A Herpes Simplex Virus Type 1 Mutant Expressing a Baculovirus Inhibitor of Apoptosis Gene in Place of Latency-Associated Transcript Has a Wild-Type Reactivation Phenotype in the Mouse (original) (raw)
Abstract
The latency-associated transcript (LAT) is essential for the wild-type herpes simplex virus type 1 (HSV-1) high-reactivation phenotype since LAT− mutants have a low-reactivation phenotype. We previously reported that LAT can decrease apoptosis and proposed that this activity is involved in LAT's ability to enhance the HSV-1 reactivation phenotype. The first 20% of the primary 8.3-kb LAT transcript is sufficient for enhancing the reactivation phenotype and for decreasing apoptosis, supporting this proposal. For this study, we constructed an HSV-1 LAT− mutant that expresses the baculovirus antiapoptosis gene product cpIAP under control of the LAT promoter and in place of the LAT region mentioned above. Mice were ocularly infected with this mutant, designated dLAT-cpIAP, and the reactivation phenotype was determined using the trigeminal ganglion explant model. dLAT-cpIAP had a reactivation phenotype similar to that of wild-type virus and significantly higher than that of (i) the LAT− mutant dLAT2903; (ii) dLAT1.5, a control virus containing the same LAT deletion as dLAT-cpIAP, but with no insertion of foreign DNA, thereby controlling for potential readthrough transcription past the cpIAP insert; and (iii) dLAT-EGFP, a control virus identical to dLAT-cpIAP except that it contained the enhanced green fluorescent protein open reading frame (ORF) in place of the cpIAP ORF, thereby controlling for expression of a random foreign gene instead of the cpIAP gene. These results show that an antiapoptosis gene with no sequence similarity to LAT can efficiently substitute for the LAT function involved in enhancing the in vitro-induced HSV-1 reactivation phenotype in the mouse.
Herpes simplex virus type 1 (HSV-1) establishes lifelong latent infections in host sensory neurons. This virus is widespread in the general population. When the eye is infected, the virus spreads from epithelial cells to the peripheral nerve endings and then travels to the trigeminal ganglia (TG) via retrograde axonal transport and establishes latent infection in sensory neurons of the TG. During neuronal latency, HSV-1 has no apparent impact on the infected individual. However, the latent virus can reactivate sporadically throughout the life of the individual. This occurs through a mechanism or mechanisms that are currently not fully understood. HSV-1 reactivation in the TG results in the virus returning to the eye via anterograde axonal transport. At the eye, reactivated HSV-1 can replicate, and infectious virus is shed in tears. Recurrent ocular HSV-1 infection may cause corneal disease leading to corneal scarring and loss of vision. Consequently, HSV-1 is one of the most common infectious causes of corneal blindness in the developed world.
During HSV-1 neuronal latency, the latency-associated transcript (LAT) is the only abundantly transcribed viral gene (39, 45). The primary LAT transcript is approximately 8.3 kb long (11, 53) and partially or completely overlaps three viral genes, those encoding AL, ICP0, and ICP34.5, in an antisense direction (33, 39, 45). A very stable intron, the 2-kb LAT, is spliced from the primary transcript (12) and is the major LAT RNA detected during latency (11, 42, 44, 50-52). LAT− mutants have impaired reactivation phenotypes in small-animal models (3, 9, 17, 23, 29, 36, 40, 43), indicating that a LAT function enhances the HSV-1 reactivation phenotype. However, the nature of this LAT function remains unresolved.
It has been proposed that LAT may affect the latency reactivation cycle by (i) antisense regulation of the important immediate-early genes for ICP0 and/or ICP4 (via an extended LAT transcript of approximately 15 kb) (6, 13, 39, 45), (ii) association of the stable 2-kb LAT with ribosomes (26), and/or (iii) expression of a LAT protein that can substitute for an ICP0 function (46). However, a LAT function capable of fully supporting the wild-type reactivation phenotype maps to within the first 1.5 kb of the primary LAT transcript (i.e., LAT nucleotides [nt] 1 to 1,499) (4, 32), which contains only LAT exon 1 and the first 42% (837 nucleotides) of the stable 2-kb LAT intron. This region of LAT does not overlap with the ICP0 or ICP4 gene, does not contain the open reading frame (ORF) encoding the putative LAT protein, and does not retain the stability of the 2-kb LAT (32). Thus, LAT can support the wild-type reactivation phenotype in rabbits (32) and mice (30) by a mechanism that does not involve antisense regulation of ICP0 or ICP4, does not require expression of the entire 2-kb LAT intron, does not require the presence of a stable LAT RNA, and does not require expression of the putative LAT protein with ICP0-like functions.
Although there has been some controversy regarding LAT's antiapoptosis activity (47), it has been reported that (i) plasmids expressing LAT have antiapoptosis activity in the absence of other viral genes (1, 19, 20, 28), (ii) LAT has antiapoptosis activity in the context of the virus in infected tissue culture cells (16, 21), and (iii) LAT− mutants appear to have increased apoptosis in rabbit and mouse TG in vivo compared to LAT+ viruses (1, 4a, 28). We have mapped LAT's antiapoptosis activity to the first 1.5 kb of the primary LAT transcript (19, 20), the same region to which we previously mapped LAT's ability to enhance the HSV-1 reactivation phenotype (32). Collectively, these observations support our hypothesis that LAT's antiapoptosis activity is crucial for the latency reactivation cycle.
Recently, we showed that the bovine herpesvirus 1 (BHV-1) LAT homologue, the LR gene, can efficiently substitute for the LAT function involved in supporting the wild-type HSV-1 reactivation phenotype (34). This ability was abrogated by inhibiting expression of the LR protein encoded by ORF-2 (25). Although this LR protein has antiapoptosis activity (25), it also has additional activities that have not been ruled out for LAT. For example, the LR gene can inhibit cell growth, interact with cyclin-dependent kinase 2/cyclin complexes, inhibit transcription of the BHV-1 immediate-early 1 promoter, and inhibit bICP0 RNA expression (5, 10, 14, 41). Since LAT and LR are functional homologues that could share a non-antiapoptosis-related function involved in the wild-type reactivation phenotype, these studies supported, but did not prove, the hypothesis that LAT enhances the reactivation phenotype via its antiapoptosis activity. Therefore, we constructed and analyzed dLAT-cpIAP. In this HSV-1 mutant, both copies of the region of LAT that supports the reactivation phenotype (LAT nt 76 to 1667) were removed and replaced by the baculovirus inhibitor of apoptosis gene cpIAP (from Cydia pomonella granulosis virus) (2, 7, 8, 18, 48). cpIAP functions as an inhibitor of the caspase family of apoptotic proteases. Since LAT and the cpIAP gene are unrelated, it is extremely unlikely that they share an undetermined non-antiapoptosis-related function. We report here that dLAT-cpIAP had a wild-type reactivation phenotype in the TG explant reactivation mouse model. This strongly suggests that LAT's antiapoptosis activity is sufficient to account for LAT's ability to support the wild-type HSV-1 reactivation phenotype.
MATERIALS AND METHODS
Cell lines.
Rabbit skin (RS) cells, CV-1 cells, and Neuro2A cells (CCL 131; American Type Culture Collection) were maintained in Eagle minimal essential medium (MEM) with 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 10% fetal bovine serum (Promega Scientific), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Sigma, St. Louis, MO).
Viruses.
