Rab9 GTPase Is Required for Replication of Human Immunodeficiency Virus Type 1, Filoviruses, and Measles Virus (original) (raw)
Abstract
Rab proteins and their effectors facilitate vesicular transport by tethering donor vesicles to their respective target membranes. By using gene trap insertional mutagenesis, we identified Rab9, which mediates late-endosome-to-_trans_-Golgi-network trafficking, among several candidate host genes whose disruption allowed the survival of Marburg virus-infected cells, suggesting that Rab9 is utilized in Marburg replication. Although Rab9 has not been implicated in human immunodeficiency virus (HIV) replication, previous reports suggested that the late endosome is an initiation site for HIV assembly and that TIP47-dependent trafficking out of the late endosome to the _trans_-Golgi network facilitates the sorting of HIV Env into virions budding at the plasma membrane. We examined the role of Rab9 in the life cycles of HIV and several unrelated viruses, using small interfering RNA (siRNA) to silence Rab9 expression before viral infection. Silencing Rab9 expression dramatically inhibited HIV replication, as did silencing the host genes encoding TIP47, p40, and PIKfyve, which also facilitate late-endosome-to-_trans_-Golgi vesicular transport. In addition, silencing studies revealed that HIV replication was dependent on the expression of Rab11A, which mediates _trans_-Golgi-to-plasma-membrane transport, and that increased HIV Gag was sequestered in a CD63+ endocytic compartment in a cell line stably expressing Rab9 siRNA. Replication of the enveloped Ebola, Marburg, and measles viruses was inhibited with Rab9 siRNA, although the nonenveloped reovirus was insensitive to Rab9 silencing. These results suggest that Rab9 is an important cellular target for inhibiting diverse viruses and help to define a late-endosome-to-plasma-membrane vesicular transport pathway important in viral assembly.
Most developed antiviral drugs have been designed to inhibit the function of viral proteins. In the case of human immunodeficiency virus (HIV), an unfortunate consequence of using drugs that target viral proteins has been the emergence of drug-resistant virions having compensatory genetic mutations (8, 20). An alternative approach is to identify cellular genes essential for the viral life cycle, but not essential for the more genetically diverse host cell, and then to develop agents that inhibit their expression or function. In principle, such an approach may pose an insurmountable barrier against viral replication, as resistance would arise from a complex adaptation to use a different cellular protein for viral replication. There are several examples where a partial or complete resistance to pathogens results from the loss of expression or function of a critical host gene. Examples include the high degree of protection against HIV transmission afforded to individuals homozygous for a dysfunctional allele of CCR5 (a major HIV coreceptor) and the complete resistance to Plasmodium vivax malaria infection of individuals lacking expression of the erythrocyte receptor DARC (26, 38).
We have used gene trap insertional mutagenesis (58) as a high-throughput forward genetics approach to randomly trap host genes and discover cellular genes that are required for viral replication but not for host cell survival. This methodology is based on two important considerations: (i) the viral infection is toxic to the chosen host cell line and (ii) disrupting a gene critical for the viral life cycle confers survivability during subsequent viral selection, provided that the host cell can survive the reduced or abolished expression of the gene. In this approach, cells are infected with a recombinant Moloney murine leukemia virus (MMLV)-derived vector that randomly integrates into the host chromosome, using a low multiplicity of infection (MOI = 0.1) to favor only one gene disruption per cell. Genes trapped in clonal, virus-resistant cell lines are later identified based on cell survival following viral infection. This approach favors the discovery of either imprinted genes, where only a single allele is normally expressed, or genes whose down-regulation or inhibition need not be complete in order to control viral replication. Using this approach, our group has identified over 200 candidate host genes that potentially play critical roles in the life cycles of Marburg virus, Ebola virus, HIV type 1 (HIV-1), HIV-2, influenza A virus, and reovirus. We previously showed that disrupting the Ctcf gene that regulates the expression of Igf2 confers resistance to lytic reoviral infection (53). Gene trapping by our group also identified two cellular genes previously implicated in HIV replication (the annexin II and phosphodiesterase 4 genes) (34, 55) as well as 16 gene products utilized in the life cycles of 19 other viruses (46).
Rab proteins are small GTP-binding proteins that form the largest family within the Ras superfamily (36). Rab proteins are thought to regulate vesicular transport pathways by incorporating into transport vesicles and binding their cognate Rab effectors on the target membrane prior to SNARE (soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor)-catalyzed membrane fusion. The Rab9 GTPase was disrupted in a clonal Vero cell line surviving the cytopathic effects of Marburg virus infection, suggesting that Rab9 may be utilized by Marburg virus. Rab9 facilitates late-endosome-to-_trans_-Golgi-network transport by pairing with its effector, p40, at the _trans_-Golgi network (11, 33). Membrane anchoring of p40 is thought to depend upon serine phosphorylation by PIKfyve (27). Interestingly, Rab9 also binds the vesicle cargo selection protein TIP47 (19), whose interaction with the cytoplasmic tail of the HIV envelope glycoprotein (Env) subunit gp41 was found to be critical for the incorporation of Env into mature virions (5). Recent reports provide evidence for a pathway of viral egress used by HIV-1, Marburg virus, and murine leukemia virus whereby viral assembly initiates at the late endosome and budding is completed at the plasma membrane, although the mechanism of endosome membrane transport to the plasma membrane is not defined (2, 28, 42).
Here we show that small interfering RNA (siRNA)-directed silencing of host genes involved in late-endosome-to-_trans_-Golgi transport, i.e., Rab9, TIP47, p40, and PIKfyve, and in _trans_-Golgi-to-plasma-membrane transport, i.e., Rab11A, dramatically impairs HIV replication. HIV Gag showed increased colocalization with a CD63+ endocytic compartment in cells having Rab9 expression stably down-regulated with siRNA, suggesting that the observed defect in HIV replication may result from impaired trafficking of endocytosed viral proteins out of the late endosome. Reducing Rab9 expression also inhibited the replication of the enveloped Ebola and Marburg filoviruses as well as that of measles virus, but not that of the nonenveloped reovirus. These results suggest a transport pathway used for enveloped viral egress and implicate Rab9 as an important cellular target for inhibiting several unrelated viruses.
MATERIALS AND METHODS
Cell culture.
Sup-T1 human lymphoblastic leukemia cells were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin, streptomycin, and amphotericin B (Fungizone). JC53 HeLa cells (Tranzyme, Birmingham, AL), rat intestinal 1 (RIE-1) cells, and MDCK normal canine kidney cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, penicillin, and streptomycin. Vero African green monkey kidney cells were cultured in DMEM supplemented with 10% FBS, amphotericin B, streptomycin, and glutamine. L cells were maintained in DMEM with 5% FBS, penicillin, streptomycin, and amphotericin B. All cultures were grown in 5% CO2. Selection was done in the presence of either 1 mg/ml G418 (Geneticin; Invitrogen, Carlsbad, CA) for Sup-T1, RIE-1, and MDCK cells or 400 μg/ml G418 for Vero cells.
