LIM Protein KyoT2 Negatively Regulates Transcription by Association with the RBP-J DNA-Binding Protein (original) (raw)
Abstract
The RBP-J/Su(H) DNA-binding protein plays a key role in transcriptional regulation by targeting Epstein-Barr virus nuclear antigen 2 (EBNA2) and the intracellular portions of Notch receptors to specific promoters. Using the yeast two-hybrid system, we isolated a LIM-only protein, KyoT, which physically interacts with RBP-J. Differential splicing gave rise to two transcripts of the KyoT gene, KyoT1 and KyoT2, that encoded proteins with four and two LIM domains, respectively. With differential splicing resulting in deletion of an exon, KyoT2 lacked two LIM domains from the C terminus and had a frameshift in the last exon, creating the RBP-J-binding region in the C terminus. KyoT1 had a negligible level of interaction with RBP-J. Strong expression of KyoT mRNAs was detected in skeletal muscle and lung, with a predominance of KyoT1 mRNA. When expressed in F9 embryonal carcinoma cells, KyoT1 and KyoT2 were localized in the cytoplasm and the nucleus, respectively. The binding site of KyoT2 on RBP-J overlaps those of EBNA2 and Notch1 but is distinct from that of Hairless, the negative regulator of RBP-J-mediated transcription in Drosophila. KyoT2 but not KyoT1 repressed the RBP-J-mediated transcriptional activation by EBNA2 and Notch1 by competing with them for binding to RBP-J and by dislocating RBP-J from DNA. KyoT2 is a novel negative regulatory molecule for RBP-J-mediated transcription in mammalian systems.
Transcription of mRNA-encoding genes in eukaryotic cells involves RNA polymerase II in conjunction with a set of basal transcription factors. Increased levels of gene-specific transcription are achieved by transcriptional activators that bind regulatory cis elements and stimulate the basal rate of transcription initiation (38). In addition, there are other sets of transcription factors that lower the rate of transcription initiation. Expression of genes is influenced by these opposing elements and regulated to obtain the optimum concentration of gene products temporally and spatially (13, 25).
RBP-J/RBP-Jκ/Su(H), a 60-kDa DNA-binding protein recognizing the core sequence C/TGTGGGAA (27, 33, 49), is highly conserved from humans to Drosophila and is expressed in embryos and all adult tissues in the mouse (3, 19, 24). The targeted disruption of mouse RBP-J revealed that homozygous null mutants die before 10.5 days postcoitum (dpc) with various abnormalities, including growth retardation and defects in the central nervous system and somites, suggesting a role of RBP-J in development of the central nervous system and somites in the mouse (37).
The Notch gene of Drosophila encodes a large transmembrane protein required for segregation of neural precursor cells from neuroectodermal cell clusters through the process called lateral specification (4). Genetic analyses as well as in vitro biochemical studies suggested that Suppressor of Hairless [Su(H)], the Drosophila homolog of RBP-J, is a component of the Notch signaling pathway in Drosophila (6, 18, 21, 31, 45). Transcription from the Drosophila Enhancer of split [_E(spl)_] m8 promoter in S2 cells is enhanced by the transfection of Su(H) and reduced by cotransfection of Hairless, a gene that is known to counteract the function of Su(H) in vivo (8, 20). Likewise, mammalian RBP-J has been shown to associate physically with the intracellular portion of Notch1 (47) and to activate transcription from the mouse Hairy enhancer of split (HES-1) promoter in HeLa cells (29). The phenotype of mice carrying a disrupted RBP-J gene is reminiscent of that of Notch1 null mutant mice (12, 37, 46). Detailed analyses of Notch1−/− and RBP-J−/− mutants indicate the functional interaction between RBP-J and Notch in mouse embryogenesis (15).
Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) is a transcriptional activator encoded by EBV and is essential for immortalization of primary human B lymphocytes by the viral infection in vitro. EBNA2 is responsible for upregulation of the viral latent membrane protein, terminal proteins (TP) 1 and 2, and cellular antigens such as CD21 and CD23 (2). Although EBNA2-responsive cis elements have been identified upstream of most of these genes, EBNA2 does not bind to DNA by itself (48, 52, 54). Subsequently, RBP-J was shown to bind to these elements and to interact physically with EBNA2, thus mediating the transcriptional activation of EBNA2-regulated genes (22, 26, 51, 55).
RBP-J has been shown to act as a repressor as well from a study of the transcriptional regulation of the adenovirus capsid protein polypeptide IX (pIX) (16). RBP-J binds to its cognate sequence present in the pIX promoter and downregulates its transcription. RBP-J fused with the DNA binding domain of yeast GAL4 also repressed, in HeLa cells, the basal transcription level from a herpes simplex virus thymidine kinase promoter that contained GAL4 binding sites upstream of the TATA box. The domain of RBP-J responsible for this repression function has been identified (28).
It is thus clear that RBP-J binds to DNA in a sequence-specific manner and acts as a transcription factor. Although several proteins have been identified as interacting with RBP-J and modulating transcription, the mechanisms by which RBP-J mediates transcription regulation are not fully understood. It is likely that several other proteins interacting with RBP-J are required to accomplish the appropriate transcriptional regulation of target genes. Here we show that a novel LIM protein, KyoT, interacts with RBP-J and negatively regulates transcription by competing with other RBP-J-binding transcriptional activators and displacing RBP-J from DNA.
MATERIALS AND METHODS
Cells and transfections.
Simian COS-7 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin sulfate per ml, and 2 mM l-glutamine. G418 (470 μg/ml) was added to the medium for F9 embryonal carcinoma cells stably transfected with KyoT or vector plasmid. For expression of KyoT and RBP-J proteins in mammalian cells, a Myc epitope tag was attached to the amino or carboxy terminus of the KyoT or RBP-J open reading frame, respectively, and they were cloned into the pEFBOSneo vector (35). T7-tagged RBP-J was described previously (47). Cells were transfected by using Lipofectamine as recommended by the manufacturer (GIBCO BRL).
Yeast two-hybrid screening.
The cDNA library from mouse 9.5-dpc embryos was screened as previously described (47). For screening of a HeLa cDNA library, a yeast reporter strain containing the plasmid pEG202-mRBP2, which encoded the entire mouse RBP2 open reading frame fused in frame to the LexA DNA-binding domain (23), was generated. Approximately 106 transformants were screened for the ability to grow on plates with medium lacking leucine and for LacZ expression (β-galactosidase activity). Plasmids were rescued from about 60 positive clones and sequenced. cDNA sequences and their deduced amino acids were compared with the GenBank and SwissProt databases.