All parental and mutant viruses were triply plaque purified and passaged only one or two times in rabbit skin (RS) cells prior to use. The wild-type McKrae (wt), dLAT2903, dLAT1.5, and dLAT-EGFP viruses have been previously described (29, 30, 37).
Apoptosis in tissue culture cells.
As previously described (21), Neuro2A cells (1 × 106 to 2 × 106) were seeded into six-well plates 2 days prior to infection and maintained throughout in medium with 0.5% fetal bovine serum. Cells were infected with 5 PFU/cell. Apoptosis was examined at 24 h postinfection using a modified DNA laddering assay as previously described (21).
Construction of dLAT-cpIAP.
Plasmid pLAT5.6 was constructed by cloning the SwaI-to-MluI LAT locus (corresponding to 800 bp of the LAT promoter and the first 2,850 bp of the primary 8.3-kb LAT) into the BamHI site of pNEB193. The plasmid was digested with StyI and HpaI to remove a StyI-HpaI region corresponding to LAT nt 76 to 1667. BamHI linkers were added, followed by ligation. The resulting plasmid, pNEBLAT, thus contains HSV-1 DNA corresponding to LAT nt −800 to +76, followed by a BamHI site, followed by LAT nt +1667 to +2850. A plasmid containing the cpIAP gene (IAP-3 gene from Cydia pomonella granulosis virus) was a gift from L. K. Miller (Department of Genetics, University of Georgia, Athens). The complete cpIAP ORF, including 36 bp upstream of the ATG initiation codon and 104 bp downstream of the TAA stop codon, was PCR amplified, cloned into pCRII-TOPO (Invitrogen, Inc.), and subsequently cloned into the EcoRI site of plasmid pTRE (Clontech, Inc.). BglII linkers were added to the beginning of the cpIAP ORF and the end of the simian virus 40 (SV40) poly(A) signal (nt 488 to 1087 of pTRE) using the PCR-TOPO cloning method. The entire cpIAP ORF sequence and SV40 poly(A) signal sequence were isolated following BglII digestion and cloned into the BamHI restriction site of pNEBLAT between LAT nt 76 and 1667. The resulting plasmid, pNEBLAT-IAP, was verified by DNA sequencing. It was then cotransfected with infectious dLAT2903 genomic DNA into RS cells to generate the mutant dLAT-cpIAP by homologous recombination, as previously described for the construction of other HSV-1 mutants (29, 30, 35). Briefly, the cotransfection mix was plated on RS cells, individual viral plaques were analyzed by restriction digestion and Southern analysis, and plaques containing mixtures of wt and mutant virus were repeatedly replated and analyzed until all of the plaques appeared free of wt virus. Those plaques were then triply plaque purified, and the structure of the virus was confirmed as described above. In the resulting chimeric virus, dLAT-cpIAP, LAT nt 76 to 1667 are replaced by the complete cpIAP ORF followed by the SV40 poly(A) signal sequence, thus placing cpIAP under control of the intact LAT promoter.
Southern analyses.
Briefly, viral DNA was digested with BamHI; the restriction fragments were separated in a 1% agarose gel, transferred to Zeta paper, rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min, and cross-linked to the membrane by UV light; and DNA-DNA hybridization was performed with 32P-labeled probes as previously described (29, 30, 35).
Mice.
Eight- to 10-week-old Swiss-Webster female mice (Jackson Labs) were used for all experiments. Viral infections were done without corneal scarification as we previously described (29, 30, 35).
RNA isolation.
Total RNA was isolated from individual cell monolayers, each from one well of a six-well plate. Isolations were performed using Trizol followed by a treatment with 0.1 U of DNase I (Roche) in Tris-EDTA (pH 8.0) containing 100 mM MgCl2, 10 mM dithiothreitol, and 0.01 U of RNase inhibitor at 37°C for 30 min. The total RNA was then isolated using a GenElute Mammalian Total RNA isolation kit (Sigma, St. Louis, MO).
Probes and primers.
The LAT primers were OPUSQ (LAT nt 470 to 500; CCACAACGGCCCGGCGCATGCGCTGTGGTT) for the LAT coding strand and OPURS (LAT nt 646 to 615; TCTTTGTTGAACGACACCGGGCGCCCTCGA) for the LAT anticoding strand. The cpIAP primers were cpIAPF2 (cpIAP ORF nt 141 to 161; AGTGGAGATAATGCGTTGGA) for the cpIAP coding strand and cpIAPR2 (cpIAP ORF nt 517 to 497; GCAACGGTCGAACCATCTTA) for the cpIAP anticoding strand. The DNA IAP probe used for Southern analysis of dLAT-cpIAP corresponds to a StyI-to-StyI restriction fragment within the cpIAP ORF (IAP nt 77 to 481). The LAT DNA probe corresponds to a NotI-to-AlwNI LAT restriction fragment (LAT nt −363 to +1677).
RT-PCR.
First-strand cDNAs were synthesized from 0.5 μg of total RNA using the ThermoScript reverse transcription-PCR (RT-PCR) system (GIBCO) according to the manufacturer's recommendations. PCRs were performed in a volume of 50 μl consisting of 1× PCR buffer containing DNA polymerase, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each primer, 1 mM MgSO4, 0.5 U of Platinum Pfx DNA polymerase (GIBCO), and 2.5 μl of completed RT reaction product. The reactions were cycled 30 times at 94°C for 30 s, 60°C for 30 s, and 68°C for 30 s and then extended once at 68°C for 7 min in a DNA thermal cycler (GeneAmp PCR system 2700; PE Applied Biosystems). Ten microliters of each of the RT-PCR products was separated in a 1.5% agarose gel in Tris-acetate-EDTA buffer and then visualized by UV illumination after staining with 1 μg/ml ethidium bromide.
Western blots.
Total cell extracts were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane. The membrane was incubated with polyclonal rabbit anti-IAP antibody at a 1:200 dilution (Abcam Inc., Cambridge, MA) and then washed, and the antibody bound to the blots was visualized by chemiluminescence with anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (Chemicom, Inc.).
Statistical analysis.
Analyses were performed using the personal computer program GraphPad Prism, version 4.00, for Windows (GraphPad Software, San Diego, California).
RESULTS
Construction and genomic structure of dLAT-cpIAP.
