On the sequencing of the human genome (original) (raw)
Abstract
Two recent papers using different approaches reported draft sequences of the human genome. The international Human Genome Project (HGP) used the hierarchical shotgun approach, whereas Celera Genomics adopted the whole-genome shotgun (WGS) approach. Here, we analyze whether the latter paper provides a meaningful test of the WGS approach on a mammalian genome. In the Celera paper, the authors did not analyze their own WGS data. Instead, they decomposed the HGP's assembled sequence into a “perfect tiling path”, combined it with their WGS data, and assembled the merged data set. To study the implications of this approach, we perform computational analysis and find that a perfect tiling path with 2-fold coverage is sufficient to recover virtually the entirety of a genome assembly. We also examine the manner in which the assembly was anchored to the human genome and conclude that the process primarily depended on the HGP's sequence-tagged site maps, BAC maps, and clone-based sequences. Our analysis indicates that the Celera paper provides neither a meaningful test of the WGS approach nor an independent sequence of the human genome. Our analysis does not imply that a WGS approach could not be successfully applied to assemble a draft sequence of a large mammalian genome, but merely that the Celera paper does not provide such evidence.
Two scientific papers (1, 2) recently appeared reporting “draft” sequences of the human genome. One was the product of the international Human Genome Project (HGP), and the other was the product of the biotechnology firm Celera Genomics. The two groups set out by using different methodologies, and each collected independent data sets.
In principle, the availability of two papers on the human genome has much potential scientific benefit. In addition to the comparison of two independently derived genome sequences, it should also allow methodological analysis of the sequencing strategies used for insights concerning the design of future genome-sequencing efforts.
Here, we focus on the methodological issues of genome sequence assembly. In general, genomic sequencing projects employ the same basic technique of shotgun sequencing developed by Sanger and others shortly after the invention of DNA sequencing around 1980 (e.g., see ref. 3). To determine the sequence of a large DNA molecule, the method begins by breaking up the DNA into smaller random overlapping fragments, obtaining sequence “reads” from these fragments, then using computer analysis to reassemble the random reads into “contigs”. Because of cloning biases and systematic failures in the sequencing chemistry, the random data alone are usually insufficient to yield a complete, accurate sequence. Instead, it is usually more cost-effective to supplement the random data with the collection of sequence data directed to close the gaps and solve remaining problems. The technique has been refined over the ensuing two decades. The initial version, for example, involved sequencing from one end of each fragment. Ansorge and others (4) extended this approach in 1990 to include the sequencing of both ends (paired-end shotgun sequencing), thereby obtaining linking information that could be used to connect contigs separated by gaps into “scaffolds”.
The shotgun sequencing technique can be directly applied to genomes with relatively few repeat sequences. The assembly problem is straightforward, because reads with overlapping sequence can typically be merged together without risk of misassembly. The relatively few gaps and problems can be solved to produce complete sequences. The approach has been applied successfully to produce complete sequences of simple genomes such as plasmids, viruses, organelles, and bacteria. Whole-genome shotgun data alone also has been applied with an almost 15-fold redundancy (5) to produce a draft sequence of the euchromatic portion of the Drosophila genome (3% repeat content), although a clone-based strategy is being applied to convert this to a finished sequence.
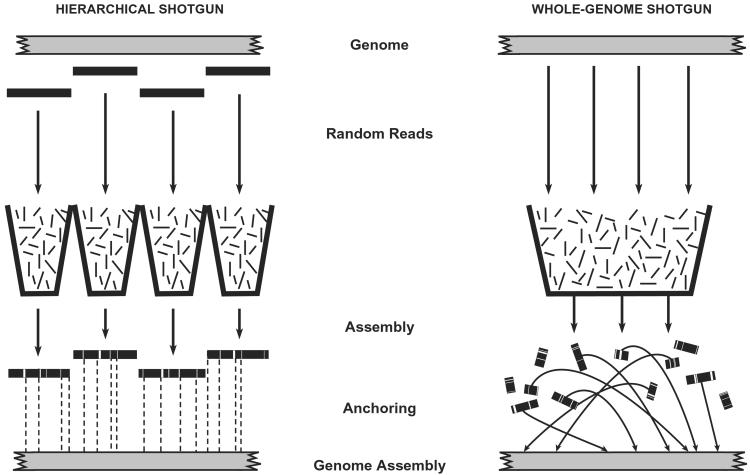
A greater challenge arises in tackling complex genomes with a large proportion of repeat sequences that can give rise to misassembly. Two alternative approaches (Fig. 1) can be taken.
Figure 1.
