05g05780 | N | |||||
| Antisilencing factor 1-related protein (SGA2) | 2.7 | At1g66740 | 1 | Os05g48030 | Y | |
| DNA repair | ||||||
| Uracil DNA glycosylase family protein | 17.4 | At3g18630 | 1 | Os04g57730 | Y | |
| DNA mismatch repair protein (MSH6-1) | 16.2 | At4g02070 | 1 | Os09g24220 | Y | |
| RAD1-like protein | 10.0 | At4g17760 | 1 | Os06g04190 | Y | |
| DNA mismatch repair protein (PMS2) | 7.9 | At4g02460 | 1 | Os02g37920 | N | |
| 6-4 Photolyase (UVR3) | 6.8 | At3g15620 | 1 | Os02g10990 | N | |
| Cell cycle checkpoint protein (RAD17) | 6.6 | At5g66130 | 1 | Os03g13850 | N | |
| DNA mismatch repair MutS family (MSH1) | 6.1 | At3g24320 | 1 | Os04g42780 | N | |
| UMUC-like DNA repair protein | 5.2 | At5g44740 | 1 | Os01g55300 | N | |
| DNA repair protein (RAD5) | 4.7 | At5g22750 | 2 | Os04g09800 | Y | |
| Os02g32570 | N | |||||
| DNA mismatch repair protein (MSH3) | 4.3 | At4g25540 | 1 | Os04g58630 | Y | |
| DNA nick sensor | 4.2 | At3g14890 | 1 | Os01g53560 | Y | |
| DNA-(apurinic or apyrimidinic site) lyase | 4.0 | At2g41460 | 2 | Os01g58680 | N | |
| Os01g58690 | N | |||||
| Transcription factors | ||||||
| PHD finger family protein | 26.0 | At1g33420 | 2 | Os11g12650 | Y | |
| Os01g65600 | N | |||||
| Zinc knuckle (CCHC-type) family protein | 16.1 | At3g02820 | 1 | Os02g52460 | Y | |
| Zinc finger (C3HC4-type RING finger) family protein | 9.9 | At4g14365 | 1 | Os07g26490 | Y | |
| Zinc finger (CCCH-type) family protein | 9.2 | At1g68200 | 2 | Os01g45730 | N | |
| Os05g50080 | N | |||||
| PHD finger protein-related | 3.6 | At4g23860 | 1 | Os06g33810 | Y | |
| Others | ||||||
| WD-40 repeat family protein | 22.7 | At3g27640 | 1 | Os03g49200 | Y | |
| ATP-dependent RNA helicase A (nuclear DNA helicase II) | 20.6 | At2g47680 | 1 | Os01g15300 | N | |
| Kinesin heavy chain | 11.6 | At3g63480 | 1 | Os08g02380 | N | |
| MAGE-8 antigen-related | 11.3 | At1g34770 | 1 | Os07g05650 | Y | |
| F-box family protein (FBL17) | 10.1 | At3g54650 | 2 | Os03g43390 | Y | |
| Os12g40860 | N | |||||
| Timeless family protein | 10.1 | At5g52910 | 1 | Os05g11980 | Y | |
| AAA-type ATPase family protein | 8.7 | At1g04730 | 1 | Os03g15810 | Y | |
| DnaJ protein | 6.9 | At5g06910 | 1 | Os02g10220 | Y | |
| Mitochondrial glycoprotein | 6.4 | At1g80720 | 2 | Os08g34130 | N | |
| Os09g38500 | N | |||||
| DnaJ protein | 5.9 | At3g12170 | 1 | Os02g10220 | Y | |
| WD-40 repeat family protein | 5.8 | At5g14530 | 2 | Os07g40030 | Y | |
| Os05g46570 | N | |||||
| WD-40 repeat family protein | 5.6 | At3g42660 | 1 | Os09g06560 | Y | |
| WD-40 repeat family protein | 5.3 | At5g19920 | 1 | Os03g63470 | Y | |
| UDP-Gal/UDP-Glc transporter | 5.3 | At2g02810 | 1 | Os06g39260 | N | |
| TSO1-like protein 1 (SOL1) | 4.8 | At4g14770 | 2 | Os03g43730 | Y | |
| Os12g41230 | N | |||||
| Strictosidine synthase | 4.2 | At3g51450 | 2 | Os07g42250 | N | |
| Os10g39710 | N | |||||
| WD-40 repeat family protein | 4.1 | At4g30840 | 1 | Os08g16600 | N | |
| Tetrapyrrole methylase family protein | 4.0 | At1g45110 | 1 | Os10g27450 | Y | |
| Pentatricopeptide (PPR) repeat-containing protein | 3.9 | At3g23020 | 2 | Os07g28900 | Y | |
| Os03g06710 | N | |||||
| WD-40 repeat family protein | 3.1 | At1g49540 | 1 | Os08g38570 | N | |
| PAN domain-containing protein | 3.0 | At5g03700 | 2 | Os02g32620 | Y | |