Screening of cDNA and genomic libraries.
KyoT1 and KyoT2 cDNAs were isolated from a mouse brain cDNA library (Clontech) by using the cDNA fragments recovered from the two-hybrid screen as probes, and both strands were sequenced with an Applied Biosystems automated sequencing apparatus. Full-length cDNAs were generated by ligating two cDNA fragments at _Bst_XI sites.
To isolate genomic clones, a mouse genomic DNA library (Stratagene) was screened by using the 643-bp _Eco_47III fragment from KyoT1 cDNA as a probe. Five exons contained in the cDNA and their introns were sequenced to determine the exon-intron structure of the KyoT gene. The putative first exon containing the 5′ untranslated sequence was not determined.
Northern blotting and RT-PCR.
For Northern blotting, 18 μg of total RNA from different adult mouse tissues was loaded on each lane. The 800-bp _Eco_RI-_Bst_XI fragment isolated from KyoT1 cDNA was radiolabelled by the random priming method and used as a probe. The hybridization was carried out for 20 h at 65°C, and the membrane was subjected to stringent washes with 2× SSPE (360 mM NaCl, 20 mM sodium phosphate, 2 mM EDTA, pH 7.4)–0.1% sodium dodecyl sulfate (SDS) for 20 min at room temperature followed by 0.1× SSPE–0.1% SDS for 30 min at 65°C. The blot was exposed to the imaging plate overnight and was detected with a Fuji BioImage analyzer BAS1500.
Reverse transcription-PCR (RT-PCR) was performed essentially as described previously (34) except that PCR was carried out for 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The primers used for PCR were 5′-GACCAGAACGTGGAGTACAA-3′ and 5′-AGTCAGGGCAATACACCT GC-3′. As an internal standard, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ were used to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in a parallel reaction.
Immunofluorescence.
Stably transfected F9 cells grown overnight on glass coverslips were rinsed in phosphate-buffered saline, fixed in 2% paraformaldehyde for 30 min at room temperature, and permeabilized in ethanol for 10 min at −20°C. To detect KyoT, permeabilized cells were incubated with an anti-c-Myc antibody (Santa Cruz) followed by fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G antibody (Southern Biotechnology Inc.). Slides were mounted in glycerol and viewed on a Zeiss fluorescence microscope.
Yeast interaction assay.
For yeast interaction assays, the plasmids were subcloned into either pGBT9 or pGAD424, and all procedures were carried out according to the instructions of the manufacturer (Clontech). For construction of R1, R2, and R3, the cDNA encoding RBP-J was digested with the restriction enzymes indicated at the top in Fig. 4A and subcloned into pGBT9. All other RBP-J mutants were described previously (11) and were subcloned into pGBT9.
FIG. 4.
KyoT2 competes with Notch1 for binding to RBP-J. (A) KyoT2 was fused with the GAD and tested for interaction with various RBP-J deletion mutants by using the two-hybrid system. The wild-type RBP-J protein is shown at the top. The restriction enzyme sites with amino acid positions are shown above. Solid boxes, integrase motif; slashed boxes, DNA-binding regions; +++, colonies turning blue within 15 min; ++, colonies turning blue within 30 min; +, colonies turning blue after 60 min of incubation. (B) RBP-J-binding regions of EBNA2 encoded by two strains of EBV, B95-8 and AG876 (32), and RAM domains of murine Notch (mNotch) family members (30) were compared with the RBP-J-binding region of KyoT2 (residues 168 to 194). The conserved amino acids are shaded, and the conserved tryptophan and proline are boxed. The amino acids which have been shown to be critical for the interaction with RBP-J are marked with dots. (C) COS-7 cell lysates containing Myc-tagged RBP-J protein were mixed with increasing amounts of GST or GST-KyoT proteins for 1 h at 4°C prior to incubation with a saturating amount of GST-Notch1[1751–2170]. Subsequently, the mixtures were immunoprecipitated with anti-Myc antibody, washed three times with solubilization buffer, and analyzed by Western blotting with anti-GST antibody. The membrane was reprobed with T6709 to detect precipitated RBP-J.
Production of GST fusion proteins and GST pull-down assays.
The full-length KyoT1, KyoT2, and hRAM8 cDNA fragments were subcloned into the pGEX-4T vector. Glutathione _S_-transferase–RAM23 (GST-RAM23) and GST-Notch1[1751–2170] were described previously (47). GST fusion proteins were produced and used for interaction assays as described previously (47). GST-KyoT1 and GST-KyoT2 were eluted from beads with 20 mM glutathione at pH 8.5, dialyzed against phosphate-buffered saline, and semiquantified by Coomassie brilliant blue staining of SDS-polyacrylamide gels.
EMSA.
Electrophoretic mobility shift assays (EMSA) were carried out essentially as described previously (49). Two nanograms of radiolabelled oligonucleotides encoding the EBV C promoter (Cp) was used as a probe for each reaction mixture. The anti-RBP-J monoclonal antibody K0043 was described previously (40). The sequences of Cp and mCp probes are as follows (the RBP-J cognate sequences are underlined): Cp, 5′-GATCTGGTGTAAACACGCCGTGGGAAAAAATTTATG-3′; mCp, 5-GATCTGGTGTAAACACGCCGTCCCAAAAAATTTATG-3′.
Production of antibody and immunoprecipitation.
The purified GST-KyoT2 protein was injected into a rabbit to produce the polyclonal antibody against KyoT proteins. The specificity of the antiserum was confirmed by Western blotting analysis. The immunoprecipitation was carried out essentially as described previously (47) with either anti-Myc (Santa Cruz) or T7 (Novagen) antibody. The precipitates were washed five times with the solubilization buffer. Anti-RBP-J monoclonal antibody T6709 (40) and the antiserum raised against GST-KyoT2 were used for detection of RBP-J and KyoT proteins, respectively. For Fig. 4C, the appropriate amount of GST, GST-KyoT1, or GST-KyoT2 protein was added to the precleared COS-7 cell lysate containing Myc-tagged RBP-J protein. The mixture was incubated on ice for 1 h prior to addition of a saturating amount of GST-Notch1[1751–2170]. The incubation was continued on ice for 1 h, followed by immunoprecipitation with anti-Myc antibody (Santa Cruz). The proteins were detected with anti-GST antibody (Calbiochem) or T6709.
Transcription activity assays.
The procedures for luciferase assays, the expression constructs for EBNA2 and RAMIC, and the pGa981-6 reporter plasmid were all described previously (30).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper will appear in the GenBank, EMBL, and DDBJ nucleotide sequence databases with the accession number U41739.