Sequencing strategies. (Left) The hierarchical shotgun (HS) strategy involves decomposing the genome into a tiling path of overlapping BAC clones, performing shotgun sequencing on and reassembling each BAC, and then merging the sequences of adjacent clones. The method has the advantage that all sequence contigs and scaffolds derived from a BAC belong to a single compartment with respect to anchoring to the genome. (Right) Whole-genome shotgun (WGS) strategy involves performing shotgun sequencing on the entire genome and attempting to reassemble the entire collection. With the WGS method, each contig and scaffold is an independent component that must be anchored to the genome. In general, many scaffolds may not be anchored without directed efforts. (Contigs are contiguous blocks of sequence; scaffolds are sets of contigs joined by paired reads from both ends of a plasmid insert.)
Hierarchical shotgun (HS) assembly.
In this approach, the genome is first broken up into an overlapping collection of intermediate clones such as bacterial artificial chromosomes (BACs). The sequence of each BAC is determined by shotgun sequencing, and the sequence of the genome is obtained by merging the sequences of the BACs. The HS approach provides a guaranteed route for producing an accurate finished genome sequence, because the sequence assembly is local and anchored to the genome. But it requires some additional preliminary work, including selecting overlapping BACs and preparing shotgun libraries from each BAC.
Whole-genome shotgun (WGS) assembly.
In this approach, the genome is decomposed directly into individual random reads. One then attempts to assemble the genome as a whole. The WGS approach avoids the preliminary work but has potential disadvantages: there is a greater risk of long-range misassembly. The resulting sequence components must be individually anchored to the genome, and the resulting assembly may be difficult to convert to a finished sequence.
Whether to tackle the sequencing of the human genome with the HS or WGS approach was extensively debated in the scientific literature in 1996 and 1997 (6, 7). There was no doubt that the WGS approach could yield a large amount of the human sequence, but there was serious concern that the ultimate cost of producing a finished human reference sequence would be much greater. In fact, the potential cost savings in producing a draft sequence was unclear, because map construction and library production account for a minor fraction (<10%) of the total sequencing costs. For these reasons, the HGP elected to use the HS approach.
In 1998, Celera Genomics was formed with the goal of applying the alternative WGS approach to the human genome. Differing opinions were expressed concerning the likely product. Venter (8) projected that the WGS approach would suffice to assemble the entire genome in a small number of pieces. He estimated that an assembly based on 10-fold coverage would have fewer than 5,000 contigs separated by gaps of ≈60 bp. Such an assembly would include the nucleotide sequence of 99.99% of the human euchromatic genome and would consist of a small number of components that could then be positioned, or anchored, in the genome. Relatively little work would be required to produce a finished sequence. Olson (9) foresaw a different outcome, projecting there would be more than 100,000 components. That number of components would be far too many to anchor in the genome by using sequence-tagged sites (STS) as landmarks, resulting in a significant portion of the genome being unanchored. Moreover, he argued that it would be difficult or impossible to use such results as a foundation for producing a finished sequence.
With the recent publications on the human genome sequence (1, 2), it is possible to examine the reported results for insight concerning sequencing methodology. In particular, the Celera authors reported that their paper constituted a successful application of the WGS approach to a mammalian genome and that they had provided a genome sequence based primarily on their own data. Here, we examine the validity of these conclusions.
Analysis and Results
HGP Data and HS Assembly.
The HGP strategy is based on the sequencing of overlapping BACs (≈170 kb) with known locations in the human genome. BACs are subjected to increasing levels of sequence coverage and completion: draft at ≈5-fold coverage, deep shotgun at ≈10-fold coverage, and finished resulting from directed gap closure.
At the time of publication (1), the BACs sequenced to at least draft status covered ≈94% of the euchromatic genome. The merged sequence itself had fairly large contigs (half the sequence resided in contigs of >80 kb) and represented ≈90% of the euchromatic portion of the human genome, roughly equally divided among draft, full shotgun, and finished status. The total sequence coverage was 7.5-fold.
Celera Data and WGS Assembly.
The Celera strategy was based on assembling the genome from random sequences generated from both ends of whole-genome plasmid libraries with 2-, 10-, and 50-kb inserts. The authors generated a total sequence coverage of 5.1-fold.
The Celera authors presented no genome assembly based on their own WGS data (2). Thus, their paper provides neither a direct experimental test of the WGS method nor a direct assessment of the Celera data.
Joint Assemblies.
The Celera paper presented only joint analyses based on a combined data set including both the HGP data (which had been made available on the world wide web, in keeping with the HGP's policy of free and immediate data release) and Celera's own data. The paper reported two joint analyses: a “faux” WGS assembly and a compartmentalized sequence assembly (CSA).