| Os11g05240 | Y | |||||
| Thioredoxin o (TRXO2) | 2.7 | At1g31020 | 1 | Os06g45510 | N | |
| Unknown | ||||||
| Expressed protein | 20.0 | At2g24970 | 1 | Os01g56090 | N | |
| Expressed protein | 15.9 | At3g60660 | 1 | Os07g30110 | N | |
| Expressed protein | 11.6 | At5g06590 | 1 | Os12g42840 | N | |
| Expressed protein | 8.1 | At2g40550 | 1 | Os01g07260 | Y | |
| Hypothetical protein | 6.6 | At1g78650 | 1 | Os01g10690 | Y | |
| Expressed protein | 4.2 | At3g17030 | 1 | Os03g61700 | Y | |
| Hypothetical protein | 4.2 | At2g46380 | 2 | Os02g15820 | Y | |
| Os06g33320 | Y | |||||
| Expressed protein | 3.4 | At4g31400 | 2 | Os04g42120 | N | |
| Os05g31230 | N | |||||
| Expressed protein | 3.1 | At1g80190 | 1 | Os05g37980 | N | |
| Expressed protein | 3.1 | At2g46200 | 1 | Os01g12670 | Y | |
| Putative protein | 3.0 | At5g15540 | 1 | Os07g01940 | Y | |
| Expressed protein | 2.3 | At5g07950 | 1 | Os03g11450 | Y |
Because of the observed high conservation of the E2F boxes between orthologous genes of Arabidopsis and rice, one might wonder if applying solely an evolutionary filter, omitting the microarray expression data, might result in the similar selection of putative E2F target genes. Sequence analysis revealed that in total 3,788 genes on the ATH1 array harbor an E2F box in the first 400 bp upstream of the start of translation (Table I). Among those, 1,137 Arabidopsis genes (representing 1,063 gene families) have one or two putative orthologous rice genes. Of these, for 440 Arabidopsis genes (401 gene families), an E2F element was found within the first 300 bp upstream of the start of translation in the rice ortholog(s). Apart from the 70 conserved targets represented in Table II, only 29 of the remaining 370 displayed a significantly up-regulated expression level in the E2F-DPaOE plants. This finding indicates that applying an evolutionary filter in the absence of expression data would yield a high number of false-positive targets, emphasizing the need in combining both wet and dry science techniques for identifying true target genes of the transcription factor of interest.
DISCUSSION
Ectopic expression of E2Fa-DPa severely affects plant development. In comparison to wild-type plants, E2Fa-DPaOE transgenic plants are small and display curled leaves and cotyledons (De Veylder et al., 2002). These dramatic phenotypes are reflected in large differences in the transcriptomes of wild-type versus E2Fa-DPaOE plants. Nevertheless, among the 412 genes that were transcriptionally >2-fold induced, almost 44% (181 genes) harbored an E2F-binding site in close proximity to their start of translation. Among these, all previously characterized E2F target genes were present, illustrating the strength of the approach followed for identifying potential E2F targets genes. Nevertheless, still some true E2Fa-DPa targets with an E2F-binding motif in their promoter might have been missed in our analysis because of the 400-bp selection criterion applied. The genes that were significantly up-regulated in the E2Fa-DPaOE seedlings and that lack an E2F-binding site could represent genes that are downstream of the E2F-DP regulatory circuit or could represent genes altered in expression by the wholesale disruption of the cell cycle and consequent developmental abnormalities caused by E2Fa-DPa overexpression.