RESULTS
Identification of an RBP-J-binding protein with multiple LIM domains.
To identify RBP-J-interacting proteins, we performed yeast two-hybrid screens of cDNA libraries from mouse 9.5-dpc embryos and HeLa cells by using a reporter strain expressing a LexA-mouse RBP2 fusion protein. One million transformants of each library were screened, and about 60 clones that were positive for both nutritional selection and β-galactosidase activity were recovered. Nucleotide sequence determination and comparison with the GenBank and SwissProt databases revealed that one clone from the mouse embryo library, termed RAM23, encoded a portion of murine Notch1 between the transmembrane region and the ankyrin repeat. Subsequent analyses defined the RAM domain of Notch1, which is responsible for the direct interaction with RBP-J (47). In addition, multiple independent copies of cDNA fragments were identified. One of these clones, designated KyoT, was isolated from both mouse embryo and HeLa cDNA libraries, was characterized extensively, and is described in this paper. The other clones identified will be described elsewhere.
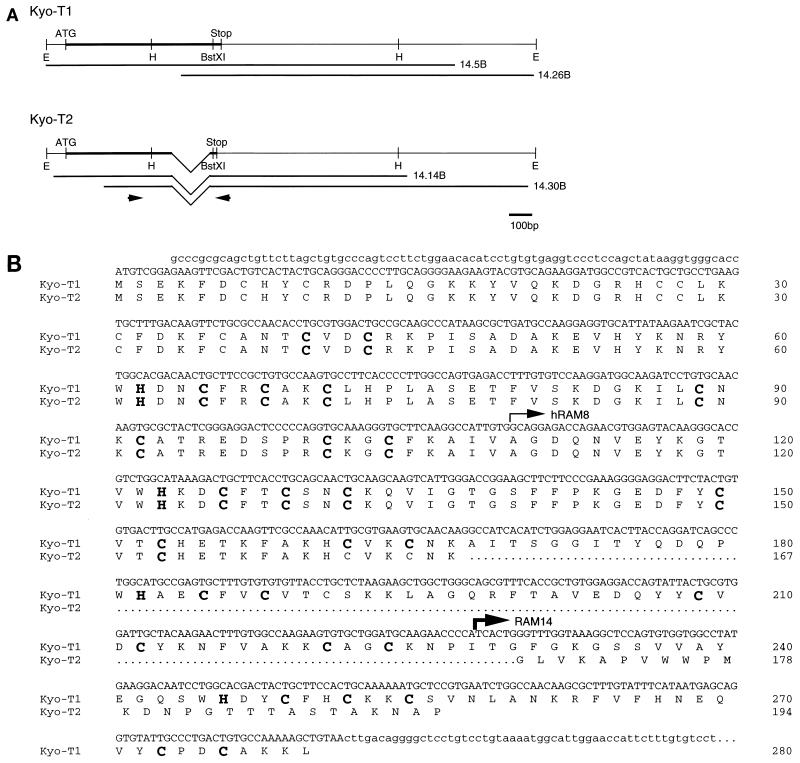
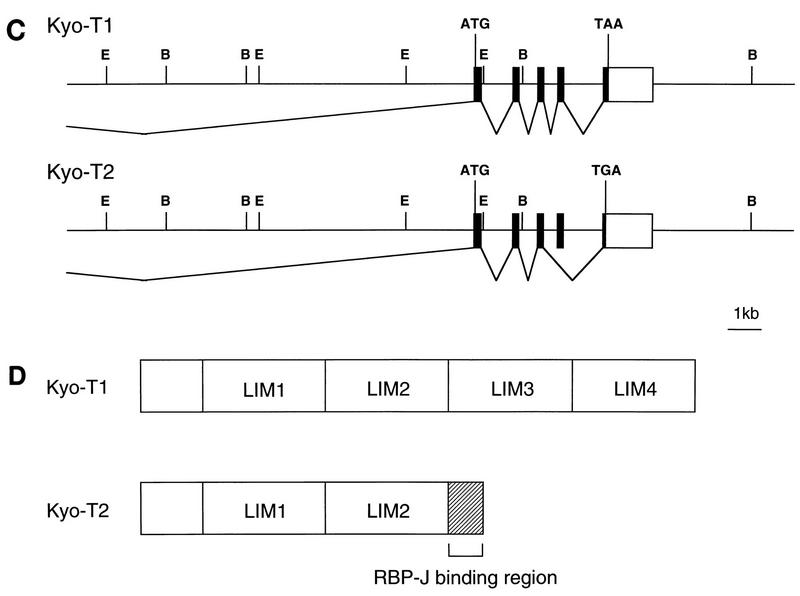
The KyoT partial cDNA recovered from the two-hybrid screens was used to screen a mouse brain cDNA library. Two types of cDNAs were identified during screening and were designated KyoT1 and KyoT2. The full-length cDNA sequences of KyoT1 and KyoT2 were constructed by combining sequences of two overlapping cDNA fragments (Fig. 1A). The two sequences are identical except for the 3′ portion of the coding region. The open reading frames of KyoT1 and KyoT2 encode predicted proteins of 280 and 194 amino acids, respectively. They have the first 167 amino acids in common and differ in the C terminus, suggesting a differential splicing mechanism (Fig. 1B). To determine the structure of the KyoT gene, a mouse genomic library was screened with the KyoT1 cDNA fragment as a probe. One of the positive clones obtained spanned about 20 kb and encoded five exons extending from just upstream of the first methionine of the coding region to the 3′ untranslated region. The promoter region and most of the 5′ untranslated region were not obtained. Comparison of sequences of complementary and genomic DNAs supports the alternative splicing mechanism for synthesis of KyoT1 and KyoT2, by which one of the exons is spliced out, causing a frameshift in the following exon in KyoT2 cDNA (Fig. 1C). The cDNA fragments isolated in the two-hybrid screens of the mouse embryo and the HeLa cDNA libraries (designated RAM14 and hRAM8, respectively, in Fig. 1B) encode the most C-terminally located 27 and 85 amino acids of KyoT2, respectively. This implies that 27 amino acids in the carboxy terminus of KyoT2, which are missing in KyoT1, are sufficient for the interaction with RBP-J (see Discussion).
FIG. 1.