DNA plasmids.
pGag-EGFP (21) was a gift from Marilyn Resh (Sloan-Kettering Institute, NY). pCD63-mRFP was a gift from George Patterson (National Institutes of Health, MD).
Gene trap library construction.
The U3neoSV1 retrovirus vector (22) was obtained from H. Earl Ruley (Vanderbilt University). Sup-T1, Vero, MDCK, and RIE-1 cells served as suitable parental cell lines for the preparation of gene trap libraries, as they are efficiently killed by infection with HIV-1 and HIV-2, filoviruses, influenza A virus, and reovirus, respectively. Parental, virus-sensitive cells were infected in T75 flasks at 37°C for 1 h with U3neoSV1 (MOI = 0.1) in the presence of 4 μg/ml Polybrene (Sigma, St. Louis, MO). Subsequently, the medium was changed and the cells were grown overnight at 37°C. On the following day, G418 selection was initiated (at the concentrations indicated above), and cells were grown to confluence.
Generation of filovirus-resistant, HIV-resistant, and reovirus-resistant cells from gene trap libraries.
Vero gene trap library cells were infected (MOI, >1) with Ebola virus (Zaire species, 1976 Mayinga strain) or Marburg virus (1967 Voege strain). After a cytopathic effect of 4+ was observed, i.e., >95% of cells were rounded and detached, survivors were harvested and expanded in culture medium containing 400 μg/ml G418 for 10 days. Surviving cells were reinoculated into T12.5 flasks, expanded, and cloned by limiting dilution.
HIV-resistant Sup-T1 library cells were generated by infecting Sup-T1 cells three times with the HIV-1 strain LAV (MOI = 10). Survivors were expanded for 2 weeks in G418 (1 mg/ml) before reinfection. Next, HIV-1-resistant cells were exposed to three successive rounds of HIV-2 infection by coculturing Sup-T1 cells (1:100) with BC7 T cells expressing the HIV-2 strain 3BX (obtained from James Hoxie, University of Pennsylvania) and expanding the survivors for 2 weeks. CD4+ Sup-T1 cells resistant to HIV-1 and HIV-2 infection were reinfected with LAV (MOI = 10) and cloned by limiting dilution.
Reovirus-resistant RIE-1 library cells were prepared as described previously (53). Briefly, 20 RIE-1 gene trap libraries, each harboring approximately 104 gene entrapment events, were expanded until approximately 103 daughter cells represented each clone. The RIE-1 cells were plated at a subconfluent density and incubated in serum-free medium for 3 days until the cells became quiescent. The cells were then infected overnight with reovirus serotype 1 (MOI = 35). At 18 h postinfection, RIE-1 cells were detached with trypsin and subcultured in DMEM supplemented with 10% FBS. After 6 h, the medium was changed to serum-free DMEM, and cells were maintained in serum-free medium until only a few cells remained attached to the flask. Cells surviving selection were expanded in DMEM supplemented with 10% FBS, expanded, and cloned by limiting dilution.
U3neoSV1 shuttle vector rescue and sequencing.
Cellular DNAs from clonal virus-resistant cell lines were extracted using a QIAamp DNA Blood Mini kit (QIAGEN, Inc., Valencia, CA). Shuttle vectors and genomic DNA flanking the integration site were recovered by digestion of the genomic DNA (150 mg/ml) with EcoRI or BamHI, self-ligation, transformation into Escherichia coli, and selection on Luria broth agar containing 100 μg/ml carbenicillin (Sigma). Individual colonies were amplified, and recovered genomic DNAs were sequenced using primers annealing to the shuttle vector. Sequencing reactions were performed using an ABI BigDye Terminator cycle sequencing kit, and sequences were analyzed using an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The sequences of the sequencing primers are available upon request.
Sequence analysis.
Genomic sequences obtained from shuttle clones were analyzed by the RepeatMasker web server, followed by nucleotide-nucleotide BLAST searches against the National Center for Biotechnology Information databases. Virtually all genes identified matched murine and human sequences with probability scores (P) of <10−10 and <10−20, respectively.
siRNA transfections.
SMARTpool siRNAs directed against candidate genes were synthesized by Dharmacon (Lafayette, CO). In the case of human Rab9, the siRNA oligonucleotides targeted the Rab9 gene at positions 55 to 73 (GGGAAGAGUUCACUUAUGA), 147 to 165 (GGAAGUGGAUGGACAUUUU), 274 to 292 (UCACAAAGCUUCCAGAACU), and 368 to 386 (GUAACAAGAUUGACAUAAG). The simian Rab9 gene (cloned for this study) was 100% homologous at the chosen target sites. siRNA SMARTpool sequences directed against the TIP47, p40, PIKfyve, and Rab11A genes are available upon request. A siRNA against green fluorescent protein (GFP) (QIAGEN, Inc.) was used as a negative control. All siRNA transfections were performed with Lipofectamine 2000 (Invitrogen), using a 50 nM siRNA concentration in the culture medium for 48 h before viral infections.
Real-time quantitative PCR.
Total mRNAs were isolated from parental and siRNA-transfected cells using RNeasy Mini and QIAshredder kits (QIAGEN, Inc.) and then were reverse transcribed using random hexamers (Applied Biosystems). Real-time PCR was performed as described previously (24), using an Mx4000 multiplex quantitative PCR system (Stratagene). Real-time PCR detection assays for all siRNA target genes described in this study were purchased from Applied Biosystems. Target gene mRNA expression levels were normalized to hypoxanthine-guanine phosphoribosyltransferase (HGPRT) mRNA expression levels, using a human HGPRT TaqMan assay kit (Applied Biosystems). Relative gene expression levels in silencing studies were determined using cycle threshold (CT) numbers for detecting the expression of both the targeted gene and HGPRT in each siRNA transfectant. Quantitation was based on CT numbers from at least three independent experiments; for each siRNA, the standard deviations of CT numbers were <3% of the mean.
HIV-1 p24 and luciferase assays.
CD4+ CCR5+ CXCR4+ JC53 cells were infected overnight with HIV LAV (MOI = 1) 2 days following transfection with siRNA and then seeded into 24-well plates (25,000 cells per well) or T75 flasks (1.5 million cells per flask). p24 production was assayed in culture supernatants at 3 days postinoculation using the p24 antigen enzyme-linked immunosorbent assay system (Beckman/Coulter/Immunotech, Brea, CA). Luciferase assays were performed (in quadruplicate) with detergent lysates at 1 day postinfection with LAV (MOI = 1), using the Steady-Glo luciferase assay system (Promega, Madison, WI) and an EL 312e microplate biokinetics reader (Bio-Tek Instruments, Winooski, VT).
HIV Gag/CD63 colocalization studies.
The JC53 Rab9 siRNA cell line was prepared using the pSIRENretroQ retroviral vector (BD Biosciences, Franklin Lakes, NJ), which encodes a short hairpin RNA (shRNA) against Rab9 mRNA bases 336 to 354 with a downstream termination sequence (TTTTT) and the puromycin resistance gene (Puror). The Rab9 shRNA vector was transfected into the PA317 retroviral packaging cell line (Nature Technology Corporation, Lincoln, NE), resulting in the secretion of replication-deficient amphotropic MMLV particles carrying the Rab shRNA and the Puror gene. JC53 Rab9 siRNA cells were generated by infecting cells with the resultant MMLV vector, followed by selection in 1 μg/ml puromycin.