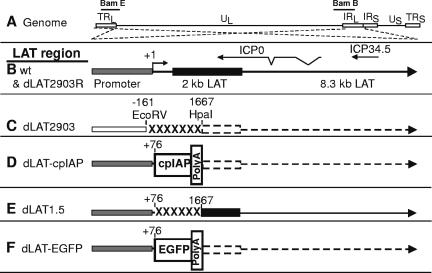
The genomic structures of dLAT-cpIAP and the other viruses used for this study are shown schematically in Fig. 1. All viruses were derived from HSV-1 strain McKrae. Figure 1A shows the prototypic structure of the wild-type HSV-1 McKrae genome. The relative locations of the BamHI E (Bam E) and BamHI B (Bam B) genomic restriction fragments are shown for reference. The viral long repeats (TRL and IRL) are expanded (dashed lines) to show the relative locations and statuses of the LAT gene and LAT region in each of the panels below. The dashed lines cross to indicate that the repeats are inverted (i.e., the DNA sequences are in opposite orientations). The wild-type and dLAT2903R LAT region is shown in panel B. (dLAT2903R is marker-rescued dLAT2903, which is shown in panel C.) The locations of the ICP0 and ICP34.5 mRNAs are shown for reference. The primary LAT transcript in wild-type virus and marker-rescued dLAT2903R is approximately 8.3 kb (49, 50) (large arrow). A very stable and easily detected 2-kb LAT (black rectangle) is an intron derived by splicing of the primary LAT (12). The LAT region of dLAT2903 (panel C) contains a deletion in both copies of LAT from −161 to +1,667 relative to the start of the primary LAT transcript (indicated by “XXXXX”). This virus is missing key promoter elements, makes no LAT RNA (indicated by a dashed line), is a true LAT null mutant, and has a low reactivation phenotype compared to wt HSV-1 (29). The construction of dLAT2903 and dLAT2903R has been previously described (29). The LAT region of dLAT-cpIAP (panel D) contains the complete ORF of the baculovirus cpIAP gene preceded by the entire LAT promoter, including the portion missing in dLAT2903 (i.e., LAT nt −161 to +76). The cpIAP ORF is followed by the SV40 polyadenylation signal sequence. This poly(A) signal sequence was cloned from and corresponds to nt 1066 to 1266 of a BD Biosciences Clontech plasmid. dLAT-cpIAP thus contains two complete copies of the cpIAP ORF (one in each viral long repeat), each in place of LAT nt 76 to 1667 and driven by the LAT promoter. The poly(A) site decreases the transcription of LAT downstream of the cpIAP insertion to undetectable levels (represented by a dashed arrow; data not shown). The structures of dLAT1.5 and dLAT-EGFP (panels E and F) have been previously published (30, 37) and are discussed below.
FIG. 1.
Structure of the LAT region of dLAT-cpIAP and other viral mutants. (A) HSV-1 genomic structure. TRL and IRL indicate the viral long repeats (terminal and internal, respectively). IRS and TRS indicate the viral short repeats. UL and US indicate the long and short unique regions, respectively. The dashed lines indicate that the regions of the TRL and IRL are expanded below, with the TRL inverted relative to the IRL so that both identical regions can be represented by a single image. (B) Wild-type (wt) and dLAT2903R (marker-rescued dLAT2903; see panel C) LAT region. The intact LAT promoter is indicated by the shaded rectangle. The solid black rectangle indicates the stable 2-kb LAT. The relative locations of ICP0 and ICP34.5 are shown for reference. (C) LAT− dLAT2903 has a deletion of LAT nt −161 to +1667 (XXXXXX). dLAT2903 is a true LAT null mutant that is missing the primary LAT promoter elements between −161 and +1. dLAT2903 is also deleted for a putative secondary LAT promoter, LAP2, located within the 5′ end of the primary LAT transcript prior to the start of the 2-kb LAT (15). This mutant is therefore not capable of expressing any LAT RNA (dashed lines) (29). (D) dLAT-cpIAP contains the antiapoptosis gene cpIAP followed by a poly(A) signal inserted in place of LAT nt 76 to 1667. The entire LAT promoter is present. No LAT RNA is transcribed past the poly(A) site. (E) dLAT1.5 is identical to dLAT-cpIAP, but with no inserted ORF or polyadenylation signal. The region of LAT past the deletion is transcribed normally. (F) dLAT-EGFP is identical to dLAT-cpIAP, except that the irrelevant gene EGFP is inserted instead of cpIAP.
Because the cpIAP gene has no sequence homology to the HSV-1 LAT gene, the construction of dLAT-cpIAP by homologous recombination required that the cpIAP gene was first cloned into a plasmid containing appropriate HSV-1 flanking sequences. This and additional details of the construction of dLAT-cpIAP are given in Materials and Methods.
Confirmation of the dLAT-cpIAP genomic structure by Southern blot analysis.
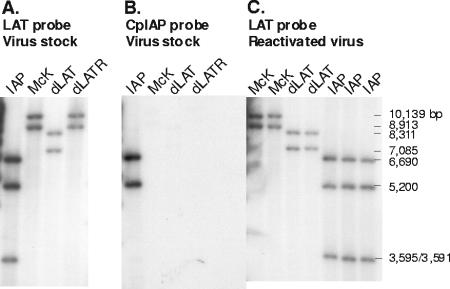
dLAT-cpIAP viral DNA was purified and digested with BamHI. Restriction digestion fragments were separated by agarose gel electrophoresis, transferred to a Zeta membrane, and hybridized to a 32P-labeled NotI-AlwNI (LAT nt −363 to +1677) restriction fragment (Fig. 2A). As expected, only two BamHI restriction fragments are seen in the McKrae (McK) and dLAT2903R (dLATR) lanes. The larger band is BamHI B (10,139 bp; genomic nt 113,322 to 123,461, corresponding to LAT nt −5479 to +4660) (also see Fig. 1A). This restriction fragment spans the junction between the internal long repeat and the long unique region. The smaller band is BamHI E (8,913 bp; genomic nt 2,905 to 11,818, corresponding to LAT nt −4253 to +4660), which spans the junction between the terminal long repeat and the long unique region (Fig. 1A). Thus, although the BamHI B and E restriction fragments have one common restriction site at LAT nt +4660 within the long repeats, they are different sizes because the other restriction site of each fragment is in a different location in the unique long region of the virus genome. The same two bands are seen with dLAT2903 (dLAT), except that each is 1,828 bp shorter due to the deletion of LAT nt −161 to +1667 (BamHI B, 8,311 bp; BamHI E, 7,085 bp). dLAT-cpIAP has an additional BamHI site between the end of the cpIAP ORF and the beginning of the poly(A) insertion signal. Therefore, four BamHI restriction fragments are predicted, two from each of the modified BamHI B and E fragments (5,200 bp and 3,595 bp from BamHI E and 6,690 bp and 3,591 bp from BamHI B). However, the predicted sizes of two of these bands are very similar (3,595 and 3,591 bp) and appear as one band. Thus, only three bands are seen in the dLAT-cpIAP (IAP) lane. To further confirm the genomic structure of dLAT-cpIAP in the LAT region, the probe was stripped from the membrane shown in Fig. 2A, and the membrane was reprobed with a cpIAP-specific probe. This probe corresponds to a region within the cpIAP ORF (nt 77 to 481) and therefore should hybridize only to the 5,200-bp and 6,686-bp IAP-containing fragments. As seen in Fig. 2B, the cpIAP probe hybridized only to the expected bands (lane IAP) and did not hybridize to any bands in the McKrae (McK), dLAT2903 (dLAT), and dLAT2903R (dLATR) lanes. These analyses confirmed the correct genomic structure of dLAT-cpIAP in the LAT region. Panel C shows analyses of viruses that were reactivated following mouse TG explantation and is discussed below.
FIG. 2.
Southern hybridization analysis of dLAT-cpIAP's genomic structure. dLAT-cpIAP viral DNA was purified and digested with BamHI. Restriction digestion fragments were separated by agarose gel electrophoresis, transferred to a Zeta membrane, and hybridized to a 32P-labeled NotI-AlwNI (LAT nt −363 to +1667) restriction fragment (A and C). The membrane from panel A was stripped and reprobed using a 32P-labeled probe specific for nt 141 to 517 of the cpIAP ORF. Panels A and B show DNAs prepared from the viral stocks used for this report. Panel C shows DNAs from reactivated viruses recovered from latently infected mouse TG following explantation.
Tissue culture characterization of dLAT-cpIAP replication, cpIAP mRNA expression, and cpIAP protein expression.