The methods are discussed below, and their output is summarized in Table 1. Notably, the joint assemblies do not contain dramatically more total sequence than the HGP assemblies that were used as input. Both the HGP assembly and the joint assembly based on the HGP and Celera data contain ≈90% of the human euchromatic genome. To be sure, the joint assembly contains some additional sequence (estimated to be a few percent) and adds additional ordering information. But the differences in the results using the combined data sets and the HGP data alone are relatively slight.
Table 1.
Reported statistics for genome assemblies in the HGP and Celera papers
| Category | Celera | HGP | ||
|---|---|---|---|---|
| WGS | Faux WGS | Faux CSA | ||
| Sequence coverage | 5.1 × Celera | 5.1 × Celera | 5.1 × Celera | 7.5 × HGP |
| + 7.5 × HGP | + 7.5 × HGP | |||
| 12.6 × total | 12.6 × total | |||
| Length (in Gb) of draft genome assembly, counting only bases with known sequence* | NR | 2.587 | 2.654 | 2.693 |
| Length (in Gb) of draft genome assembly, including unknown nucleotides in gaps† | NR | 2.848 | 2.906 | 2.916 |
| Proportion of sequence in euchromatic genome present in draft genome assembly, %‡ | NR | 89 | 91 | 92 |
| Number of contigs§ | NR | 221,036 | 170,033 | 149,821 |
| Number of scaffolds§ | NR | 118,968 | 53,591 | 87,757 |
| Number of components, to be anchored in genome¶‖ | NR | 118,968 | 3,845 | 942 |
Faux WGS Reads.
The manner in which the joint assemblies used the HGP data are noteworthy. The Celera authors stated that they combined 2.9-fold coverage from the HGP with 5.1-fold coverage from Celera. However, a close reading of the paper shows that the 2.9-fold coverage was derived in an unusual manner that is very unlike shotgun data and implicitly preserved much of the HGP assembly information.
The authors “shredded” the HGP's assembled sequence data into simulated reads of 550 bp, which they termed “faux reads”. Each BAC was shredded to yield 2-fold coverage; given the overlaps between BAC clones, this yielded a total of 2.9-fold coverage. The authors then fed these faux reads into their assembly program, together with their own WGS reads. The stated purpose of shredding the HGP data was to break any misassemblies; this goal is a reasonable one. However, the shredding was done in such a manner that the resulting assembly bore no relation to a WGS assembly. Specifically, the faux reads were not a random 2-fold sampling, but instead comprised perfect 2× coverage across the assembled HGP contigs. In other words, the faux reads were perfectly spaced, with each overlapping the next by half its length, thereby completely avoiding the problems of gaps, small overlaps, and errors that arise in realistic data (Fig. 2).¶
Figure 2.
Random vs. perfect spacing in 100 kb. (Upper) Two-fold coverage in perfectly spaced reads. (Lower) Two-fold coverage in randomly selected reads. There are frequent regions in which the adjacent reads either fail to overlap or the overlap is too small to allow reliable detection (< 40 bp). These breaks in continuity are indicated by vertical lines.
Faux WGS Assembly.
The first joint assembly (faux WGS) was modeled on the WGS approach in the sense that the sequence reads (both actual and faux) were fed into a genome assembly program without additional information. In particular, the genome assembly program did not have explicit information about the overlaps or location of the faux reads in the HGP sequence contigs.
We were curious, however, whether the HGP assembly information was implicitly preserved through use of a perfect 2-fold tiling path. We tested this hypothesis by creating simulated data sets from the finished sequence of human chromosome 22 and performing genome assemblies by using a WGS assembly program, ARACHNE (10, 11).
- Set A (2× perfect tiling path) consisted of a perfect tiling path of 550-bp reads, each overlapping the next by 275 bp, across the finished sequence.
- Set B (2× random coverage) consisted of randomly chosen reads of 550 bp, providing a total of 2-fold coverage.
- Set C (5× random coverage) consisted of randomly chosen reads of 550 bp, providing a total of 5-fold coverage.