Rab9 immunostaining was performed by fixing JC53 cells grown on coverslips in 4% formalin (15 min), followed by blocking in 1% bovine serum albumin-phosphate-buffered saline (PBS) containing 0.1% saponin (10 min) and sequential incubations first with mouse anti-Rab9 antibody (Mab9; Calbiochem, San Diego, CA), followed by a wash with 1% bovine serum albumin-PBS containing 0.1% saponin, and then with anti-mouse antibody conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, OR). Stained cells were washed with PBS and mounted on slides with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL) in preparation for microscopy. JC53 and JC53 Rab9 siRNA cells were prepared for colocalization studies by seeding cells overnight in Lab-Tek chambers (Nalge Nunc, Rochester, NY) and cotransfecting them with pGag-EGFP and pCD63-mRFP, using Lipofectamine 2000. Cells were imaged live at 36 h posttransfection using a Zeiss LSM510 confocal microscope with a 63×, 1.2-numerical-aperture oil immersion lens. Healthy cells expressing both proteins were selected for z sectioning. z stacks were taken using a pinhole of 1.1 airy units for both channels. Single confocal sections were exported as TIFF files and processed in Adobe Photoshop.
Ebola and Marburg virus immunofluorescence, electron microscopy, and viral replication assays.
At 2 days posttransfection with siRNA, Vero cells were transported to a maximum containment laboratory and infected for 6 to 7 days with Ebola virus (Zaire species, 1976 Mayinga strain; MOI = 0.01) or Marburg virus (1967 Voege strain; MOI = 0.01 or 0.1). Subsequently, supernatants were clarified by centrifugation and inactivated by gamma irradiation on dry ice (>3 Mrad). Cells spotted onto glass slides were gamma irradiated and fixed with acetone for 10 min. Indirect staining was performed using polyclonal antisera against Ebola or Marburg proteins (prepared at the Centers for Disease Control and Prevention), followed by washing and staining with an appropriate concentration of fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit immunoglobulin G (IgG) (Kirkegaard & Perry Laboratories, Inc.). Cells were processed following Ebola and Marburg virus infection for electron microscopic examination by standard methods (16). The detection of viral antigens (VP40 from Ebola virus or NP/VP40 from Marburg virus) in clarified supernatants was performed (in duplicate) by solid-phase antigen-capture assays at 6 days postinoculation, as described previously (30).
Measles virus in vitro assays.
Vero cells were infected with the Edmonston strain of measles virus (MOI = 0.1) at 2 days post-siRNA transfection. To determine the efficacy of siRNA silencing, virus titers in the infected cells were determined. The virus inoculum was removed at 1 h postinfection, and cells were incubated for 48 h with fresh medium. Culture supernatants were harvested daily, and the secreted virus was titrated by standard plaque assays on Vero cell monolayers. Measles virus syncytium formation was detected using appropriate dilutions of the anti-measles virus nucleoprotein antibody mab-KK2 (Chemicon International, Temecula, CA) and an Alexa Fluor-conjugated goat anti-mouse secondary antibody (Molecular Probes).
Reovirus assays.
Vero cells were seeded in 12-well plates (5 × 105 cells/well) and transfected with GFP, Rab9, or Rin2 siRNA. siRNA transfectants were inoculated with reovirus type 1 (Lang strain; MOI = 35). At 0, 24, and 48 h postinoculation, cells were frozen and stored at −20°C until virus titers could be simultaneously determined by standard plaque assays as described previously (51).
Flow cytometry.
Flow cytometry was used to determine Rab9 protein expression in siRNA-transfected JC53 cells. Briefly, the cells were prepared for analysis by permeabilization using a Cytofix/Cytoperm kit (BD Biosciences) and then stained with mouse anti-Rab9 antibody (Mab9; Calbiochem) or an isotype-matched negative control. The cells were washed and then subsequently stained using phycoerythrin-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA). Measles virus nucleoprotein (MV-NP) staining was performed by first infecting siRNA-transfected cells with measles virus (MOI = 0.1) and then preparing cells for cytofluorimetry as previously described (60). Internal staining for MV-NP was carried out using FITC-labeled KK2 (anti-MV-NP) or a FITC-conjugated isotype-matched negative control (clone MOPC-21; BD Biosciences). Stained cells were analyzed using a Beckman Coulter EPICS-XL flow cytometer with Beckman Coulter EXPO32 ADC software.
Cloning simian Rab9 cDNA.
Simian Rab9 cDNA was amplified using the forward primer 5′-ATTAACAATGGCAGGAA-3′ and the reverse primer 5′-GTGTCCCTTCTCCCACCAACTAATGA-3′. Simian Rab9 cDNA was cloned using a Topo TA cloning kit (Invitrogen) and sequenced with manufacturer-supplied primers.
Nucleotide sequence accession number.
The nucleotide sequence for the Rab9 cDNA is available at GenBank under accession number AY601640.
RESULTS
Determination of candidate host genes required for viral replication by gene trap mutagenesis and viral selection.
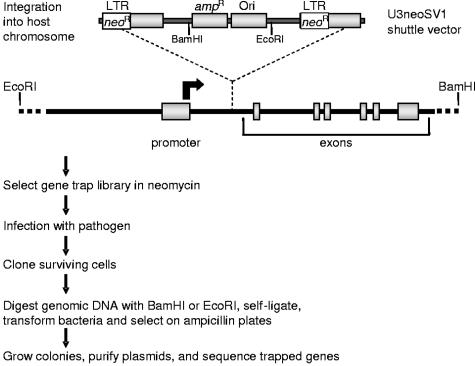
Gene trap insertional mutagenesis (58) involves infection with a replication-deficient Moloney murine leukemia virus that randomly integrates its genetic material into the host cell chromosome, carrying with it a promoterless neomycin resistance gene (_neo_r) (Fig. 1). We have adapted this approach towards the determination of host genes whose disruption confers survival following infection with HIV-1, HIV-2, Marburg virus, Ebola virus, influenza A virus, or reovirus, normally a lethal event in the chosen host cell lines. Cells surviving both neomycin and viral selection are cloned, and the identity of disrupted (trapped) genes is determined as outlined in Fig. 1. Sequence analysis identified over 200 candidate host genes potentially required in the life cycles of HIV-1 and -2, Marburg virus, Ebola virus, influenza A virus, or reovirus.
FIG. 1.
Identification of cellular genes required for viral replication by gene trap mutagenesis. Susceptible host cell lines are chosen that are normally killed by the virus being studied. Cells are infected with the U3neoSV1 retrovirus (MOI = 0.1) carrying promoterless neomycin resistance genes (_neo_r) in each LTR. Integration of the U3neoSV1 shuttle vector between an active promoter and an early exon results in gene disruption (trapping), with concomitant expression of the _neo_r gene from the upstream (5′) LTR. The promoterless 3′ LTR is transcriptionally inactive. Following neomycin selection, the resultant gene trap library is infected with the virus of choice. Cell survival following viral infection results from trapping genes that are necessary for viral replication. Surviving cells are expanded and cloned. The shuttle vector is then rescued from genomic DNA as indicated above, and the recovered chromosomal DNA flanking the shuttle vector integration site is sequenced to identify the trapped gene.