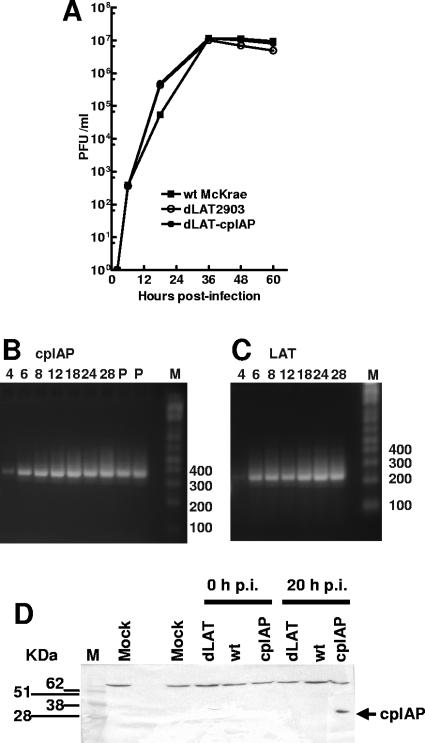
CV-1 cells were infected at a multiplicity of infection (MOI) of 0.01 with dLAT-cpIAP, dLAT2903 (the immediate parental virus), or wild-type McKrae (the parental virus for dLAT2903). Replication appeared similar for all three viruses (Fig. 3A), indicating that the insertion of the cpIAP gene followed by the SV40 poly(A) signal sequence in place of LAT nt 76 to 1667 did not significantly alter virus replication in tissue culture and that the ICP0 gene was functioning correctly.
FIG. 3.
Characterization of dLAT-cpIAP in tissue culture. (A) Replication in CV-1 cells. Cell monolayers were infected with virus at a low MOI of 0.01. At the indicated times, infected cell monolayers with culture medium were harvested and freeze-thawed to release virus, and the amount of total infectious virus was determined using standard plaque assays. (B and C) cpIAP RNA expression in dLAT-cpIAP-infected CV-1 cells. CV-1 cell monolayers were infected with dLAT-cpIAP (B) or wild-type McKrae (C) at an MOI of 5. Total RNAs were isolated at the indicated times p.i. (hours), and RT-PCRs were performed using primers specific for cpIAP or LAT (see Materials and Methods and the text). Lanes: P, PCR of cloned DNA as marker; M, DNA size markers (numbers represent base pairs). (D) Western blot detection of cpIAP expressed by dLAT-cpIAP. RS cells were infected at an MOI of 5, and cell extracts were prepared immediately following infection (0 h p.i.) and at 20 h p.i. Extracts were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed using a cpIAP-specific antibody (Abcam, Inc.). Lanes: dLAT, dLAT2903-infected cells; wt, wild-type McKrae; cpIAP, dLAT-cpIAP; M, molecular size marker; mock, mock-infected cells. The lane between the mock lanes is empty. The arrow indicates the location of the cpIAP band.
CV-1 cells were infected at an MOI of 5, and total RNA was prepared at various times postinfection (p.i.). RT-PCR was performed using cpIAP-specific primers corresponding to cpIAP nt 141 to 160 and 517 to 497 or primers specific for LAT exon 1 (the region prior to the stable 2-kb LAT intron) corresponding to LAT nt 470 to 500 and 652 to 621. The RT-PCR products were analyzed by agarose gel electrophoresis as described in Materials and Methods (Fig. 3B and C). Lanes labeled P are cpIAP products generated from cloned (plasmid) DNA. A faint cpIAP RT-PCR product with the expected mobility (376 bp) was detected at 4 h p.i. (Fig. 3B). The cpIAP RT-PCR product was readily detectable at 6 h, and the intensity of this band remained fairly constant until at least 28 h p.i. A faint LAT RT-PCR product with the expected mobility (182 bp) was detected at 4 h p.i., while a more intense band was seen from 6 to 28 h p.i. Thus, cpIAP RNA was expressed with similar kinetics as LAT. As expected, no LAT or cpIAP RT-PCR product was seen with RNAs from mock-infected cells or when reverse transcriptase was left out of the reaction mix (data not shown). Note that the LAT primers used generated a LAT product from outside the stable 2-kb LAT. This was done to eliminate any potential artifacts due to accumulation of the stable 2-kb LAT.
RS cells were infected at an MOI of 5. Immediately after infection (Fig. 3D; 0 h p.i.) or at 20 h p.i., total cell extracts were subjected to SDS-PAGE, proteins were transferred to a membrane, and Western analysis was performed using a cpIAP-specific antibody. A band with the correct mobility for cpIAP (33 kDa) was recognized by the cpIAP antibody in the dLAT-cpIAP (lane cpIAP)-infected cells at 20 h p.i. (arrow), but not in dLAT-cpIAP-infected cells immediately after infection or in dLAT2903 (dLAT)- or wt McKrae-infected cells at either time p.i. The band migrating more slowly than the 62-kDa molecular size marker is an unidentified cellular band that also cross-reacts with the polyclonal rabbit serum in uninfected RS cells (not shown). In summary, cpIAP was expressed during productive infection and did not impair viral replication in tissue culture.
The cpIAP protein expressed by dLAT-cpIAP is functional.
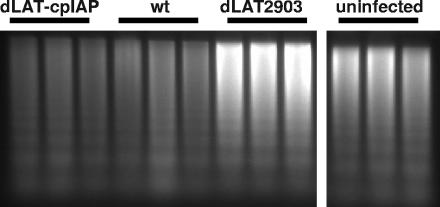
We previously developed a modified DNA ladder assay that allows us to examine the ability of LAT, in the context of the virus, to block apoptosis in infected Neuro2A cells (21). We used this approach to confirm that the cpIAP protein seen in the above Western blot was functional. Neuro2A cells were infected with 5 PFU/cell of dLAT-cpIAP, wt McKrae, or dLAT2903. Apoptosis was examined at 24 h postinfection as we previously described (21). For this assay, the harvested cells are incubated in hypotonic buffer for 2 h at 4°C. This lyses the cells without disrupting the nuclei and allows fragmented DNA to be eluted from the nuclei while leaving the large unfragmented chromosomal DNA trapped inside the nucleus. The nuclei (and unfragmented chromosomal DNA) are removed by centrifugation, and equal aliquots are subjected to electrophoresis. This results in both more “laddering” and more total (fragmented) DNA in samples from apoptotic cultures. Triplicate independent samples were used to confirm the assay's reproducibility. Neuro2A cells infected with dLAT-cpIAP (Fig. 4, lanes 1, 2, and 3) or the wild type (lanes 4, 5, and 6) had less fragmented DNA than Neuro2A cells infected with dLAT2903 (lanes 7, 8, and 9) or than uninfected cells (lanes 10, 11, and 12). Both LAT in Neuro2A cells infected with wt virus and cpIAP in Neuro2A cells infected with dLAT-cpIAP virus therefore appeared to decrease both the background level of apoptosis seen in uninfected Neuro2A cell cultures and the additional apoptosis induced following infection of Neuro2A cells with the LAT− mutant dLAT2903. It is extremely unlikely that LAT nt −161 to +76 (present in dLAT-cpIAP but not in dLAT2903) somehow contributed to dLAT-cpIAP's antiapoptosis activity, since we previously showed that this LAT region does not have any detectable antiapoptosis activity (19). In addition, DNA fragmentation in dLAT1.5 (which contains these LAT sequences; Fig. 1E)-infected cells was indistinguishable from that in dLAT2903-infected cells (not shown). Thus, in dLAT-cpIAP, the cpIAP ORF followed by the SV40 poly(A) signal sequence and driven by the LAT promoter appeared capable of substituting for the deleted LAT region in terms of blocking apoptosis in the context of the virus. This indicated that the cpIAP protein expressed by dLAT-cpIAP was functional.