We first compared the results of assembling sets A and B (Table 2). The perfect 2× tiling path (set A) yields an assembly covering essentially the entire chromosome in huge sequence contigs. More than 99% of the sequence lies in contigs >10 kb, and more than 43% in contigs >500 kb. The N50 contig length (the length L such that 50% of the sequence lies in contigs of at least L) is 421 kb. The huge size of the contigs is not surprising, because the underlying data have no true gaps, and the reads have large overlaps that are readily detected by a shotgun assembly algorithm. By contrast, the random 2× coverage (set B) yields tiny contigs. The N50 contig length is <2 kb for the random coverage compared with 421 kb for the perfect tiling path. This result is also not surprising, because the random 2-fold coverage necessarily leaves many true gaps and small overlaps (12). Thus, shotgun assembly programs cannot assemble long contigs from such data.
Table 2.
Implications on assembly using perfect tiling paths of faux data based on simulated data from finished sequence from human chromosome 22*
| 2× perfect tiling path | 2× random coverage | 5× random coverage | |
|---|---|---|---|
| N50 contig length† (kb) | 421 kb | <2 kb | 11 kb |
| Sequence in contigs > 5 kb, % | >99 | 6.4 | 80 |
| Sequence in contigs > 10 kb, % | 99 | 0.2 | 55 |
| Sequence in contigs > 20 kb, % | 98 | ‡ | 20 |
| Sequence in contigs > 50 kb, % | 94 | ‡ | 0.5 |
| Sequence in contigs > 100 kb, % | 87 | ‡ | ‡ |
| Sequence in contigs > 500 kb, % | 43 | ‡ | ‡ |
| Sequence in contigs > 1000 kb, % | 29 | ‡ | ‡ |
We next compared the results for sets A and C (Table 2). The 2× perfect tiling path yields dramatically larger contigs than 5× random shotgun data. The N50 length is almost 40-fold larger for the perfect tiling path than for the 5× random data (421 vs. 11 kb).
These results show that a fundamental limitation in genome sequence assembly is the random nature of the data. A perfect tiling path avoids this issue and implicitly preserves the underlying assembly information.
The results have several clear implications for the faux WGS assembly. First, dissection of the HGP contigs into a 2× perfect tiling path suffices to allow their nearly complete reconstruction (even up to hundreds of kilobases) without the explicit need for positional information. Second, a 2× perfect tiling path contains much more inherent assembly information than 5× random coverage. It is not surprising, then, that the combined assembly quite faithfully (although not completely) reproduced the finished sequence of chromosome 22.
Thus, for the portions of the genome where sequence was available from the HGP, it is impossible to learn from the paper the respective contributions of the two data sets. On the one hand, the WGS reads can fill gaps, link contigs, and correct misassemblies. On the other hand, sequence from the BAC contigs was used directly in a final assembly step to fill gaps (in a process the authors referred to as “external gap walking”). In any case, the fact is that there is very little difference in the coverage and continuity of the faux WGS assembly using the two data sets and the HGP genome sequence.
Anchoring the Faux WGS Assembly.
Although the faux WGS assembly is not a meaningful test of a true WGS assembly, it is nonetheless worth examining for insight about other issues pertinent to genome assembly. In particular, a key measure of the faux WGS assembly is the proportion of the genome sequence contained in large, anchored components. Although all sequence produced by the HS strategy can be localized to a small region of the genome (namely, a mapped BAC clone), the sequence components produced by the WGS assembly are initially free-floating islands that do not contribute to a genome assembly until they have been anchored. In the current case, the Celera authors aimed to anchor their sequence components by using the HGP's STS maps.
The faux WGS assembly yielded 221,036 contigs linked together into 118,986 scaffolds (sets of contigs joined by paired reads from both ends of a plasmid insert) and contained a total of 88.6% of the euchromatic genome (Table 1). If one focuses only on scaffolds larger than 30 kb, their number is small enough to allow them to be anchored. But these scaffolds contain only ≈2.334 × 109 bases (see table 3 in ref. 2) or ≈79.9% of the euchromatic genome, leaving ≈20% of the euchromatic genome unanchored.
To anchor more of the genome, the full set of nearly 119,000 scaffolds must be used. But this quantity vastly exceeds the number that can be anchored by using existing STS maps. Localizing so many scaffolds would require a large directed mapping effort, which could only commence after the assembly was completed.
Moreover, these 119,000 scaffolds still contain only 88.6% of the euchromatic genome. The remaining 11.4% lies in the 25% of the reads that remain unassembled or is missing entirely from the WGS coverage. Clearly, it is not feasible to anchor this portion of the genome.
Compartmentalized Sequence Assembly.
The second joint assembly (CSA) used a local clone-based rather than a genome-wide WGS approach. In fact, the CSA was conceptually identical to the HGP's HS approach, and it explicitly used all of the HGP's clone-based sequence and map data.