Screening candidate genes with siRNAs reveals a role for Rab9 in HIV-1 infection.
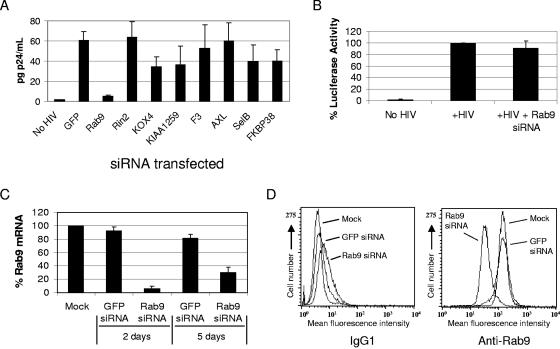
As an independent approach to test the role of candidate trapped genes in viral replication, we have used siRNAs to study the effect of inhibiting candidate gene mRNA expression on viral replication. This approach has proven useful for degrading mRNA and controlling HIV-1 replication by reducing the message coding for essential viral or cellular proteins (41). An initial panel of siRNAs was developed that targeted candidate genes implicated by insertional mutagenesis in the replication of Marburg virus (Rab9, hypothetical protein KIAA1259, Axl, and SelB), HIV-1 and -2 (F3), and reovirus (Rin2, KOX4, and FKBP38) and was tested in JC53 cells for antiviral activity against the CXCR4-tropic LAV strain of HIV (Fig. 2A). Among the siRNAs examined, Rab9 siRNA showed the strongest effect in inhibiting HIV-1 replication (∼80 to 90% inhibition of p24 release into the culture supernatant) compared to cells transfected with an irrelevant siRNA control against GFP.
FIG. 2.
Inhibition of HIV-1 replication with Rab9 siRNA. The JC53 cells used for HIV p24 assays are CD4+ CCR5+ CXCR4+ HeLa-derived indicator cells that express luciferase following HIV infection. Luciferase expression from a stably integrated HIV-LTR-luciferase construct is regulated by the early HIV gene product Tat. (A) JC53 HeLa cells were transfected with the indicated siRNAs and inoculated with HIV-1 LAV. HIV p24 secreted into culture supernatants was quantitated in p24 assays at 3 days postinoculation (n = 6). (B) Luciferase assays (n = 5) were performed to quantitate the infection of JC53 cells following Rab9 siRNA transfections. (C) Total mRNA isolated from JC53 cells (at 2 and 5 days posttransfection with GFP or Rab9 siRNA) was reverse transcribed, and the relative levels of Rab9 message originally present were determined by real-time PCR and normalized to HGPRT expression (n = 4). (D) Flow cytometric analysis of Rab9 protein expression following the indicated siRNA transfections. Staining was performed using a mouse anti-Rab9 antibody or a mouse IgG1 negative control.
JC53 cells are HeLa-derived indicator cells that produce luciferase following HIV infection, due to a stably integrated HIV-long terminal repeat (LTR)-luciferase construct regulated by the early HIV gene product Tat. Although HIV replication was significantly decreased in Rab9 siRNA transfectants (P < 0.001 by Student's t test; Fig. 2A), no significant difference was observed in luciferase activities between Rab9 siRNA-transfected and parental JC53 cells (Fig. 2B), indicating that the observed defect in HIV replication occurs downstream of viral entry and proviral gene expression. Rab9 siRNA specifically inhibited HIV replication and resulted in ∼90% degradation of Rab9 mRNA and an ∼75% decrease in Rab9 mean fluorescence intensity, as determined by flow cytometry prior to infection, compared to GFP siRNA controls (Fig. 2A, C, and D).
Inhibiting HIV with siRNAs directed against TIP47, p40, PIKfyve, and Rab11A.
Rab9 facilitates the vesicular transport of cargo proteins from the late endosome to the _trans_-Golgi network (33). Evidence suggests that HIV Env and Gag are endocytosed (6, 32, 59) and that HIV initiates assembly at the late endosome (42) and then employs a cell-type-dependent pathway for viral egress (see Fig. 6) (43). In macrophages, HIV buds into the lumen of the late endosome to form multivesicular bodies, which can fuse with the plasma membrane and release nascent virions as exosomes (40, 43, 45, 47, 48), whereas in T cells and cultured cell lines (including HeLa cells), HIV primarily buds from the plasma membrane, although budding into multivesicular bodies has been observed (21, 39, 42, 43, 54). The late-endosome-to-_trans_-Golgi transport vesicle cargo selection protein TIP47 has been shown to be important in HeLa cells for trafficking Env out of the late endosome and for the incorporation of Env into virions budding from the plasma membrane (5). To further characterize the egress pathway used by HeLa cells, host genes involved in late-endosome-to-_trans_-Golgi transport (TIP47, p40, and PIKfyve) and _trans_-Golgi-to-plasma-membrane transport (Rab11A) were silenced with siRNA and then infected with HIV.
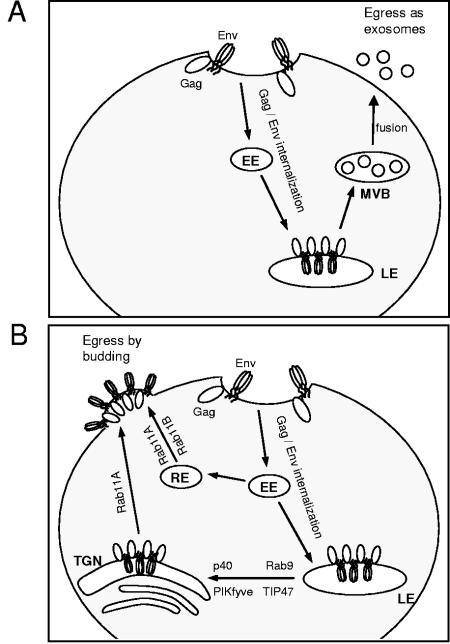
FIG. 6.
Alternative pathways for HIV viral egress. HIV Env and Gag are expressed on the cell surface before internalization to the late endosome (LE), an important site of assembly initiation. (A) In macrophages, HIV buds into the lumen of the late endosome to form multivesicular bodies (MVBs), which can be released as exosomes by multivesicular body fusion with the plasma membrane. (B) In T cells and cultured cell lines, budding occurs primarily at the plasma membrane. Vesicular trafficking of viral proteins out of the late endosome to the _trans_-Golgi network (TGN) and plasma membrane may be aided, as indicated, by the host proteins Rab9, Tip47, p40, PIKfyve, and Rab11A. Rab11A may also favor plasma membrane budding by taking part in the recycling of viral proteins back to the plasma membrane via early endosomes (EEs) and recycling endosomes (REs).