FIG. 4.
Protection against DNA fragmentation by dLAT-cpIAP. Neuro2A cells were infected with 5 PFU/cell of dLAT-cpIAP, wt McKrae, or dLAT2903. At 24 h p.i., apoptotic DNA ladder assays were performed in triplicate as previously described (21). All lanes are from the same gel, but irrelevant lanes between lanes 9 and 10 were deleted from the final image.
Replication of dLAT-cpIAP in mouse eyes.
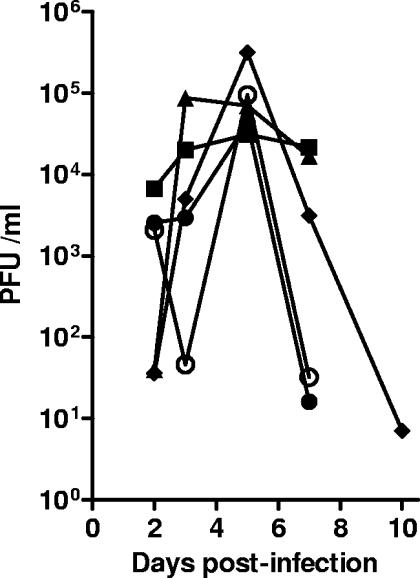
Eight-week-old Swiss Webster mice were infected with 2 × 105 PFU of dLAT-cpIAP, dLAT2903, wild-type McKrae, dLAT1.5, or dLAT-EGFP/eye as described in Materials and Methods. Tear swabs were collected from 10 eyes/group at various times, and the amount of infectious virus in each swab was determined by a standard plaque assay on RS cell monolayers (Fig. 5). All of the viruses replicated to similar peak titers, suggesting that dLAT-cpIAP replicated normally in mouse eyes.
FIG. 5.
Replication of dLAT-cpIAP in mouse eyes. Mice were ocularly infected with 2 × 105 PFU/eye. Tear swabs were collected on days 3, 5, 7, and 10 p.i. Each symbol represents the average titer for each group (n = 10). Solid circles, wt McKrae; open circles, dLAT-cpIAP; squares, dLAT2903; triangles, dLAT1.5; diamonds, dLAT-EGFP.
cpIAP RNA is expressed in TG of mice latently infected with dLAT-cpIAP.
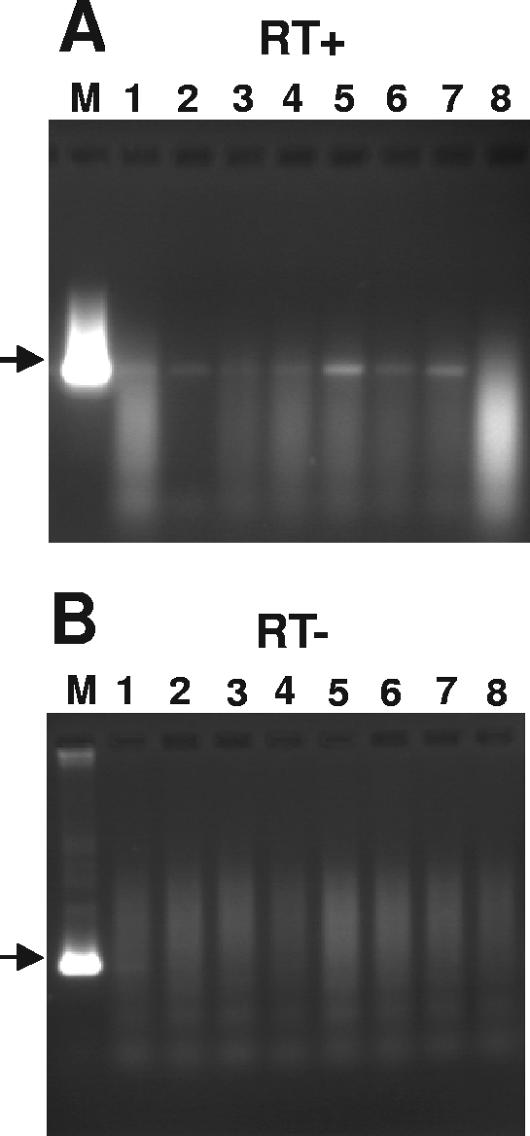
Mice were infected with 2 × 105 PFU of dLAT-cpIAP/eye as described above. At 30 days p.i., the mice were euthanized, TG were removed, and the total RNA from individual TG was subjected to RT-PCR analysis using cpIAP-specific primers as described above for cpIAP RNA in infected tissue culture cells (Fig. 6). Lane M shows the PCR product obtained from a cpIAP-containing plasmid amplified using identical primers to those used for RNAs isolated from TG and acts as a positive control and a size marker. An RT-PCR product corresponding in size to the marker in lane M was detected using extracts from seven of the eight latently infected TG (lanes 1 to 7). The variation in intensity of the cpIAP RT-PCR products among lanes 1 to 7 is consistent with the variation in HSV-1 genome copy numbers seen among individual latently infected TG (40). Thus, as expected, the LAT promoter in dLAT-cpIAP appeared to express cpIAP RNA during latency.
FIG. 6.
Expression of cpIAP RNA in TG of mice latently infected with dLAT-cpIAP. The total RNA was isolated from individual TG (lanes 1 to 8) at 30 days p.i. with dLAT-cpIAP (initial infectious dose, 2 × 105 PFU/eye). RT-PCR was performed using the same cpIAP-specific primers as those described for Fig. 3B, and the products were run in a 2% agarose gel. Lane M, PCR of cloned cpIAP DNA, used as a size marker. (A) Reverse transcriptase was included. (B) Reverse transcriptase was not included.
Wild-type explant TG reactivation of dLAT-cpIAP.