Briefly, the CSA analysis began by assigning Celera's WGS reads to individual HGP BAC clones (by matching their sequences to the assembled sequence contigs) then to overlapping sets of HGP BACs, dubbed “compartments” (there was a total of 3,845 HGP compartments). The CSA then performed separate local sequence assembly on each of the compartments (using both the Celera WGS reads and faux HGP reads corresponding to the compartment). The number of compartments was small enough that nearly all could be readily anchored to the chromosomes by using the available HGP map resources.
The construction of compartments closely mirrored the HGP's construction of clone contigs and used the HGP clone sequences, STS content, and fingerprint clone maps.‖ The local sequence assembly was similarly straightforward. The compartments had an average size of ≈760 kb (only a few times larger than a BAC clone) and were readily assembled by using standard computer programs such as PHRAP (by P. Green, available at http://www.phrap.org/). The resulting scaffolds were anchored and then further ordered and oriented by using the HGP's STS content and fingerprint clone maps.
The CSA thus provides a revised version of the HGP assembly based on the addition of WGS reads to the individual clone contigs. All of the biological analyses reported in the Celera paper were based on the CSA sequence.
Discussion
Here, our primary purpose is to examine whether the recently published paper from Celera Genomics (2) provides insight into the performance of the WGS approach on mammalian genomes. Our analysis indicates that it is not possible to draw meaningful conclusions about the WGS approach because the authors did not perform an analysis of their own data by itself. Instead, they used an unorthodox approach to incorporate simulated data from the HGP. In particular, the paper presented only joint assemblies of Celera's 5.1-fold WGS data together with a perfect tiling path of faux reads that implicitly retained the full information inherent in the HGP's 7.5-fold coverage. Furthermore, the joint assemblies were anchored to the genome by using the HGP's clone and marker maps.
We should emphasize that our analysis does not imply that a WGS approach cannot, in principle, produce a valuable draft sequence of a mammalian genome. To the contrary, we are optimistic that this approach will be possible with improved computational algorithms. Indeed, we expect that the WGS approach can play a useful role in obtaining a draft sequence from various organisms, including the mouse (13). For the mouse, with 6-fold WGS coverage (13) and the use of improved algorithms (11), it has yielded initial assemblies with N50 scaffold lengths exceeding 1 Mb and N50 contig lengths of 16 kb. Conceivably, such improved algorithms could yield similar results from human WGS data.
Whether the WGS approach alone can provide an efficient technique for producing a finished genome sequence, however, remains an open question. To obtain the complete sequence of the mouse genome (13), the WGS data are being used in conjunction with clone-based data. The WGS assembly has been anchored to an extensive BAC-based physical map by matching the sequence contigs to BAC-end sequences, thereby localizing numerous small contigs and providing longer range continuity. Moreover, the BACs serve as substrates for producing a finished sequence, as is being done in the human and fly. Current experience suggests that clone-based sequencing will remain an essential aspect of producing finished sequence from large, complex genomes.
Although the Celera paper leaves open many methodological issues, it does demonstrate one of the HGP's core tenets, the value of making data freely accessible before publication. As the analysis above shows, the availability of the HGP data contributed to Celera's ability to assemble and publish a human genome sequence in a timely fashion. When speed truly matters, openness is the answer.
Abbreviations
HGP
Human Genome Project
HS
hierarchical shotgun
BACs
bacterial artificial chromosomes
WGS
whole-genome shotgun
STS
sequence-tagged sites
CSA
compartmentalized sequence assembly
Footnotes
¶
See ref. 2. The nature of “shredding” is mentioned in passing on p. 1309, column 3, paragraph 4; the implications for assembly are not discussed in the paper.
‖
See ref. 2, p. 1313, column 2, paragraph 3 and p. 1314, column 2, paragraph 2.
References
- 1.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 2.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 4.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmermann J, Erfle H, Caskey C T, Ansorge W. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 5.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 6.Weber J L, Myers E W. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- 7.Green P. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- 8.Marshall E, Pennisi E. Science. 1998;280:994–995. doi: 10.1126/science.280.5366.994. [DOI] [PubMed] [Google Scholar]
- 9.Marshall E. Science. 1999;284:1906–1909. doi: 10.1126/science.284.5422.1906. [DOI] [PubMed] [Google Scholar]
- 10.Batzoglou S. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 2000. [Google Scholar]
- 11.Batzoglou S, Jaffe D B, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov J, Lander E S. Genome Res. 2001;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander E S, Waterman M S. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 13.Mouse Genome Sequencing Consortium. Genesis. 2001;31:137–141. doi: 10.1002/gene.10016. [DOI] [PubMed] [Google Scholar]