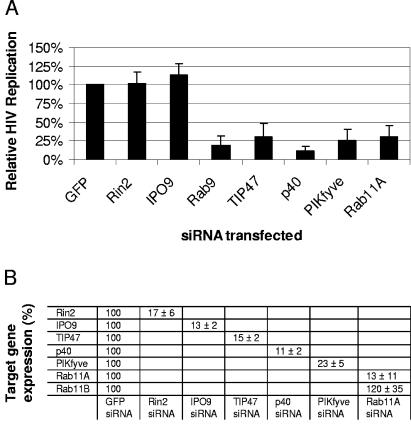
Our results confirmed that HIV utilizes TIP47, and we show for the first time that HIV replication depends on the expression of the Rab9 effector p40, PIKfyve (which promotes p40 membrane attachment), and Rab11A (Fig. 3A). HIV replication was significantly inhibited (P < 0.001) by silencing the expression of Rab9, TIP47, p40, PIKfyve, or Rab11A compared to control transfections with siRNA directed against the irrelevant target GFP and the host genes Rin2 and IPO9 (Fig. 3A). IPO9 mRNA (trapped with Marburg virus) was included as a negative control, as targeting this message with siRNA was consistently found not to affect HIV replication in a separate study (not shown). Rab11A siRNA specifically reduced the expression of Rab11A (facilitating transport to the plasma membrane from both the _trans_-Golgi and recycling endosomes) (9, 18, 57) but not that of Rab11B (used for recycling endosome-to-plasma-membrane transport) (52). The inhibition of target gene expression was quantitated by real-time PCR. Figure 3B reveals that target gene mRNA expression levels were generally reduced >80% relative to the expression levels of the same gene in GFP siRNA-transfected control cells at the time of infection. Together, these results suggest the importance of late-endosome-to-_trans_-Golgi-network vesicular transport in HIV replication and indicate that silencing Rab9 expression may result in impaired trafficking of internalized HIV proteins out of the late endosome.
FIG. 3.
HIV utilization of host genes facilitating vesicular transport. (A) JC53 cells were transfected with the indicated siRNAs for 48 h, inoculated with HIV (LAV strain; MOI = 1), and expanded in duplicate T75 flasks. Culture supernatants were harvested at 3 days postinoculation, and HIV p24 assays were performed. Values were normalized to HIV p24 secretion from GFP siRNA-transfected cells and represent the results of at least four independent experiments. (B) JC53 cells were transfected with siRNAs against the target genes specified, and the effect on target gene expression was determined as described in the legend to Fig. 2C.
Internal Gag sequestration in Rab9-depleted cells.
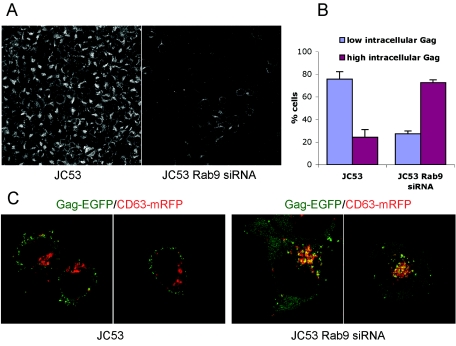
To study the internal sequestration of HIV Gag, a stable cell line stably expressing Rab9 siRNA was derived from JC53 cells. Rab9 expression was reduced ∼80 to 90% in the JC53 Rab9 siRNA cell line, based on Rab9 immunofluorescence (Fig. 4A) and Rab9 Western blots using lysates from JC53 and JC53 Rab9 siRNA cells (not shown). Both parental JC53 and JC53 Rab9 siRNA cells were transfected with Gag-EGFP and CD63-mRFP. For both cell lines, intracellular Gag expression was classified arbitrarily as low intracellular Gag expression (<10 internal Gag-EGFP puncta) or high intracellular Gag expression (≥10 internal Gag-EGFP puncta). High intracellular Gag-EGFP expression was observed in 75% of JC53 Rab9 siRNA cells, compared to 25% of parental JC53 cells, and this difference was statistically significant (P < 0.001) (Fig. 4B). Representative examples of the Gag-EGFP expression patterns in parental and Rab9 siRNA cells are shown in Fig. 4C, with internalized Gag-EGFP showing strong colocalization with CD63 in the Rab9 siRNA cells. These results indicate that Rab9 depletion results in internal Gag sequestration, which may impair budding from the plasma membrane.
FIG. 4.
Colocalization of CD63 and HIV Gag in Rab9-depleted cells. (A) Rab9 expression was visualized in parental JC53 cells (left panel) and JC53 Rab9 siRNA cells (right panel) by sequential incubations with a primary anti-Rab9 antibody and an AF488-conjugated secondary antibody. (B) JC53 cells and JC53 Rab9 siRNA cells were transfected with pGag-EGFP and pCD63-mRFP plasmids, and 20 to 60 cells per experiment (n = 3) were optically _z_-sectioned live at 36 h posttransfection. Cells having <10 or ≥10 intracellular Gag-EGFP puncta colocalizing with CD63-mRFP were classified as having low or high intracellular Gag, respectively. (C) Representative confocal sections of JC53 cells (left panels) and JC53 Rab9 siRNA cells (right panels) cotransfected with pGag-EGFP (green) and pCD63-mRFP (red).
Inhibition of Ebola, Marburg, and measles virus replication with Rab9 siRNA.
Rab9 was originally implicated by gene trap insertional mutagenesis in Marburg virus replication. Interestingly, the late endosome has also been implicated as an assembly initiation site for Marburg virus (28). To determine if Rab9 is also utilized in the replication of Marburg virus or other unrelated viruses, Rab9 siRNA-treated Vero cells were infected with Marburg virus, Ebola virus, measles virus, and reovirus. Replication assays for Marburg virus, Ebola virus, measles virus, and reovirus were performed in Vero cells. The simian Rab9 gene was cloned from Vero cells (accession number AY601640) to aid in designing the appropriate primers for the quantitation of mRNA expression. Sequence analysis revealed that simian Rab9 is 98% homologous to human Rab9 at the mRNA level, with 100% homology at the chosen siRNA target sites (see Materials and Methods for target sites).
The treatment of Vero cells with Rab9 siRNA effectively reduced Rab9 mRNA expression by ∼80 to 90%, while GFP siRNA-treated cells showed mRNA expression levels similar to those of parental cells (Fig. 5A). Figure 5B shows a dramatic decrease in the immunofluorescence staining of viral proteins at 6 days postinfection in Ebola virus-infected cells (and to a lesser degree in Marburg virus-infected cells) transfected with Rab9 siRNA compared to that in mock or GFP-siRNA transfectants. Electron microscopic examination confirmed the decrease in the number of infected cells, with an absence of cytoplasmic nucleocapsids and mature virions in most Ebola virus-infected cells, and to a lesser extent, in Marburg virus-infected cells (data not shown). These results were further corroborated by antigen-capture assays (Fig. 5C), which showed an ∼70 to 75% decrease in the amount of viral antigen released into the culture medium at 6 days postinfection.
FIG. 5.