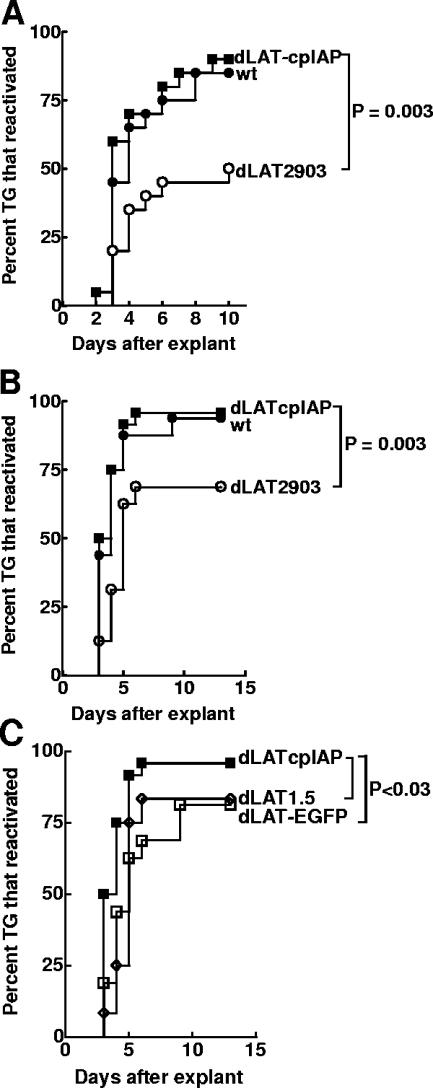
Mice were infected with 2 × 105 PFU of dLAT-cpIAP, wt, or dLAT2903/eye. On day 30 p.i., surviving mice were sacrificed, and individual TG were cultured in tissue culture medium. Aliquots of medium were removed from each culture daily for up to 10 days and plated on indicator cells (RS cells) to look for the presence of reactivated virus. The results are shown in Fig. 7A. The decreased reactivation phenotype of LAT− viruses compared to that of wt viruses in the mouse explant TG model is often only apparent when the time to the reactivation event is examined rather than just determining the percentage of TG from which reactivated viruses are detected at some arbitrary late time point after explantation (43). Since survival analysis (Kaplan-Meier) is appropriate for any kind of experiment where the result is expressed as the time to a well-defined end point, we feel that this analysis is preferable to chi-square (or Fisher's exact) analysis. The reactivation phenotypes of dLAT-cpIAP and wt were similar (P = 0.5), and both were significantly higher than that of LAT− dLAT2903 (P = 0.003 for dLAT-cpIAP and 0.015 for wt). Thus, the substitution of cpIAP for LAT nt 76 to 1667 appeared to have “rescued” the LAT− low-reactivation phenotype of dLAT2903 to the LAT+-like high-reactivation phenotype in the mouse model. This strongly suggests that the LAT function involved in supporting the wt reactivation phenotype in mice can be replaced by an alternative antiapoptosis gene. To confirm that the virus recovered from the explanted TG was the correct input virus, reactivated viruses from some of the cultures were analyzed by Southern blotting (Fig. 2C). As expected, the results shown in the lanes labeled IAP confirmed that the virus recovered from the explanted TG of latently dLAT-cpIAP-infected mice was dLAT-cpIAP.
FIG. 7.
Explant reactivation of dLAT-cpIAP from mouse TG. (A) Experiment 1. Ten-week-old Swiss Webster mice were infected with 2 × 105 PFU/eye. On day 30 p.i., individual TG (n = 20/group) were explanted into tissue culture medium. Aliquots were removed daily and plated on indicator cells (RS cells) to look for the presence and time of first appearance of reactivated virus. For dLAT-cpIAP versus dLAT2903, P = 0.003 by Kaplan-Meier survival analysis; for dLAT-cpIAP versus wt, P = 0.5. (B and C) Experiment 2. The experiment was identical to experiment 1 except that mice were 8 weeks old at the time of infection. For dLAT-cpIAP, n = 24; for wt, n = 16; for dLAT2903, n = 16; for dLAT1.5, n = 12; for dLAT-EGFP, n = 16. For dLAT-cpIAP versus wt, P = 0.7; for dLAT-cpIAP versus dLAT2903, P = 0.003; for dLAT-cpIAP versus dLAT1.5, P = 0.01; for dLAT-cpIAP versus dLAT-EGFP, P = 0.02.
Low-reactivation phenotype of dLAT1.5 and dLAT-EGFP in mice.
A second, independent experiment was done in which two additional mutants, dLAT1.5 and dLAT-EGFP, were included as controls (Fig. 7B and C). dLAT1.5 contains the same LAT deletion as dLAT-cpIAP (LAT nt 76 to 1667) but does not contain any foreign DNA insert (30) (Fig. 1E). In addition, dLAT1.5 transcribes all of LAT past nt 1667 (i.e., past the location of the cpIAP insert in dLAT-cpIAP). dLAT-EGFP is identical to dLAT-cpIAP, except that it contains the enhanced green fluorescent protein (EGFP) gene in place of cpIAP (37) (Fig. 1F). dLAT1.5 acts as a control for the unlikely possibility that LAT nt −161 to +76 (present in dLAT-cpIAP and dLAT1.5 but absent from dLAT2903) or a small amount of LAT transcription past the cpIAP-SV40 poly(A) signal sequence inserted in dLAT-cpIAP might account for the LAT+-like wt reactivation phenotype of dLAT-cpIAP, while dLAT-EGFP serves as a control for the insertion of an irrelevant gene or DNA sequence into the LAT locus used for the insertion of cpIAP into dLAT-cpIAP.
Mice were infected as described above, and on day 30 p.i. TG explant reactivations were performed. As in the first mouse experiment, dLAT-cpIAP had a reactivation phenotype significantly higher than that of dLAT2903 (Fig. 7B; P = 0.003) and similar to that of the wt (P = 0.69). As expected, both dLAT1.5 and dLAT-EGPF had reactivation phenotypes significantly lower than that of dLAT-cpIAP (Fig. 7C; P = 0.01 and P = 0.02, respectively) and not significantly different from that of dLAT2903 (P = 0.63 and P = 0.48, respectively). These results are identical to those of studies with a rabbit ocular model in which dLAT1.5 and dLAT-EGFP had reduced spontaneous reactivation (compared to the wild type) that was indistinguishable from that of dLAT2903 (19, 37). As described above, these statistical analyses are based on a Kaplan-Meier survival curve analysis rather than just the final percentage of TG from which virus was reactivated. Thus, neither readthrough LAT transcription nor the insertion of irrelevant DNA can account for the LAT+-like reactivation phenotype of dLAT-cpIAP. These findings therefore strongly support our hypothesis that LAT's antiapoptosis activity is sufficient for LAT's ability to support the wt reactivation phenotype.
DISCUSSION
We report here that replacing the HSV-1 LAT gene with the inhibitor of apoptosis gene (cpIAP) resulted in a recombinant virus with a reactivation phenotype similar to that of the wild-type virus. This supports the hypothesis that LAT's antiapoptosis activity is a key LAT function involved in enhancing the reactivation phenotype. cpIAP is a well-characterized inhibitor of apoptosis from the baculovirus Cydia pomonella granulovirus (2, 7, 8). It can block apoptosis by binding to caspase-9 (18, 48). The dLAT-cpIAP mutant inhibited apoptosis as efficiently as the wild-type virus, as judged by DNA ladder assays of extracts from acutely infected tissue culture cells, and fully supported the wt TG-explant reactivation phenotype in the mouse.
To eliminate both the influence of potential readthrough transcription past the location of the inserted cpIAP gene (LAT nt 1667) and the expression of a random foreign gene at the location of the inserted cpIAP gene, in a second independent experiment we compared dLAT-cpIAP to two additional control viruses, namely, dLAT1.5, which contains the identical LAT deletion with no foreign insertion and therefore controls for potential readthrough transcription past LAT nt 1667, and dLAT-EGFP, which is identical to dLAT-cpIAP except that it contains the EGFP gene ORF in place of the cpIAP ORF and therefore controls for expression of a random foreign gene at the location of cpIAP. Again, dLAT-cpIAP had a significantly higher reactivation phenotype than did dLAT2903 (P = 0.003). dLAT-cpIAP also had a significantly higher reactivation phenotype than those of both dLAT1.5 and dLAT-EGFP (P = 0.01 and P = 0.02, respectively), which were similar to that of dLAT2903 (P > 0.4). The wild-type-like reactivation phenotype of dLAT-cpIAP was also not due to LAT nt −161 to +76, which are present in dLAT-cpIAP but not in dLAT2903, because dLAT1.5, which had a reactivation phenotype similar to that of dLAT2903, contains this region. In addition, we previously showed that LAT1.8A, a mutant expressing LAT nt 1 to 76 (and containing the entire LAT promoter and thus LAT nt −161 to +76), and LAT2.5A, a mutant expressing just LAT nt 1 to 661 (and also containing the LAT promoter), both have a dLAT2903 (LAT null) reactivation phenotype in rabbits (19). Thus, cpIAP efficiently substituted for LAT's ability to enhance the HSV-1 reactivation phenotype in the mouse model, supporting our hypothesis that LAT's antiapoptosis activity is critical for LAT's ability to enhance the reactivation phenotype.