Of the 181 putative E2F target genes we identified in our analysis, only 34 could be found among the 183 targets genes predicted by an in silico sequence analysis approach, as reported by Ramirez-Parra et al. (2003). There are several experimental differences that could explain this small overlap between the two data sets. First, Ramirez-Parra et al. (2003) only used the TTTCCCGCC motif to search promoter regions in their genome-wide analysis. Although this motif is found most frequently among genes up-regulated during S phase, our analysis has shown that other E2F elements are also enriched in genes up-regulated in E2Fa-DPaOE seedlings. Second, Ramirez-Parra et al. (2003) analyzed 800-bp promoter regions for the TTTCCCGCC motif. We restricted our analysis to 400-bp upstream of the translation start site because we found that this was the only region to be significantly enriched for E2F-binding sites. Third, the small overlap of both data sets could also be explained in part by the existence of different functional E2F complexes that could be formed by the three E2F and two DP proteins encoded in the Arabidopsis genome (Mariconti et al., 2002). Arabidopsis E2Fa and E2Fb are potent transcriptional activators, but E2Fc appears to function as a repressor. Microarray analysis in Drosophila melanogaster has revealed that the number of target genes shared between activating and repressing E2F proteins is very small (Dimova et al., 2003). The activating E2F mainly controls the expression of genes that encode proteins with roles in cell cycle progression and DNA metabolism. In contrast, the repressing E2F seems to target genes involved in differentiation. Thus, it will now be important to verify experimentally whether E2F target genes predicted by Ramirez-Parra et al. (2003) that were not induced by E2Fa-DPa in E2Fa-DPaOE seedlings are controlled by other E2F-DP complexes.
Mapping of the 5′ UTR sequences with all available expressed sequence tags and cDNA sequences revealed that 28 of the 181 Arabidopsis E2F target genes have an E2F motif in their 5′ UTR sequence. Comparison of orthologous rice genes, however, indicates that the presence of an E2F motif in the 5′ UTR is not necessarily conserved between plant species. In only 2 out of the 10 gene pairs for which 5′ UTR sequence was available, the E2F motif was found in the UTR of both the rice and Arabidopsis genes. Similarly, whereas in the RNR large subunit gene of tobacco (Nicotiana tabacum) the E2F motif is located in the 5′ leader (Chabouté et al., 2002), this is not the case for the othologous Arabidopsis gene.
It has been demonstrated that mammalian promoter activation by E2Fs relies on the concerted function of an E2F-binding site with other cis-acting elements (Schlisio et al., 2002; Giangrande et al., 2003, 2004). A similar mechanism is most likely required in plants because only a subset of all Arabidopsis genes that have an E2F motif close to the translation start site are transcriptionally induced in E2Fa-DPaOE seedlings. To identify putative cis-acting elements that cooperate with E2F-binding sites, we searched the promoter regions of the 181 putative E2F target genes for the presence of a significantly overrepresented promoter element. Only one 12-bp-long motif (consensus sequence nTTssCGssAAn, with n being predominantly A or T) was found to be overrepresented in the data set, representing the E2F motif itself (Fig. 6). Interestingly, this motif extends the length of the previously recognized canonical E2F element from 8 to 12 bp. The extension of the motif to include the 3′ adenine residues results in a previously unrecognized palindrome-like structure. Failure to identify other possible motifs for cis-acting elements that cluster near the E2F motifs, using a variety of motif detection methods (AlignACE and CONSENSUS), suggests that such motifs might be too degenerated to be detected with currently available bioinformatics tools.
Figure 6.
Sequence logo of the overrepresented motif found in the set of 181 putative E2F target genes. The logo was created based on 272 motif instances using WebLogo (Crooks et al., 2004). The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding nucleic acid at that position.
Analysis of cell-cycle-regulated expression profiles of mammalian E2F target genes revealed that E2F activity appears to be highest at the G1-to-S transition. However, genes normally activated at G2 of the cell cycle were also under E2F control (Ishida et al., 2001; Ren et al., 2002). The recently reported genome-wide expression data from synchronized Arabidopsis MM2d cells (Menges et al., 2003) allowed us to perform a similar analysis. Among all significantly induced genes, we identified several G2-to-M-specific genes (e.g. CYCA2;2 and CYCB1;1). However, almost all putative E2F target genes with a cell-cycle-modulated expression profile were expressed in G1 or S, illustrating a specific role for the E2Fa-DPa complex at the G1-to-S transition. This observation should be interpreted with caution, though, because the timing of gene expression does not necessarily correlate with the activity peak of the protein encoded by these genes. For example, it has been reported that CDKB1;1 transcription is activated during S phase in an E2F-dependent manner, but CDKB1;1 protein activity peaks at the G2-to-M transition (Porceddu et al., 2001; Boudolf et al., 2004), suggesting that S phase activity can contribute to later events in the cell cycle.