Effects of Rab9 silencing on Ebola virus, Marburg virus, measles virus, and reovirus replication. All assays were performed in Vero cells. (A) Rab9 silencing in Vero cells. Rab9 mRNA expression in mock- or siRNA-transfected Vero cells was determined at 2 or 5 days posttransfection, as described in the legend to Fig. 2C (n = 3). (B) At 2 days posttransfection, mock- or siRNA-transfected Vero cells were inoculated with Ebola virus (EBO) (left panels) or Marburg virus (MBG) (right panels), and viral antigen production was visualized at 6 days postinoculation using polyclonal antisera against Ebola virus or Marburg virus antigens and a FITC-labeled secondary antibody. Magnification, ×20. (C) Antigen-capture assays. Quantitation of viral antigens released into culture supernatants (at 6 days postinoculation) was performed using an enzyme-linked immunosorbent assay-based antigen-capture assay (n = 2). (D) Viral titers of measles virus strain Edmonston (Edm-MV) released into the supernatants of GFP and Rab9 siRNA Vero cell transfectants (n = 3). (E) Measles virus Edmonston immunofluorescence staining. Measles virus syncytium formation was visualized by immunostaining of the measles virus nucleoprotein (green) at 2 and 6 days postinoculation. Cell nuclei were stained with propidium iodide (red). Magnification, ×200. (F) Reovirus assays. Vero cell GFP, Rab9, and Rin2 siRNA transfectants were inoculated for 1 h with reovirus type 1, washed, and grown for 0 to 2 days postinoculation before freezing at −20°C. Cell lysates were titrated by standard plaque assays on L-cell monolayers. Results from a single experiment are shown and are representative of three independent experiments.
Targeting Rab9 mRNA for degradation with siRNA also inhibited measles virus replication. Both GFP and Rab9 siRNA transfectants were readily infectible based on fluorescence-activated cell sorting detection of the measles virus nucleoprotein in 66% and 83% of cells, respectively, at 2 days postinfection with the Edmonston strain of measles virus (data not shown). Although viral entry was uninhibited, there was an obvious delay in the downstream events of viral particle assembly and syncytium formation in Rab9 siRNA transfectants compared to GFP siRNA transfectants. Figures 5D and E show that syncytium formation and the release of viral particles into the culture supernatant were undetectable 2 days following infection in Rab9 siRNA transfectants and that measles virus production in Rab9 transfectants was ∼90% lower (statistically significant; P < 0.001) than that in GFP siRNA transfectants 5 days after infection. The inhibition of measles virus replication by Rab9 siRNA was overcome by 6 days postinfection, which may reflect the transient nature of mRNA knockdown using siRNA (Fig. 2C and 5A).
In contrast, replication of the nonenveloped reovirus was uninhibited by Rab9 silencing at either 1 or 2 days postinfection, as determined by viral titer plaque assays (Fig. 5F). Similarly, no decrease in reovirus immunostaining was observed in Rab9 siRNA-transfected cells relative to GFP or Rin2 siRNA-transfected controls (data not shown). The results of this study suggest that host genes mediating late-endosome-to-plasma-membrane vesicular transport, i.e., Rab9, TIP47, p40, PIKfyve, and Rab11A, are required for HIV replication and that Rab9 expression is also required for replication of the enveloped Ebola, Marburg, and measles viruses but not for that of the nonenveloped reovirus. These results provide insights into viral budding mechanisms as well as potential targets for intervention.
DISCUSSION
For this study, we identified the Rab9 GTPase by gene trap insertional mutagenesis as a candidate cellular gene utilized in Marburg virus infection. Following siRNA-directed Rab9 silencing, we observed inhibition with the enveloped viruses HIV, Ebola virus, Marburg virus, and measles virus, but not with the nonenveloped reovirus. HIV was also inhibited by reducing the mRNA expression of host genes involved in the same vesicular transport pathway as Rab9 (TIP47, p40, and PIKfyve) and Rab11A, which facilitates plasma membrane trafficking of cargo proteins from either the _trans_-Golgi network or recycling endosomes. An important role for late-endosome-to-_trans_-Golgi vesicular transport is further suggested by the observation that HIV Gag is sequestered in an endocytic CD63+ compartment in HeLa cells with depleted Rab9 protein expression.
The HIV Pr55Gag and Ebola virus matrix VP40 proteins alone have been shown to be sufficient for the release of virus-like particles in the absence of other viral proteins (14, 56); thus, the subcellular location of Gag proteins is thought to influence the pathway of viral release. Several HIV studies have shown two cell-type-dependent pathways for viral egress (Fig. 6). In macrophages, HIV Gag is restricted to CD63-positive late endosomes, and the prevailing evidence indicates that viral egress proceeds through TSG101-dependent budding into the lumen of late endosomes to form multivesicular bodies, followed by the export of viral particles as exosomes (40, 43, 45, 47, 48).
In contrast to the case for macrophages, studies with T cells and HeLa cells (used in this study) have shown that HIV Gag is either localized predominantly at the plasma membrane (21, 39, 43) or partitioned between the plasma membrane and the late endosome (42, 54). Viral assembly was observed to occur at both subcellular locations, although the relative importance of multivesicular body budding in HeLa cells is unclear (54). Both Gag and Env contain endocytosis motifs that promote redirection from the plasma membrane to the late endosome, and in the case of Gag, internalization is functionally important for virus-like particle production (6, 32, 59). HIV-1 Gag colocalized with the late endosomal marker CD63, irrespective of whether budding occurred in multivesicular bodies or at the plasma membrane (42). Therefore, it was suggested that viral assembly originates at the late endosome and that viral egress is gated through endosomally derived membranes even if budding ultimately occurs at the plasma membrane (42). Other studies have shown that Marburg virus and murine leukemia virus employ a similar strategy of beginning assembly at the late endosome and budding at the plasma membrane (2, 28). An important mechanistic question has been how endosomal membranes are transported to the cell surface during the course of viral maturation and budding.
Our results confirm the previously reported dependence of HIV replication on TIP47 and show for the first time that Rab9, p40, and PIKfyve are also critical for the HIV life cycle. Interestingly, murine leukemia virus potentially utilizes Rab11-positive recycling endosomes en route to the plasma membrane, and Rab11 is incorporated into the Marburg viral coat in secreted virions, although the effect of reducing Rab11 expression was not tested for either virus (2, 29). We showed that Rab11A expression is functionally important for HIV, although its precise function(s) requires further clarification, as Rab11A is involved in plasma membrane trafficking from both the _trans_-Golgi network and recycling endosomes.
Although major advances have been made in recent years towards understanding the host component of HIV and Ebola virus budding, several issues remain unresolved. Initiating viral assembly in an internal compartment may offer HIV and Ebola virus several advantages, including (i) the avoidance of immune surveillance, (ii) the concentration of viral proteins in one subcellular location, and (iii) hijacking of the endosomal sorting machinery (which normally sorts ubiquitinated proteins into multivesicular bodies) for the topologically similar process of viral budding (reviewed in reference 49). The respective p6 and VP40 structural proteins of HIV and Ebola virus are ubiquitinated as a prerequisite to TSG101-dependent recruitment of the endosomal sorting machinery (13, 15, 31, 37). Whether or not endocytosis of HIV and Ebola virus proteins from the plasma membrane precedes their ubiquitination, the compartment where ubiquitinated HIV and Ebola virus cargo proteins first interact with TSG101, and the mechanism used to recruit TSG101 to the plasma membrane at sites of viral egress are not yet clear.