Sequence and structure of KyoT. (A) cDNA structure of KyoT. The coding regions are represented by thick lines. The cDNA fragments obtained from the mouse brain cDNA library are shown below the putative full-length cDNAs. The positions of primers used for RT-PCR are shown by arrows. E, _Eco_RI; H, _Hin_dIII; ATG, putative initiation codon; Stop, termination codon. (B) Nucleotide and deduced amino acid sequences of KyoT. Nucleotide sequences of coding and noncoding regions are shown as upper- and lowercase letters, respectively. The first 167 amino acids are shared between two alternatively spliced isoforms, KyoT1 and KyoT2. The amino acids removed by alternative splicing in KyoT2 are represented by dots. Four LIM domains are boxed, and the conserved cysteines and histidines within the LIM motif are shown as boldface letters. The 5′ ends of the fragments obtained by two-hybrid screening from mouse embryo (RAM14) and HeLa (hRAM8) cDNA libraries are shown by thin and thick arrows, respectively. (C) Genomic structure of KyoT. The functional gene is composed of at least five exons and spans at least 20 kbp. In KyoT2 mRNA, one of the exons is removed by alternative splicing. Most of the 5′ untranslated region was not cloned. Solid boxes, coding region; open boxes, untranslated region; ATG, first methionine of the coding region; TAA and TAG, termination codons of KyoT1 and KyoT2, respectively; E, _Eco_RI; B, _Bam_HI. (D) Simplified diagram showing the structures of the KyoT1 and KyoT2 proteins. The LIM domains are numbered from the N terminus.
A computer search for homologous proteins by using BLAST showed that KyoT1, the longer transcript, is identical to the murine counterpart of SLIM (skeletal muscle LIM protein), the LIM-only protein of unknown function originally cloned from the human skeletal muscle cDNA library (36). Computer analysis using PROSITE revealed that the predicted KyoT1 protein possesses four copies of the LIM motif with the sequence CX2CX17HX2CX2CX2CX17CX2C but that the KyoT2 protein has only two such copies, due to the frameshift 3′ of the second LIM motif (Fig. 1D). Neither KyoT1 nor KyoT2 contained homeodomains, indicating that they are LIM-only proteins. The comparison of sequences of various LIM proteins has led to the phylogenic classification of LIM domains into five discrete clusters, namely, groups, A to E (14). While LIM domains of all the LIM homeodomain proteins are classified in group A or B in the dendrogram, all four LIM domains of KyoT1 were classified in either group C, D, or E. The further classification into each group by using the neighbor-joining method (39) was ambiguous (data not shown).
Expression profile of KyoT.
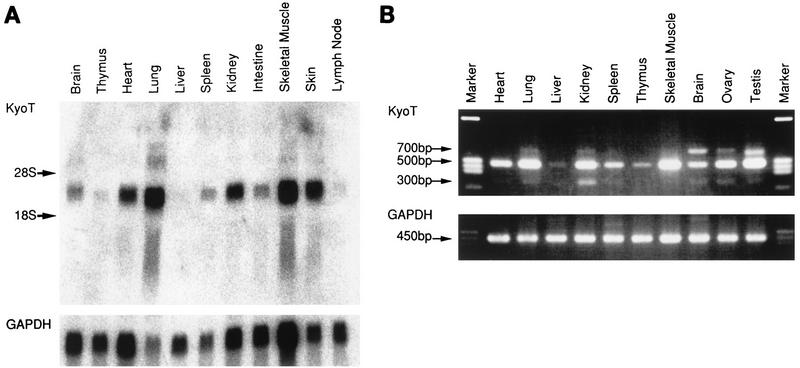
To investigate the expression pattern of KyoT, Northern blot analysis was performed. KyoT mRNA was detected by using the coding region of KyoT1 as a probe. Expression was observed in a variety of tissues, including strong expression in skeletal muscle and lung. Few transcripts were seen in the thymus, lymph nodes, and liver (Fig. 2A). In agreement with the tissue expression profile, KyoT mRNAs were highly expressed in myogenic C2C12 cells but were not detectable in any of the B-cell lines tested (data not shown). Since the tissue expression of SLIM was reported to be confined to skeletal muscle, RT-PCR was carried out with an independently prepared pool of RNA. The primers were synthesized to distinguish KyoT1 and KyoT2 transcripts as shown in Fig. 1A. The results of RT-PCR generally agreed with our findings from Northern blot analysis (Fig. 2B). There seems to be abundant KyoT mRNA in genital organs as well. Interestingly, there was a larger band in some tissues in addition to the expected 493- and 306-bp bands of KyoT1 and KyoT2, respectively. A 692-bp band in brain was sequenced and proved to be another variant of KyoT cDNA (data not shown). This form was not characterized further in this study. The amount of each amplified fragment varied from tissue to tissue. Although closer studies are required to determine the exact proportion of each transcript, KyoT1 seems to be most abundant in each tissue, with KyoT2 being much less abundant. There were relatively larger amounts of KyoT2 mRNA in brain, lung, kidney, and genital organs.
FIG. 2.