Among the core cell cycle genes (Vandepoele et al., 2002) that are induced in the E2Fa-DPaOE seedlings, we identified several negative regulators of the cell cycle, including RBR1, E2Fc, DEL3, KRP3, and WEE1. E2Fc, RBR1, and DEL3 are probably under direct E2Fa-DPa control because these genes have E2F-binding sites in close proximity to the transcription start site. E2Fc and RBR1 were also up-regulated in plants, in which cell divisions were induced as a result of ectopic expression of CYCD3;1 (Dewitte et al., 2003). The transcriptional up-regulation of genes encoding negative regulators in mutants with ectopic cell divisions suggests the existence of a negative feedback mechanism, in which the activating E2Fs regulate their own inactivation through the transcriptional activation of negative regulators of the E2F pathway.
The reported rice genome sequences (Sasaki and Burr, 2000) allowed us to establish a set of evolutionarily conserved E2F plant target genes by screening the promoters of the rice orthologs for the presence of an E2F consensus motif. We found that for more than half of the identified Arabidopsis E2F target genes a conserved E2F element could be found in the promoter of the putative rice ortholog. Arabidopsis genes for which the putative rice orthologous genes did not show an E2F motif could represent dicot-specific E2F target genes. Alternatively, some of the Arabidopsis E2F target genes might encode endocycle-specific E2F targets because in contrast to Arabidopsis, rice does not display somatic endoreduplication. The majority of the 70 evolutionarily conserved E2F targets have a function related to DNA replication, nucleotide metabolism, and chromatin assembly. The observed high number of known replication genes, including those encoding proteins that recognize and establish a functional origin of replication, supports the idea that activation of DNA replication is completely under E2Fa-DPa control. Interestingly, seven genes of unknown function were found among the conserved E2F target genes, as well as other annotated genes encoding proteins with an unidentified role in plant development, such as four WD40-repeat proteins and four zinc-finger proteins. These genes represent strong candidates of components of the DNA replication machinery that function downstream in the E2Fa-DPa pathway, although this will still have to be experimentally validated.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Transgenic E2Fa-DPaOE plants were obtained as described (De Veylder et al., 2002). Plants were grown on 1× Murashige and Skoog medium (Duchefa) and 0.6% plant tissue culture agar (LabM) at 22°C and 65 _μ_E·m−2·s−1 radiation in a 16-h-light/8-h-dark photoperiod.
Microarray Hybridization and Analysis
Experimental procedures are described as follows, according to the minimum information about a microarray experiment standards (Brazma et al., 2001). Microarray results have been submitted to the public Arabidopsis microarray database Genevestigator (https://www.genevestigator.ethz.ch) and the European Arabidopsis Stock Centre (http://arabidopsis.info/).
Experimental Design
Plant lines and growth conditions were as described above. Six days after germination, complete wild-type and E2Fa-DPaOE seedlings were grown side-by-side and harvested at noon into liquid nitrogen. The entire experiment was performed four times. Each of the eight RNA samples was hybridized independently to a microarray.
Array Design
Affymetrix Arabidopsis ATH1 GeneChip microarrays were used throughout the experiment. The list of probes present on the arrays can be obtained from the manufacturer's Web site (http://www.affymetrix.com).
Samples
Total RNA was prepared from frozen tissue using Trizol and purified with RNeasy columns (Qiagen). Labeled RNA was prepared as described previously (Hennig et al., 2004).
Hybridizations
Hybridization of arrays, washing, and detection of labeled cRNA using streptavidin-phycoerythrin were performed as described previously (Hennig et al., 2004).
Measurements
The arrays were scanned with the GS 2500 confocal scanner (Agilent Technologies).