If ubiquitination and the interaction of HIV viral proteins with TSG101 depend on internalization to the late endosome, then our results may reflect the importance of host proteins mediating late-endosome-to-_trans_-Golgi transport (Rab9, TIP47, p40, and PIKfyve) and _trans_-Golgi-to-plasma-membrane transport (Rab11A) once such communication with TSG101 has occurred (Fig. 3A and 6B). The dependence of Ebola virus and Marburg virus replication on Rab9 and TSG101 (reported for Ebola virus) indicates overlapping mechanisms for viral egress between HIV and the filoviruses Ebola virus and Marburg virus. If the HIV Gag interaction with TSG101 also occurs in the early endocytic compartment, then Rab11A may serve an additional role in the redirection of Gag-TSG101 complexes back to the plasma membrane (Fig. 6B). However, it is difficult to rationalize why Rab11B expression in HeLa cells (Fig. 3B) would not compensate for blocked Rab11A-dependent recycling-endosome-to-plasma-membrane traffic, given that Rab11B also mediates transport from recycling endosomes to the cell surface (52).
Measles virus has been shown to assemble at the plasma membrane, where the viral matrix protein bridges the replicative unit to the membrane-bound hemagglutinin and fusion proteins (12, 25). No reports have yet demonstrated that endosomal assembly or vesicular transport of proteins is involved in measles virus replication, although the possibility remains that measles virus utilizes this alternative pathway for viral egress. The inhibition of measles virus replication following Rab9 knockdown may occur by a different mechanism. Several studies have implicated cholesterol- and glycosphingolipid-rich membrane rafts as gateways for viral entry and exit for a variety of viruses, including measles virus, HIV-1, Ebola virus, and Marburg virus (1, 3, 35, 44). Recently, it has been shown that Rab7, Rab9, and Rab11 regulate normal sphingolipid and cholesterol trafficking and that Rab7 or Rab9 overexpression corrects sphingolipid and cholesterol trafficking defects in Niemann-Pick C sphingolipid storage disease (10, 23). An interesting possibility is that the inhibition of Rab9 expression may result in dysregulated cholesterol and sphingolipid trafficking, with consequential impairments in the replication cycles of viruses that utilize rafts, including measles virus, HIV, Ebola virus, and Marburg virus, although such a mechanism requires further investigation.
Recent studies have indicated that the enveloped HIV-1, Marburg, and murine leukemia viruses may utilize a common pathway for egress whereby assembly can begin at the late endosome and budding is completed at the plasma membrane, although the logistics of late-endosome-to-plasma-membrane transport were not defined (2, 28, 42). In contrast, the replication and assembly of the cytolytic, nonenveloped reovirus are thought to occur within cytoplasmic viral inclusions not associated with plasma membranes or other cellular organelles (4, 7, 17, 50). Thus, Rab9's potential roles in transporting either viral proteins or lipid raft components to the _trans_-Golgi network en route to the plasma membrane are not anticipated to be important in reoviral replication. In agreement, reovirus replication was not dependent on Rab9 expression (Fig. 5F), suggesting that Rab9 may be specifically utilized by viruses associated with membranes, particularly the late endosome.
This study demonstrates the power of gene trap insertional mutagenesis and RNA interference as a combined approach for discovering new genes that may serve as targets for therapeutics and for gaining insight into viral replication mechanisms. Naturally, in vitro results showing that cultured cells can survive the disruption of one allele or the silencing of target mRNA expression may not translate to multicellular organisms, necessitating that drugs inhibiting cellular targets pass the rigors of clinical testing for safety and efficacy. Here we show that the Rab9 GTPase is required for the unrelated enveloped viruses HIV-1, Marburg virus, Ebola virus, and measles virus, but not for the cytoplasmic reovirus. HIV was also inhibited by blocking the expression of the host genes TIP47, p40, PIKfyve, and Rab11A, indicating a potential pathway of viral egress from late endosomes to the cell surface. In agreement, Gag-EGFP in Rab9 siRNA-expressing cells was dramatically sequestered in an internal CD63+ endocytic compartment.
Acknowledgments
This work was performed under a cooperative research and development agreement with Avatar BioSci, Inc., and through the support of the Hudson Alpha Research Institute.
We express our appreciation to Mark Germain of Avatar, BioSci, Inc., and Jim Hudson of the Hudson Alpha Research Institute for supporting this work. We thank Cynthia Goldsmith and Donna Rudolph of the Centers for Disease Control for technical assistance. We also thank Steve McDougal of the Centers for Disease Control and Ralph Tripp of the University of Georgia for critical reviews of the manuscript.
REFERENCES
- 1.Avota, E., N. Muller, M. Klett, and S. Schneider-Schaulies. 2004. Measles virus interacts with and alters signal transduction in T-cell lipid rafts. J. Virol. 78**:**9552-9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5**:**161-174. [DOI] [PubMed] [Google Scholar]
- 3.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195**:**593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus sigmaNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75**:**1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blot, G., K. Janvier, S. Le Panse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the _trans_-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J. Virol. 77**:**6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273**:**15773-15778. [DOI] [PubMed] [Google Scholar]
- 7.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76**:**8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castillo, R. C., S. Arango-Jaramillo, R. John, B. C. Turner, E. Zimmerman, and D. H. Schwartz. 2000. Rapid communication: development of in vitro resistance to macrophage-tropic- and T-cell-line-adapted HIV-1 strains among HIV-positive volunteers treated with highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 24**:**197-202. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W., Y. Feng, D. Chen, and A. Wandinger-Ness. 1998. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9**:**3241-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhury, A., M. Dominguez, V. Puri, D. K. Sharma, K. Narita, C. L. Wheatley, D. L. Marks, and R. E. Pagano. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Investig. 109**:**1541-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz, E., F. Schimmoller, and S. R. Pfeffer. 1997. A novel Rab9 effector required for endosome-to-TGN transport. J. Cell Biol. 138**:**283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois-Dalcq, M., and T. S. Reese. 1975. Structural changes in the membrane of Vero cells infected with a paramyxovirus. J. Cell Biol. 67**:**551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107**:**55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59**:**103-112. [DOI] [PubMed] [Google Scholar]
- 15.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77**:**9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith, C. S., T. Whistler, P. E. Rollin, T. G. Ksiazek, P. A. Rota, W. J. Bellini, P. Daszak, K. T. Wong, W. J. Shieh, and S. R. Zaki. 2003. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 92**:**89-98. [DOI] [PubMed] [Google Scholar]