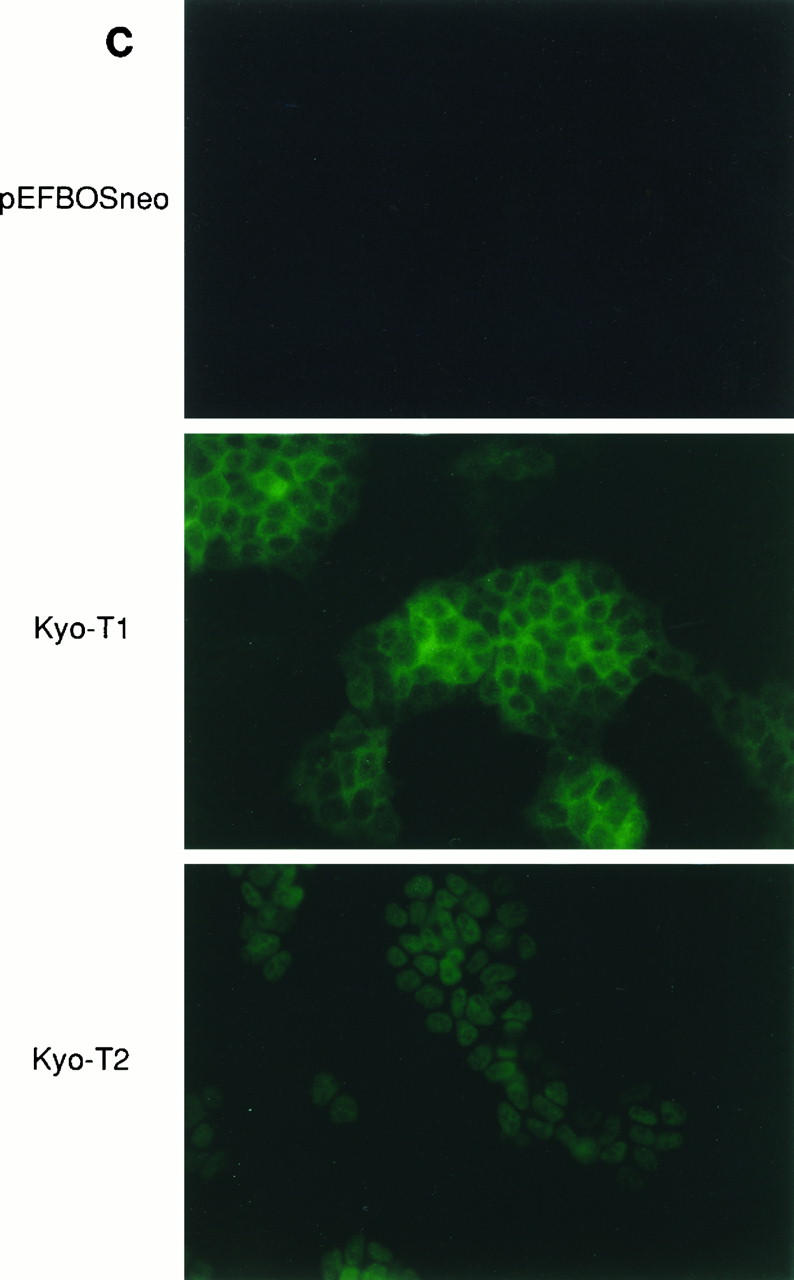
Expression profile of KyoT. (A) Northern blot analysis. Each lane contains 18 μg of total RNA prepared from various tissues of adult mice. The same membrane was reprobed with a GAPDH probe to monitor the amount of RNA. (B) RT-PCR. Equal amounts of total RNAs from various tissues were amplified by RT-PCR with primers located 5′ and 3′ of the skipped exon (Fig. 1A). GAPDH was used for an internal control. (C) Subcellular localization of KyoT1 and KyoT2 by indirect immunofluorescence. F9 cells stably transfected with either pEFBOSneo, Myc-tagged KyoT1, or Myc-tagged KyoT2 were stained with anti-Myc antibody and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G antibody.
Immunofluorescent staining was performed to examine the subcellular localization of c-Myc-tagged KyoT proteins, using an anti-Myc monoclonal antibody (9E10). KyoT2 was localized in the nuclei of the stably transfected F9 embryonal carcinoma cells, whereas KyoT1 was localized in the cytoplasm (Fig. 2C). Since we are not able to find a typical nuclear localization signal in KyoT2, it might be transported to the nucleus by modification and/or interaction with other proteins.
Physical interaction between RBP-J and KyoT2.
To confirm physical interaction with RBP-J, KyoT1 and KyoT2 were fused with either the GAL4 DNA-binding domain (GBT) or the GAL4 activation domain (GAD) and subsequently used for two-hybrid interaction analysis. When yeast was transformed with these constructs alone, no β-galactosidase activity was observed. GBT-KyoT2 showed strong β-galactosidase activity with GAD–RBP-J but not with GAD itself or GAD-p53. Very weak β-galactosidase activity was observed when GBT-KyoT1 was transformed with GAD–RBP-J. Formation of homo- or heterodimers of KyoT1 or KyoT2 could not be detected with these assays (data not shown).
GST fusion proteins containing KyoT1, KyoT2, and hRAM8 were prepared to test their interaction with RBP-J in vitro. GST fusion proteins were mixed with in vitro-translated RBP-J or luciferase that had been labelled with [35S]methionine. Proteins bound to the fusion proteins were recovered on glutathione-Sepharose beads under high-stringency conditions and analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 3A). In vitro-translated RBP-J proteins interacted strongly with the GST fusion protein containing KyoT2 or hRAM8 but not with GST per se. We estimate that about 20% of the input RBP-J was precipitated with either KyoT2 or hRAM8. A weak interaction of about one-ninth of that for KyoT2 or hRAM8 was observed for KyoT1, consistent with the results of the two-hybrid interaction assay. None of the GST fusion proteins interacted with luciferase.
FIG. 3.
KyoT2 binds to RBP-J strongly in vitro and in vivo. (A) GST pull-down assay. GST or GST fusion proteins were mixed with in vitro-translated 35S-labelled RBP-J or luciferase, recovered on glutathione-Sepharose beads, and analyzed on an SDS-polyacrylamide gel. A 1/20 volume of in vitro-translated product used for immunoprecipitation was run in lanes 1 and 6. (B) EMSA with a Cp probe. Increasing amounts of GST-KyoT proteins or excess amounts of GST and GST-RAM23 proteins were added to the RBP-J–DNA complex and analyzed by nondenaturing polyacrylamide gel electrophoresis. The amounts of GST-KyoT proteins were adjusted by Coomassie brilliant blue staining of SDS-polyacrylamide gels. (C) Immunoprecipitation (IP). The indicated expression constructs were transfected into COS-7 cells. After 48 h, cell extracts were prepared and immunoprecipitated with either anti-Myc or T7 antibody. The precipitates were washed five times with solubilization buffer and analyzed by Western blotting with either T6709 (upper panel) or antiserum raised against GST-KyoT2 (lower panel). One-tenth of the cell extract used for immunoprecipitation was run in lanes 1 to 3. The lower band of KyoT2 represents the partially degraded product.
To determine whether KyoT interacts with the RBP-J–DNA complex, EMSA were performed by using the synthetic 36-bp oligonucleotide of the EBV latency C promoter, the naturally occuring RBP-J-binding sequence containing CGTGGGAA, as a probe. The GST fusion protein containing either KyoT1, KyoT2, or RAM23 of murine Notch1 was mixed with in vitro-translated RBP-J in the presence of the Cp oligonucleotide. The excess amount of GST-RAM23 generated a shifted band when mixed with the RBP-J–DNA complex (Fig. 3B, lane 9). When increasing amounts of the KyoT2 protein were added to the reaction mixture, the RBP-J–DNA complex not only migrated more slowly but also became faint (Fig. 3B, lanes 6 to 8). No such shift or dissociation from DNA was observed when increasing amounts of the GST-KyoT1 protein or an excess amount of the GST protein was added to the reaction instead (Fig. 3B, lanes 2 to 5). None of the GST fusion proteins bound to the Cp oligonucleotide by themselves (data not shown). We next tested whether this shifted band generated by KyoT2 contains RBP-J protein. K0043, a monoclonal antibody against RBP-J, shifted the RBP-J–DNA complex (40) (Fig. 3B, lane 13). When the same amount of K0043 was added to a preincubated mixture of KyoT2 and RBP-J, they failed to form a ternary complex (Fig. 3B, lane 14). This could be due to masking of the epitope to K0043 by KyoT2, as has been shown for the association of RBP-J with Notch1 or EBNA2 (unpublished data). We therefore tried competition experiments using either wild-type or mutated unlabelled Cp oligonucleotide. As shown in Fig. 3B, lanes 15 to 19, a 10-times-greater molar amount of unlabelled Cp carrying the RBP-J-binding sequence almost completely abolished the shifted complex, while the mutated Cp to which RBP-J cannot bind did not alter the complex formation by KyoT2. This indicates that the shifted band generated by KyoT2 contains the RBP-J protein. However, the KyoT2–RBP-J complex exists mostly detached from DNA, and the amount of the KyoT2–RBP-J–DNA complex was reduced compared with that of the RBP-J–DNA complex. We also examined the formation of supershifted bands with several other oligonucleotides derived from promoters containing RBP-J-binding sites, such as the murine HES-1 and EBV TP-1 genes, and obtained similar results (data not shown).