Evaluation, Normalization, and Data Analysis
Signal values were derived from Affymetrix *.cel files using a modified version of robust multiple array normalization (GCRMA; Wu et al., 2004). Subsequent data processing was performed with the statistic package R (version 2.0.1; Ihaka and Gentleman, 1996) and the LIMMA library (Linear Models for Microarray Data; Smyth, 2004) for identification of statistically significant regulation (moderated t-statistics using empirical Bayes shrinkage of the standard errors with multiple testing correction according to Benjamini and Hochberg, 1995). To enrich for biological relevant effects, a gene was considered statistically significantly changed if (1) _P_-value ≤ 0.05 and (2) SLR ≥ 1.
Sequence Analysis
Arabidopsis (Arabidopsis thaliana) promoter sequences 1000 bp upstream of the start codon were extracted based on TIGR gene annotation release 5 (Wortman et al., 2003). Similarly, sequences 2000 bp upstream of the start codon were isolated from the rice (Oryza sativa) TIGR gene annotation version 3.0 (Yuan et al., 2005). All promoter sequences were scanned using DNA-pattern (RSA tools; Van Helden et al., 2000) for the presence of an E2F-like-binding site matching the (A/T)TT(G/C)(G/C)C(G/C)(G/C) sequence that corresponds to all the different E2F-DP-binding motifs described in plants. For all genes for which the cDNA sequence was longer than the coding sequence (CDS), the 5′ UTR was identified by mapping the CDS on the cDNA sequence using BLASTN (Altschul et al., 1997). CDS and cDNA sequences were retrieved from the TIGR annotations. Similarly, the transcription start site on the promoter sequence was defined by mapping the cDNA sequence on the corresponding genomic sequence. For all significantly >2-fold up-regulated Arabidopsis genes, rice homologs were identified with BLASTP (Altschul et al., 1997), and valid homologs were retained (Li et al., 2001). Briefly, this method considers two proteins as being homologous only when they share a substantially conserved region on both molecules with a minimum amount of sequence identity (=30%). In this manner, homology based on the partial overlap of single protein domains between two multidomain proteins, which occasionally leads to significant E-values in BLAST, is not retained. The proportion of identical amino acids in the aligned region between the query and target sequence determined by BLASTP is recalculated to I′ = I × Min(n1/L1, n2/L2), where Li is the length of sequence i, and ni is the number of amino acids in the aligned region of sequence i. This value I′ is then used in the empirical formula for protein clustering proposed by Rost (1999). The GO slim functional classification system for Arabidopsis was downloaded from www.geneontology.org.
MotifSampler (Thijs et al., 2001) was used to find overrepresented regulatory elements in promoter sequences with an Arabidopsis third-order background model. To avoid convergence to local optima, each run was repeated 25 times, and all motifs found were ranked according to their score. The prior probability of finding one motif instance was set to 0.5. AlignACE (Hughes et al., 2000) and CONSENSUS (Hertz and Stormo, 1999) were used to search for cis-acting elements that cooperate with the E2F-binding site.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining permission will be the responsibility of the requestor.
Supplementary Material
Supplemental Data
Acknowledgments
The authors thank Stephane Rombauts and Jeroen Raes for technical assistance and Martine De Cock for help in preparing the manuscript.
1
This work was supported by grants from the Interuniversity Poles of Attraction Program-Belgian Science Policy (P5/13), the European Union (European Cell Cycle Consortium QLG2–CT1999–00454), the Swiss National Science Foundation (3100–061398), the Functional Genomics Center–Zurich, the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (predoctoral fellowships to K.V. and K.V.), and the Fund for Scientific Research (Flanders; postdoctoral fellowship to L.D.V.).