The LAT locus removed from dLAT2903 and replaced by cpIAP in dLAT-cpIAP also contains AL, an HSV-1 gene antisense to the 5′ end of LAT that we recently discovered (33). Thus, it is possible that cpIAP restored the wt reactivation phenotype by substituting for an AL function rather than a LAT function. However, our previous results obtained with the mutant dLAT371 (30, 35) make this unlikely. dLAT371 is deleted for LAT nt 76 to 447 (a StyI-StyI deletion). This deletion removes the 5′ end of AL (the AL mRNA starts at LAT nt 158, and the AL ORF starts at LAT nt 98), and no AL RNA is expressed. However, all of LAT except for the deleted area is expressed normally. dLAT371 has a wild-type reactivation phenotype (35). Thus, dLAT371, a LAT+ AL− mutant, and dLAT-cpIAP, a LAT− AL− cpIAP+ mutant, both have wild-type reactivation phenotypes. This strongly suggests that in the absence of AL, cpIAP can substitute for LAT. Since the cpIAP gene is a powerful antiapoptosis gene, these results strongly argue that cpIAP can support the wt reactivation phenotype by substituting for LAT's antiapoptosis activity. This in turn strongly supports the hypothesis that LAT's antiapoptosis activity is the key LAT function responsible for its ability to support the wt reactivation phenotype.
LAT is the only HSV-1 gene abundantly transcribed during neuronal latency. LAT− mutants have a reduced or delayed reactivation phenotype in mice and a reduced reactivation phenotype in rabbits (3, 9, 17, 23, 29, 36, 40, 43). Thus, LAT plays an important role in the HSV-1 latency-reactivation cycle. However, the mechanism by which LAT enhances the reactivation phenotype remains elusive, as does the determination of whether LAT's effect on the reactivation phenotype occurs during the establishment of latency, maintenance of latency, reactivation from latency, or a combination of two or more of these time periods. Although some reports, including one from our lab, suggest that LAT− mutants establish latency less efficiently than LAT+ viruses, these findings do not demonstrate that LAT's effect on the establishment of latency is key to, or in fact plays any significant role in, LAT's ability to enhance the reactivation phenotype. In fact, a mutant that appears to establish latency significantly less well than LAT− viruses has a wild-type LAT+-like reactivation phenotype in rabbits (31). This argues that a decreased establishment of latency by LAT− mutants is unlikely to completely account for the decreased reactivation phenotype of LAT− viruses.
Although our initial report that LAT can block apoptosis both in vitro and in vivo (28) became controversial because one group suggested that our studies were flawed, LAT's antiapoptosis activity has since been confirmed by others (1). In addition, we have significantly extended our findings since our first report of LAT's antiapoptosis activity in 2000. We have shown that (i) the region of LAT that decreases apoptosis comaps with the region of LAT that supports the wild-type high-reactivation phenotype (19, 20); (ii) LAT can block apoptosis induced by the overexpression of either caspase-8 or caspase-9, suggesting that LAT can block both major apoptosis pathways (16, 20); (iii) LAT directly or indirectly affects the accumulation of Bcl-xL and Bcl-xS transcripts that encode apoptotic regulatory proteins (27); and (iv) the BHV gene LR, which has antiapoptosis activity and is a functional homologue of the HSV-1 LAT but has no sequence homology to LAT, can efficiently substitute for the LAT function responsible for enhancing the reactivation phenotype (25, 34). Most importantly, we showed here that the unrelated antiapoptosis gene cpIAP could replace LAT and support the wt reactivation phenotype. Thus, not only does LAT have antiapoptosis activity, but this antiapoptosis activity appears to be responsible for the mechanism by which LAT enhances the reactivation phenotype.
Although HSV-1 has other antiapoptosis genes, LAT is the only antiapoptosis gene that is expressed at the end of the acute infection, a time when latency is likely being established and at which it is crucial to enhance the survival of neurons that suffered damage induced by virus infection. LAT is also the only viral antiapoptosis gene expressed at high levels during the maintenance of latency and when reactivation is triggered. Thus, if preventing cell death by apoptosis is important during the latency-reactivation cycle, then LAT is likely the primary or only viral gene involved.
The mechanism by which LAT's antiapoptosis activity enhances the reactivation phenotype is not yet known. In addition to LAT blocking virus-induced apoptosis in neurons, we propose two additional potential mechanisms that we feel are worthy of investigation. First, we hypothesize that the insults that induce reactivation also lead to apoptosis. In fact, it is possible that the initiation of apoptosis is what leads directly or indirectly to HSV-1 reactivation. For instance, one or more steps in the apoptosis pathways may trigger reactivation. LAT would therefore be important in preventing an apoptosis-related reactivation trigger from killing the neuron prior to the completion of reactivation. This differs from LAT protecting the neuron from apoptosis induced by the virus, because in this scenario LAT would play a critical role in the actual reactivation mechanism by preventing the reactivation trigger from killing the neuron. In the same vein, it has previously been shown that the induction of reactivation by dexamethasone in rabbits latently infected with BHV results in significantly decreased levels of LR, the BHV functional homologue of LAT, within 24 h. By 48 h, the LR levels return to normal (38). If reactivation stimuli have a similar effect on LAT, it would suggest that reducing LAT levels may lead to reactivation. We propose that decreasing LAT levels allow apoptosis to begin, thus triggering viral reactivation. The restoration of normal LAT levels then attenuates apoptosis, allowing reactivation to go to completion.
A second potential mechanism by which LAT's antiapoptosis activity could lead to an enhanced reactivation phenotype involves protection against T-cell-induced apoptosis. Recently, it was shown that during latency some neurons are surrounded by T cells, and it has been proposed that these T cells suppress reactivation (22, 24). Cytotoxic T cells kill target cells, in part by apoptosis. Thus, the high levels of LAT in some latently infected neurons may act to prevent the elimination of these neurons by T cells. This type of LAT-related immune evasion could be important during the establishment of, maintenance of, and/or reactivation from latency.
Whether LAT's antiapoptosis activity functions to enhance the reactivation phenotype by one or more of the above proposed mechanisms or whether LAT's antiapoptosis activity functions via a different mechanism remains to be determined. Regardless, the results presented here show that the replacement of LAT by a well-known apoptosis inhibitor, cpIAP, is sufficient to restore wt (LAT+) reactivation to a LAT null virus. This confirms that LAT's antiapoptosis activity is directly or indirectly involved in the mechanism by which LAT enhances the reactivation phenotype.
Acknowledgments
This work was supported by Public Health Service grants EY13191, EY12823, and P20RR15635, USDA grant 2002-35204, The Discovery Fund for Eye Research, The Henry L. Guenther Foundation, and Research to Prevent Blindness.