- 17.Gomatos, P. J., I. Tamm, S. Dales, and R. M. Franklin. 1962. Reovirus type 3: physical characteristics and interaction with L cells. Virology 17**:**441-454. [DOI] [PubMed] [Google Scholar]
- 18.Green, E. G., E. Ramm, N. M. Riley, D. J. Spiro, J. R. Goldenring, and M. Wessling-Resnick. 1997. Rab11 is associated with transferrin-containing recycling compartments in K562 cells. Biochem. Biophys. Res. Commun. 239**:**612-616. [DOI] [PubMed] [Google Scholar]
- 19.Hanna, J., K. Carroll, and S. R. Pfeffer. 2002. Identification of residues in TIP47 essential for Rab9 binding. Proc. Natl. Acad. Sci. USA 99**:**7450-7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecht, F. M., R. M. Grant, C. J. Petropoulos, B. Dillon, M. A. Chesney, H. Tian, N. S. Hellmann, N. I. Bandrapalli, L. Digilio, B. Branson, and J. O. Kahn. 1998. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N. Engl. J. Med. 339**:**307-311. [DOI] [PubMed] [Google Scholar]
- 21.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74**:**8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks, G. G., E. G. Shi, X. M. Li, C. H. Li, M. Pawlak, and H. E. Ruley. 1997. Functional genomics in mice by tagged sequence mutagenesis. Nat. Med. 16**:**338-344. [DOI] [PubMed] [Google Scholar]
- 23.Holtta-Vuori, M., K. Tanhuanpaa, W. Mobius, P. Somerharju, and E. Ikonen. 2002. Modulation of cellular cholesterol transport and homeostasis by Rab11. Mol. Biol. Cell 13**:**3107-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoque, M. O., S. Begum, O. Topaloglu, C. Jeronimo, E. Mambo, W. H. Westra, J. A. Califano, and D. Sidransky. 2004. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 64**:**5511-5517. [DOI] [PubMed] [Google Scholar]
- 25.Horikami, S. M., and S. A. Moyer. 1995. Structure, transcription, and replication of measles virus. Curr. Top. Microbiol. Immunol. 191**:**35-50. [DOI] [PubMed] [Google Scholar]
- 26.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2**:**1240-1243. [DOI] [PubMed] [Google Scholar]
- 27.Ikonomov, O. C., D. Sbrissa, K. Mlak, R. Deeb, J. Fligger, A. Soans, R. L. Finley, Jr., and A. Shisheva. 2003. Active PIKfyve associates with and promotes the membrane attachment of the late endosome-to-trans-Golgi network transport factor Rab9 effector p40. J. Biol. Chem. 278**:**50863-50871. [DOI] [PubMed] [Google Scholar]
- 28.Kolesnikova, L., S. Bamberg, B. Berghofer, and S. Becker. 2004. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J. Virol. 78**:**2382-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolesnikova, L., H. Bugany, H. D. Klenk, and S. Becker. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 76**:**1825-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ksiazek, T. G., P. E. Rollin, P. B. Jahrling, E. Johnson, D. W. Dalgard, and C. J. Peters. 1992. Enzyme immunosorbent assay for Ebola virus antigens in tissues of infected primates. J. Clin. Microbiol. 30**:**947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77**:**1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindwasser, O. W., and M. D. Resh. 2004. Human immunodeficiency virus type 1 Gag contains a dileucine-like motif that regulates association with multivesicular bodies. J. Virol. 78**:**6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombardi, D., T. Soldati, M. A. Riederer, Y. Goda, M. Zerial, and S. R. Pfeffer. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12**:**677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma, G., T. Greenwell-Wild, K. Lei, W. Jin, J. Swisher, N. Hardegen, C. T. Wild, and S. M. Wahl. 2004. Secretory leukocyte protease inhibitor binds to annexin II, a cofactor for macrophage HIV-1 infection. J. Exp. Med. 200**:**1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74**:**305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez, O., and B. Goud. 1998. Rab proteins. Biochim. Biophys. Acta 1404**:**101-112. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7**:**1313-1319. [DOI] [PubMed] [Google Scholar]
- 38.Miller, L. H., S. J. Mason, D. F. Clyde, and M. H. McGinniss. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295**:**302-304. [DOI] [PubMed] [Google Scholar]
- 39.Nermut, M. V., W. H. Zhang, G. Francis, F. Ciampor, Y. Morikawa, and I. M. Jones. 2003. Time course of Gag protein assembly in HIV-1-infected cells: a study by immunoelectron microscopy. Virology 305**:**219-227. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278**:**52347-52354. [DOI] [PubMed] [Google Scholar]
- 41.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8**:**681-686. [DOI] [PubMed] [Google Scholar]
- 42.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4**:**902-910. [DOI] [PubMed] [Google Scholar]
- 43.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78**:**1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98**:**13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orenstein, J. M., M. S. Meltzer, T. Phipps, and H. E. Gendelman. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 62**:**2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Organ, E. L., J. Sheng, H. E. Ruley, and D. H. Rubin. 2004. Discovery of mammalian genes that participate in virus infection. BMC Cell Biol. 5**:**41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pautrat, G., M. Suzan, D. Salaun, P. Corbeau, C. Allasia, G. Morel, and P. Filippi. 1990. Human immunodeficiency virus type 1 infection of U937 cells promotes cell differentiation and a new pathway of viral assembly. Virology 179**:**749-758. [DOI] [PubMed] [Google Scholar]
- 48.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162**:**443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12**:**569-579. [DOI] [PubMed] [Google Scholar]
- 50.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17**:**342-355. [DOI] [PubMed] [Google Scholar]
- 51.Rubin, D. H., M. J. Kornstein, and A. O. Anderson. 1985. Reovirus serotype 1 intestinal infection: a novel replicative cycle with ileal disease. J. Virol. 53**:**391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlierf, B., G. H. Fey, J. Hauber, G. M. Hocke, and O. Rosorius. 2000. Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res. 259**:**257-265. [DOI] [PubMed] [Google Scholar]
- 53.Sheng, J., E. L. Organ, C. Hao, K. S. Wells, H. E. Ruley, and D. H. Rubin. 2004. Mutations in the IGF-II pathway that confer resistance to lytic reovirus infection. BMC Cell Biol. 5**:**32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4**:**785-801. [DOI] [PubMed] [Google Scholar]
- 55.Sun, Y., L. Li, F. Lau, J. A. Beavo, and E. A. Clark. 2000. Infection of CD4+ memory T cells by HIV-1 requires expression of phosphodiesterase 4. J. Immunol. 165**:**1755-1761. [DOI] [PubMed] [Google Scholar]
- 56.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 283**:**1-6. [DOI] [PubMed] [Google Scholar]
- 57.Ullrich, O., S. Reinsch, S. Urbe, M. Zerial, and R. G. Parton. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135**:**913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Melchner, H., and H. E. Ruley. 1989. Identification of cellular promoters by using a retrovirus promoter trap. J. Virol. 63**:**3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75**:**2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yilla, M., C. Hickman, M. McGrew, E. Meade, and W. J. Bellini. 2003. Edmonston measles virus prevents increased cell surface expression of peptide-loaded major histocompatibility complex class II proteins in human peripheral monocytes. J. Virol. 77**:**9412-9421. [DOI] [PMC free article] [PubMed] [Google Scholar]