[w]
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25**:** 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B 57**:** 289–300 [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16**:** 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29**:** 365–371 [DOI] [PubMed] [Google Scholar]
- Chabouté M-E, Clément B, Philipps G (2002) S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J Biol Chem 277**:** 17845–17851 [DOI] [PubMed] [Google Scholar]
- Chabouté M-E, Clément B, Sekine M, Philipps G, Chaubet-Gigot N (2000) Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12**:** 1987–1999 [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14**:** 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G (2003) Identification and characterization of E2F7, a novel mammalian E3F family member capable of blocking cellular proliferation. J Biol Chem 278**:** 42041–42049 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Menges M, Bauer U-M, Murray JAH (2001) Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol 47**:** 555–568 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inzé D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa/DPa transcription factor. EMBO J 21**:** 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Joubès J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6**:** 536–543 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14**:** 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54**:** 235–264 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JAH (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15**:** 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17**:** 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K (2003) E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22**:** 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L (2002) Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14**:** 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Robertson D, Hanley-Bowdoin L (2001) Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13**:** 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande PH, Hallstrom TC, Tunyaplin C, Calame K, Nevins JR (2003) Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol Cell Biol 23**:** 3707–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande PH, Zhu W, Rempel RE, Laakso N, Nevins JR (2004) Combinatorial gene control involving E2F and E Box family members. EMBO J 23**:** 1336–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K (1998) Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev 8**:** 28–35 [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135**:** 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15**:** 563–577 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of _cis_-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296**:** 1205–1214 [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5**:** 299–314 [Google Scholar]
- Inzé, D (2005) Green light for the cell cycle. EMBO J 24**:** 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR (2001) Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol 21**:** 4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Kel-Margoulis OV, Farnham PJ, Bartley SM, Wingender E, Zhang MQ (2001) Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J Mol Biol 309**:** 99–120 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J 29**:** 45–59 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132**:** 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavia P, Jansen-Dürr P (1999) E2F target genes and cell-cycle checkpoint control. Bioessays 21**:** 221–230 [DOI] [PubMed] [Google Scholar]
- Li W-H, Gu Z, Wang H, Nekrutenko A (2001) Evolutionary analyses of the human genome. Nature 409**:** 847–849 [DOI] [PubMed] [Google Scholar]
- Ma Y, Croxton R, Moorer RL, Jr., Cress WD (2002) Identification of novel E2F1-regulated genes by microarray. Arch Biochem Biophys 399**:** 212–224 [DOI] [PubMed] [Google Scholar]
- Maiti B, Li J, de Bruin A, Gordon F, Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W, Leone G (2005) Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 280**:** 18211–18220 [DOI] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277**:** 9911–9919 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53**:** 423–442 [DOI] [PubMed] [Google Scholar]
- Müller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev 15**:** 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H, Helin K (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta 1470**:** M1–M12 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld J-P, Segers G, De Veylder L, De Pinho Barrôco R, Casteels P, Van Montagu M, Inzé D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276**:** 36354–36360 [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3**:** 827–837 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Fründt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33**:** 801–811 [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev 16**:** 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P, Stevens R, Perennes C, Jasinski S, Cella R, Tremousaygue D, Bergounioux C (2002) AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol Genet Genomics 266**:** 995–1003 [DOI] [PubMed] [Google Scholar]
- Rost B (1999) Twilight zone of protein sequence alignments. Protein Eng 12**:** 85–94 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3**:** 138–141 [DOI] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR (2002) Interaction of YY1 and E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J 21**:** 5775–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3**:** 1–26 [DOI] [PubMed] [Google Scholar]
- Stanelle J, Stiewe T, Theseling CC, Peter M, Pützer BM (2002) Gene expression changes in response to E2F1 activation. Nucleic Acids Res 30**:** 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277**:** 32978–32984 [DOI] [PubMed] [Google Scholar]
- Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouzé P, Moreau Y (2001) A higher order background model improves the detection of regulatory elements by Gibbs sampling. Bioinformatics 17**:** 1113–1122 [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA (2002) Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3**:** 11–20 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Taylor JS, Braasch I, Meyer A (2001) The ghost of selection past: rates of evolution and functional divergence in anciently duplicated genes. J Mol Evol 53**:** 436–446 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14**:** 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Helden J, André B, Collado-Vides J (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast 16**:** 177–187 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inzé D, De Veylder L (2003) Microarray analysis of _E2Fa-DPa_-overexpressing plants reveals changes in the expression levels of genes involved in DNA replication, cell wall biosynthesis, and nitrogen assimilation. J Cell Sci 116**:** 4249–4259 [DOI] [PubMed] [Google Scholar]
- Weinmann A, Bartley SM, Zhang T, Zhang MQ, Farnham PJ (2001) Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol 21**:** 6820–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK, Jr., Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, White OR, Town CD (2003) Annotation of the Arabidopsis genome. Plant Physiol 132**:** 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99**:** 909–917 [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, Wortman J, Buell CR (2005) The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 138**:** 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data