S. L. Wechsler is an RPB Senior Scientific Investigator.
REFERENCES
- 1.Ahmed, M., M. Lock, C. G. Miller, and N. W. Fraser. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76**:**717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68**:**2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192**:**618-630. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, D. C., J. M. Hill, G. Devi-Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70**:**2249-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Branco, F. J., and N. W. Fraser. 2005. Herpes Simplex Virus Type 1 Latency-Associated Transcript Expression Protects Trigeminal Ganglion Neurons from Apoptosis. J. Virol. 79**:**9019-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratanich, A. C., N. D. Hanson, and C. J. Jones. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191**:**988-991. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71**:**5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14**:**5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67**:**2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68**:**1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devireddy, L. R., Y. Zhang, and C. J. Jones. 2003. Cloning and initial characterization of an alternatively spliced transcript encoded by the bovine herpes virus 1 latency-related gene. J. Neurovirol. 9**:**612-622. [DOI] [PubMed] [Google Scholar]
- 11.Dobson, A. T., F. Sederati, G. Devi-Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63**:**3844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88**:**790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71**:**5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiser, V., M. Inman, Y. Zhang, and C. Jones. 2002. The latency-related gene of bovine herpesvirus-1 can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 83**:**2965-2971. [DOI] [PubMed] [Google Scholar]
- 15.Goins, W. F., L. R. Sternberg, K. D. Croen, P. R. Krause, R. L. Hendricks, D. J. Fink, S. E. Straus, M. Levine, and J. C. Glorioso. 1994. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J. Virol. 68**:**2239-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, G., W. Peng, L. Jin, G. C. Perng, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2002. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 latency-associated transcript. J. Neurovirol. 8**:**103-111. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174**:**117-125. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Q., Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2000. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 97**:**1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inman, M., G. Perng, G. Henderson, H. Ghiasi, A. Nesburn, S. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75**:**3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, L., W. Peng, G. C. Perng, D. J. Brick, A. B. Nesburn, C. Jones, and S. L. Wechsler. 2003. Identification of herpes simplex virus type 1 latency-associated transcript sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J. Virol. 77**:**6556-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, L., G. C. Perng, D. J. Brick, J. Naito, A. B. Nesburn, C. Jones, and S. L. Wechsler. 2004. Methods for detecting the HSV-1 LAT anti-apoptosis activity in virus infected tissue culture cells. J. Virol. Methods 118**:**9-13. [DOI] [PubMed] [Google Scholar]
- 22.Khanna, K. M., R. H. Bonneau, P. R. Kinchington, and R. L. Hendricks. 2003. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18**:**593-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leib, D. A., C. L. Bogard, M. Kosz-Vnenchak, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63**:**2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, T., K. M. Khanna, X. Chen, D. J. Fink, and R. L. Hendricks. 2000. CD8(+) T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. J. Exp. Med. 191**:**1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott, K. R., N. Osorio, L. Jin, D. J. Brick, J. Naito, J. Cooper, G. Henderson, M. Inman, C. Jones, S. L. Wechsler, and G. C. Perng. 2003. The bovine herpesvirus-1 LR ORF2 is critical for this gene's ability to restore the high wild-type reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J. Gen. Virol. 84**:**2975-2985. [DOI] [PubMed] [Google Scholar]
- 26.Nicosia, M., J. M. Zabolotny, R. P. Lirette, and N. W. Fraser. 1994. The HSV-1 2-kb latency-associated transcript is found in the cytoplasm comigrating with ribosomal subunits during productive infection. Virology 204**:**717-728. [DOI] [PubMed] [Google Scholar]
- 27.Peng, W., G. Henderson, G. C. Perng, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2003. The gene that encodes the herpes simplex virus type 1 latency-associated transcript influences the accumulation of transcripts [Bcl-x(L) and Bcl-x(S)] that encode apoptotic regulatory proteins. J. Virol. 77**:**10714-10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perng, G., C. Jones, H. Ciacci-Zanella, G. Henderson, A. Yukht, S. Slanina, F. Hofman, H. Ghiasi, A. Nesburn, and S. Wechsler. 2000. Virus induced neuronal apoptosis blocked by the herpes simplex virus latency associated transcript (LAT). Science 287**:**1500-1503. [DOI] [PubMed] [Google Scholar]
- 29.Perng, G. C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68**:**8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perng, G. C., D. Esmaili, S. M. Slanina, A. Yukht, H. Ghiasi, N. Osorio, K. R. Mott, B. Maguen, L. Jin, A. B. Nesburn, and S. L. Wechsler. 2001. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J. Virol. 75**:**9018-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. High-dose ocular infection with a herpes simplex virus type 1 ICP34.5 deletion mutant produces no corneal disease or neurovirulence yet results in wild-type levels of spontaneous reactivation. J. Virol. 70**:**2883-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J. Virol. 70**:**976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perng, G. C., B. Maguen, L. Jin, K. R. Mott, J. Kurylo, L. BenMohamed, A. Yukht, N. Osorio, A. B. Nesburn, G. Henderson, M. Inman, C. Jones, and S. L. Wechsler. 2002. A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5′ end of the latency-associated transcript produces a protein in infected rabbits. J. Virol. 76**:**8003-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perng, G. C., B. Maguen, L. Jin, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76**:**1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perng, G. C., S. M. Slanina, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1996. A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. J. Virol. 70**:**2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perng, G. C., S. M. Slanina, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2001. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. J. Gen. Virol. 82**:**1117-1122. [DOI] [PubMed] [Google Scholar]
- 37.Perng, G. C., S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J. Virol. 74**:**1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66**:**2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rock, D. L., A. B. Nesburn, H. Ghiasi, J. Ong, T. L. Lewis, J. R. Lokensgard, and S. L. Wechsler. 1987. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J. Virol. 61**:**3820-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66**:**2157-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70**:**3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61**:**3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steiner, I., J. G. Spivack, R. P. Lirette, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, and N. W. Fraser. 1989. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 8**:**505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens, J. G. 1990. Transcripts associated with herpes simplex virus latency. Adv. Exp. Med. Biol. 278**:**199-204. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, J. G., E. K. Wagner, G. B. Devi-Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235**:**1056-1059. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, S. K., C. E. Lilley, D. S. Latchman, and R. S. Coffin. 2002. A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome. J. Virol. 76**:**4056-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75**:**6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilaplana, L., and D. R. O'Reilly. 2003. Functional interaction between Cydia pomonella granulovirus IAP proteins. Virus Res. 92**:**107-111. [DOI] [PubMed] [Google Scholar]
- 49.Wagner, E. K., G. Devi-Rao, L. T. Feldman, A. T. Dobson, Y. F. Zhang, W. M. Flanagan, and J. G. Stevens. 1988. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J. Virol. 62**:**1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wechsler, S. L., A. B. Nesburn, R. Watson, S. M. Slanina, and H. Ghiasi. 1988. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J. Virol. 62**:**4051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wechsler, S. L., A. B. Nesburn, J. Zwaagstra, and H. Ghiasi. 1989. Sequence of the latency-related gene of herpes simplex virus type 1. Virology 168**:**168-172. [DOI] [PubMed] [Google Scholar]
- 52.Zwaagstra, J., H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1989. In vitro promoter activity associated with the latency-associated transcript gene of herpes simplex virus type 1. J. Gen. Virol. 70**:**2163-2169. [DOI] [PubMed] [Google Scholar]
- 53.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64**:**5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]