To test whether KyoT interacts with RBP-J in mammalian cells, immunoprecipitation was carried out. T7-tagged RBP-J was expressed in COS-7 cells with either KyoT1, KyoT2, or vector plasmid. A Myc epitope tag was attached to the N terminus of KyoT to allow the immunoprecipitation of the proteins. The cell extracts were immunoprecipitated with the anti-Myc antibody, and the precipitates were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis with an anti-RBP-J monoclonal antibody, T6709 (40). A significant amount of RBP-J was coimmunoprecipitated with KyoT2. In contrast, the amount of RBP-J coimmunoprecipitated with KyoT1 was below the level of detection, while a comparable amount of KyoT1 was immunoprecipitated (Fig. 3C, lanes 4 to 6). Similarly, the RBP-J protein was immunoprecipitated with the anti-T7 antibody, and KyoT proteins in the precipitates were subjected to immunodetection. The antibody against the T7 tag was chosen for immunoprecipitation instead of K0043, the monoclonal antibody suitable for immunoprecipitation of RBP-J, because it was demonstrated in EMSA that KyoT2 prevents K0043 from binding to RBP-J (Fig. 3B, lane 14). For detection of KyoT proteins, antisera were raised against GST-KyoT2 and used for Western blotting, since the detection of KyoT proteins with the anti-Myc antibody was obscured due to an overlapping nonspecific band (data not shown). The expression of endogenous KyoT proteins was barely detectable in COS7 cells (Fig. 3C, lane 1). When KyoT-containing cell lysates were precipitated with the anti-T7 antibody, a small but significant fraction of the KyoT2 protein was detected in the precipitates, while the KyoT1 protein was not detected (Fig. 3C, lanes 7 to 9). A faint band detected below the immunoglobulin light chain (Fig. 3C, lanes 7 and 8) may represent the precipitated endogenous KyoT2 protein.
KyoT2 and Notch1 compete with each other for binding to RBP-J.
To locate the RBP-J domain responsible for binding to KyoT2, a series of RBP-J mutants was tested for interaction with KyoT2 by using the yeast two-hybrid system. RBP-J mutants with deletions of various extents at both ends were expressed as fusions with the GBT. A yeast reporter strain, SFY526, was transformed with these constructs and KyoT2 fused with the GAD. Subsequent β-galactosidase assays identified the interaction domain between residues 141 and 371 (Fig. 4A). This central region of RBP-J includes the integrase motif of unknown function and the adjacent N and C regions that are essential for DNA binding (11). Among molecules which have been shown to interact with RBP-J so far, Notch1 and EBNA2 bind to RBP-J in a similar region required for interaction with KyoT2, whereas the domain required for interaction with Hairless has been mapped at the more C-terminal region of RBP-J (8). Moreover, the comparison of the RBP-J-interacting region of KyoT2 with those of EBNA2 and Notch family members revealed conserved tryptophan and proline residues within this region (Fig. 4B). When these amino acids are mutated, EBNA2 and Notch proteins lose their contact with RBP-J (30, 32, 47). The interaction with the central portion of RBP-J, together with the conservation of these amino acids, suggests that the topological structures of the RBP-J-interacting domains of KyoT2, Notch, and EBNA2 may be quite similar and that they might bind to RBP-J in a similar fashion.
We employed immunoprecipitation to test whether KyoT2 and Notch1 are mutually exclusive as to binding to RBP-J. COS-7 cell lysates containing the Myc-tagged RBP-J protein were mixed with increasing amounts of GST or KyoT proteins fused with GST. After 1 h of incubation at 4°C, a constant amount of GST-Notch1[1751–2170], which contains the RAM domain and the ankyrin repeats of Notch1, was added and incubation was continued for another hour at 4°C. The mixture was immunoprecipitated with the anti-Myc antibody and was analyzed by Western blotting with the anti-GST antibody and the anti-RBP-J monoclonal antibody T6709. In the presence of the same amount of RBP-J, preincubation with increasing amounts of GST-KyoT2 inhibited the binding of GST-Notch1[1751–2170] to RBP-J, while GST or GST-KyoT1 did not have any effect (Fig. 4C). These results indicate that KyoT2 and Notch1 share at least some of the RBP-J binding sites and compete with each other for binding to RBP-J.
KyoT2, but not KyoT1, represses transcription mediated by RBP-J.
We next wanted to determine the effect of KyoT2 on transcriptional activities of promoters containing the RBP-J binding sites. It is well established that RBP-J activates transcription from promoters containing the CGTGGGAA motif in association with EBNA2 or the intracellular region of Notch1, which we designate RAMIC. To test the effects of KyoT1 and KyoT2 on transcription, we used the reporter plasmid pGa981-6, which contains six copies of the EBNA2-responsive element of the TP-1 promoter upstream of the minimal promoter of the β-globin gene flanked by the luciferase reporter gene. Expression of EBNA2 or RAMIC with pGa981-6 led to 15- or several hundredfold activation of transcription, respectively, in collaboration with endogenous RBP-J in COS-7 cells (Fig. 5). To determine whether RBP-J-mediated transcriptional activation by EBNA2 or RAMIC can be repressed by KyoT2, a series of titration experiments were performed. Addition of KyoT2 but not KyoT1 repressed transcriptional activation by EBNA2 or RAMIC in a concentration-dependent manner (Fig. 5). The expression levels of RAMIC and EBNA2 proteins were not altered by coexpression of KyoT1 or KyoT2 (data not shown). These results demonstrate that KyoT2 antagonizes the transcriptional activation by EBNA2 or RAMIC.
FIG. 5.
KyoT2, but not KyoT1, represses RBP-J-mediated transcription. Results from a representative luciferase assay, in which constant amounts of EBNA2 (A) or RAMIC (B) plasmids were titrated in the presence of increasing amounts of KyoT plasmids are shown. The amounts of plasmids used (in micrograms) are indicated at the bottom. Each transfection was performed in duplicate. The average luciferase activity for the indicated transfections, relative to that for the reporter plasmid alone, is shown above each bar. Similar results were achieved repeatedly.
DISCUSSION
The LIM domain contains a cysteine-rich zinc-binding motif that is thought to be involved in protein-protein interaction (43). The LIM proteins are classified into two main groups by the presence or absence of a homeodomain (42). The former group binds to DNA via a homeodomain and modifies transcription, while the function of the latter entity, called the LIM-only protein, is largely unknown. In this study, we have isolated and characterized the LIM-only protein that interacts with a DNA-binding transcription factor, RBP-J.
This gene, named KyoT, encodes at least two transcripts (KyoT1 and KyoT2) that differ in their C termini. KyoT1 was identical to the murine counterpart of previously reported SLIM (skeletal muscle LIM protein), a LIM-only protein of unknown function expressed in the skeletal muscle (36). The analyses of genomic as well as complementary DNAs revealed that the two transcripts are generated by the alternative splicing mechanism. One of exons is deleted in the shorter transcript, KyoT2, introducing a premature termination codon in the last exon due to a frameshift. This results in replacement of the last two LIM domains present in KyoT1 with the putative RBP-J-binding domain. A similar mechanism to create different transcripts has been reported for several genes, such as those for mouse gonadotropin-releasing hormone receptor and interleukin-12 receptor, rat insulin-like growth factor-I, and human Fas, collagen IV, and alpha interferon receptor (1, 5, 9, 10, 17, 53). The resultant transcripts carry altered C-terminal sequences, giving rise to truncated forms with or without loss of transmembrane regions and glycosylation sites. The possible functional differences between alternatively spliced transcripts are suggested from their structural differences. The two transcripts of the KyoT gene encode proteins that are functionally distinct in terms of intracellular localization, interaction with RBP-J, and transcriptional regulation.
From the results of the two-hybrid interaction assay, we postulate that the RBP-J-interacting domain is located in the C-terminal 27 residues of KyoT2, although we are not sure whether other parts of the protein are also necessary for the interaction with RBP-J. Indeed, KyoT1, which lacks the 27-residue RBP-J-binding domain, also binds weakly to RBP-J in the two-hybrid interaction assay and in vitro binding assays with the GST fusion proteins. The C-terminal portion of the protein contains two tryptophans (WW). This doublet in EBNA2 has been shown to be indispensable for the interaction with RBP-J (32). The comparison of amino acids around WW in EBNA2 and KyoT2 revealed three conserved prolines, including one next to the second tryptophan (Fig. 4B). The related sequence WXP is found in the RBP-J-binding region of the Notch family members of all species identified so far (30, 47). This motif may be necessary for the interaction with the central portion of the RBP-J protein.
We have provided evidence for strong interaction between RBP-J and KyoT2 by use of the yeast two-hybrid interaction assay, GST pull-down assay, EMSA, and immunoprecipitation. It was demonstrated by EMSA that KyoT2 can form a complex with DNA-bound RBP-J, but the DNA-binding affinity of the KyoT2–RBP-J complex is greatly weakened and it exists mostly dissociated from DNA. This is in contrast to the fact that Notch and EBNA2 bind to RBP-J without dislodging it from DNA. Notch and EBNA2 are instead tethered to the specific sequence of DNA via the RBP-J protein and activate transcription. In contrast, the Hairless protein has been shown to bind to RBP-J and to inhibit its binding to DNA as does KyoT2. It seems that the affinity of the Hairless–RBP-J complex to DNA is even lower than that of the KyoT2–RBP-J complex, because there is no visible shifted band in EMSA when Hairless is bound with RBP-J (8). KyoT2 was shown to interact with RBP-J, and the complex was effectively immunoprecipitated from COS-7 cell extracts. It is notable that a much smaller fraction of the KyoT2 protein was precipitated with RBP-J compared with the fraction of the RBP-J protein precipitated with KyoT2 (Fig. 3C). This is probably due to the presence of an excess amount of KyoT2 compared with RBP-J in transfected COS-7 cells. We failed to detect any interaction between KyoT1 and RBP-J in EMSA and immunoprecipitation with COS-7 cells. Since LIM domains have been shown to provide the surface for protein-protein interaction, KyoT1, composed mainly of LIM domains, may have another major binding protein yet to be identified.
Both EBNA2 and RAMIC are transcriptional activators known to bind RBP-J to be targeted to specific gene promoters. KyoT2, but not KyoT1, repressed the transcriptional activation of EBNA2 or RAMIC when coexpressed in COS-7 cells. We showed that KyoT2 does not bind directly to the DNA sequences used in the luciferase reporter assays but forms a complex with DNA by associating with the central portion of mouse RBP-J (residues 141 to 371). The LIM-only proteins reported so far do not bind to DNA directly but may affect transcription by binding to other DNA-binding proteins (41, 50). Rhombotin, the oncogene product which accounts for some of acute T-cell lymphoblastic leukemia, is one such example. It binds to TAL1, a basic helix-loop-helix DNA-binding protein, and upregulates the level of transcription through its activation domain, leading eventually to oncogenesis of hematopoietic cells. KyoT2 offers another such example, as it negatively regulates transcription by interacting with the RBP-J DNA-binding protein. The absence of KyoT mRNA expression in lymph nodes or B-cell lines may be important for EBV to transform B cells. Otherwise, EBNA2 might have been unable to activate genes required for the transformation of B cells.
Downregulation of transcription can be brought about by repressor molecules acting in various ways (13, 25). Hairless is known to antagonize the function of Su(H) in vivo and in vitro in Drosophila by inhibiting its DNA binding (7, 8, 44). KyoT2 mostly disrupted the DNA binding of RBP-J as Hairless does, but it displayed no sequence similarity to Hairless. KyoT2 interacts with the central portion of RBP-J, whereas Hairless interacts at the C terminus of RBP-J. It has been shown that the binding surfaces of RBP-J for EBNA2 and RAM23 also are in this central region of RBP-J (40a). From the results of competition experiments, it seems that RBP-J binds only Notch1 or KyoT2 individually, indicating that KyoT2 counteracts Notch1 by competing for binding to RBP-J. Although we did not examine the mechanisms of transcriptional repression involving EBNA2, it is tempting to think that KyoT2 also competes with EBNA2 for binding to RBP-J, because EBNA2 is similar to Notch in many aspects of RBP-J-binding and _trans_activation. Taken together, these results indicate that KyoT2 has a dual role in counteracting Notch and probably EBNA2, i.e., dissociation of RBP-J from DNA and competition for binding to RBP-J. No mammalian homolog of Hairless has been identified so far, although other molecules involved in Notch signaling are conserved among vertebrates and Drosophila. KyoT2 may substitute for the function of Hairless in mammals.
ACKNOWLEDGMENTS
We gratefully acknowledge S. Hollenberg and R. Brent for providing reagents to carry out the two-hybrid screening.
This investigation was supported by grants for the COE program from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Abramovich C, Ratovitski E, Lundgren E, Revel M. Identification of mRNAs encoding two different soluble forms of the human interferon α-receptor. FEBS Lett. 1994;338:295–300. doi: 10.1016/0014-5793(94)80287-4. [DOI] [PubMed] [Google Scholar]
- 2.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 3.Amakawa R, Jing W, Ozawa K, Matsunami N, Hamaguchi Y, Matsuda F, Kawaichi M, Honjo T. Human Jκ recombination signal binding protein gene (IGKJRB): comparison with its mouse homologue. Genomics. 1993;17:306–315. doi: 10.1006/geno.1993.1326. [DOI] [PubMed] [Google Scholar]
- 4.Artavanis Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 5.Bach M A, Roberts C T, Jr, Smith E P, LeRoith D. Alternative splicing produces messenger RNAs encoding insulin-like growth factor-I prohormones that are differentially glycosylated in vitro. Mol Endocrinol. 1990;4:899–904. doi: 10.1210/mend-4-6-899. [DOI] [PubMed] [Google Scholar]
- 6.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 7.Bang A G, Posakony J W. The Drosophila gene Hairless encodes a novel basic protein that controls alternative cell fates in adult sensory organ development. Genes Dev. 1992;6:1752–1769. doi: 10.1101/gad.6.9.1752. [DOI] [PubMed] [Google Scholar]
- 8.Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila suppressor of hairless and of its human homolog, KBF2/RBP-Jκ, by direct protein-protein interaction with Drosophila hairless. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- 9.Cascino I, Fiucci G, Papoff G, Ruberti G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J Immunol. 1995;154:2706–2713. [PubMed] [Google Scholar]
- 10.Chua A O, Wilkinson V L, Presky D H, Gubler U. Cloning and characterization of a mouse IL-12 receptor-β component. J Immunol. 1995;155:4286–4294. [PubMed] [Google Scholar]
- 11.Chung C N, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-Jκ. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlon R A, Reaume A G, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 13.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 14.Dawid I B, Toyama R, Taira M. LIM domain proteins. C R Acad Sci. 1995;318:295–306. [PubMed] [Google Scholar]
- 15.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 16.Dou S, Zeng X, Cortes P, Erdjument Bromage H, Tempst P, Honjo T, Vales L D. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng L, Xia Y, Wilson C B. Alternative splicing of the NC1 domain of the human α3(IV) collagen gene. Differential expression of mRNA transcripts that predict three protein variants with distinct carboxyl regions. J Biol Chem. 1994;269:2342–2348. [PubMed] [Google Scholar]
- 18.Fortini M E, Artavanis Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa T, Kawaichi M, Matsunami N, Ryo H, Nishida Y, Honjo T. The Drosophila RBP-Jκ gene encodes the binding protein for the immunoglobulin Jκ recombination signal sequence. J Biol Chem. 1991;266:23334–23340. [PubMed] [Google Scholar]
- 20.Furukawa T, Kobayakawa Y, Tamura K, Kimura K, Kawaichi M, Tanimura T, Honjo T. Suppressor of hairless, the Drosophila homologue of RBP-Jκ, transactivates the neurogenic gene E(spl)m8. Jpn J Genet. 1995;70:505–524. doi: 10.1266/jjg.70.505. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa T, Maruyama S, Kawaichi M, Honjo T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell. 1992;69:1191–1197. doi: 10.1016/0092-8674(92)90640-x. [DOI] [PubMed] [Google Scholar]
- 22.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 24.Hamaguchi Y, Yamamoto Y, Iwanari H, Maruyama S, Furukawa T, Matsunami N, Honjo T. Biochemical and immunological characterization of the DNA binding protein (RBP-Jκ) to mouse Jκ recombination signal sequence. J Biochem. 1992;112:314–320. doi: 10.1093/oxfordjournals.jbchem.a123898. [DOI] [PubMed] [Google Scholar]
- 25.Hanna Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 26.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 27.Honjo T. The shortest path from the surface to the nucleus: RBP-Jκ/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 29.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 30.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 31.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 32.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJκ. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the Jκ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature. 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 34.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, Zimber Strobl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan M J, Madgwick A J. Slim defines a novel family of LIM-proteins expressed in skeletal muscle. Biochem Biophys Res Commun. 1996;225:632–638. doi: 10.1006/bbrc.1996.1222. [DOI] [PubMed] [Google Scholar]
- 37.Oka C, Nakano T, Wakeham A, de la Pompa J L, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak T W, Honjo T. Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 38.Roeder R G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Sakai T, Furukawa T, Iwanari H, Oka C, Nakano T, Kawaichi M, Honjo T. Loss of immunostaining of the RBP-Jκ transcription factor upon F9 cell differentiation induced by retinoic acid. J Biochem. 1995;118:621–628. doi: 10.1093/oxfordjournals.jbchem.a124955. [DOI] [PubMed] [Google Scholar]
- 40a.Sakai, T., and T. Honjo. Unpublished data.
- 41.Sanchez Garcia I, Axelson H, Rabbitts T H. Functional diversity of LIM proteins: amino-terminal activation domains in the oncogenic proteins RBTN1 and RBTN2. Oncogene. 1995;10:1301–1306. [PubMed] [Google Scholar]
- 42.Sanchez Garcia I, Rabbitts T H. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 43.Schmeichel K L, Beckerle M C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 44.Schweisguth F, Posakony J W. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- 45.Schweisguth F, Posakony J W. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 46.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 48.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-Jκ. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadman I, Li J, Bash R O, Forster A, Osada H, Rabbitts T H, Baer R. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F, Kikutani H, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou W, Sealfon S C. Structure of the mouse gonadotropin-releasing hormone receptor gene: variant transcripts generated by alternative processing. DNA Cell Biol. 1994;13:605–614. doi: 10.1089/dna.1994.13.605. [DOI] [PubMed] [Google Scholar]
- 54.Zimber Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimber Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]