Niemann-Pick C Variant Detection by Altered Sphingolipid Trafficking and Correlation with Mutations within a Specific Domain of NPC1 (original) (raw)
Abstract
Niemann-Pick disease type C (NPC) is a fatal, autosomal recessive lipidosis characterized by lysosomal accumulation of unesterified cholesterol and multiple neurological symptoms, such as vertical supranuclear ophthalmoplegia, progressive ataxia, and dementia. More than 90% of cases of NPC are due to a defect in Niemann-Pick C1 (NPC1), a late endosomal, integral membrane protein that plays a role in cholesterol transport or homeostasis. Biochemical diagnosis of NPC has relied on the use of patient skin fibroblasts in an assay to demonstrate delayed low-density lipoprotein (LDL)–derived cholesterol esterification and a cytological technique—filipin staining—to demonstrate the intracellular accumulation of cholesterol. A small percentage of patients, referred to as “NPC variants,” present with clinical symptoms of NPC but show near-normal results of these biochemical tests, making laboratory confirmation of NPC disease problematic. Here, we demonstrate that NPC-variant fibroblast samples can be detected as sphingolipid storage disease cells, using a fluorescent sphingolipid analog, BODIPY-lactosylceramide. This lipid accumulated in endosomes/lysosomes in variant cells preincubated with LDL cholesterol but targeted to the Golgi complex in normal cells under these conditions. The reproducibility of this technique was validated in a blinded study. In addition, we performed mutation analysis of the NPC1 gene in NPC variant and “classical” NPC cell samples and found a high incidence of specific mutations within the cysteine-rich region of NPC1 in variants. We also found that 5 of the 12 variant cell samples had no apparent defect in NPC1 but were otherwise indistinguishable from other variant cells. This is a surprising result, since, in general, ∼90% of patients with NPC possess defects in NPC1. Our findings should be useful for the detection of NPC variants and also may provide significant new insight regarding NPC1 genotype/phenotype correlations.
Introduction
Niemann-Pick C (NPC [MIM 257220]) disease is a fatal, autosomal recessive, neurovisceral disorder with an estimated prevalence of ∼1:150,000 in Western Europe and a much higher prevalence in some geographically isolated populations (e.g., French Acadians of Nova Scotia and Hispanics of southern Colorado) (Winsor and Welch 1978; Carstea et al. 1997; Millat et al. 1999; Patterson et al. 2001). Biochemically, NPC is characterized by extensive lysosomal accumulation of unesterified cholesterol in many tissues, as well as lysosomal storage of sphingolipids in some tissues (e.g., liver and brain) (Vanier 1999; Patterson et al. 2001). Most individuals with NPC possess a defect in Niemann-Pick C1 (NPC1), a 1,278–amino acid, integral membrane protein which has been localized to late endosomes (Carstea et al. 1997; Neufeld et al. 1999). Although the function of the NPC1 protein is not fully understood, it is believed to be involved in the intracellular transport of low-density lipoprotein (LDL)–derived free cholesterol (Neufeld et al. 1999; Cruz et al. 2000; Lange et al. 2000). Fewer than 10% of patients with NPC are not defective in NPC1 but can be shown, by cell fusion and/or linkage studies, to fall into a second complementation group, referred to as “NPC2” (MIM 601015). NPC2 has recently been shown to be a result of mutations in HE1, a ubiquitously expressed, soluble lysosomal protein which binds cholesterol (Naureckiene et al. 2000). Patients with NPC1 and those with NPC2 are indistinguishable, both clinically and in terms of the standard biochemical tests for NPC (see below) (Vanier et al. 1996; Patterson et al. 2001).
NPC has protean clinical presentations, ranging from fetal ascites to slowly progressive neurodegeneration in adult life (reviewed by Patterson et al. [2001]). The nervous system, bone marrow, liver, and lungs may all be directly impaired by the disease process. Although the disease may present in any fashion along this temporal and phenotypic continuum, it is helpful for the clinician to distinguish some common patterns. The classic presentation is with variable hepatosplenomegaly and vertical supranuclear gaze palsy, ataxia, dystonia, and dementia in late childhood (“juvenile NPC”), culminating in death before age 20 years (Patterson et al. 2001). A second group (“infantile NPC”) presents with cholestasis and/or liver failure early in infancy, and patients die before age 2 years. Some patients present primarily with cognitive or psychiatric disturbances in adulthood (“adult-onset NPC”) with a slower progressive neurodegeneration. Patients with atypical presentation (i.e., those expressing only fragments of the classical phenotype) may be the most difficult to recognize (e.g., see Uc et al. 2000) but also may be the most amenable to potential therapies and may, therefore, benefit substantially from early, accurate diagnosis.
The standard biochemical assays for NPC utilize patient fibroblasts and consist of (i) a demonstration of impaired esterification of LDL-derived cholesterol and (ii) characteristic intense, punctate, cytoplasmic staining of cells with filipin, a fluorescent polyene antibiotic which binds to free cholesterol, to show the accumulation of free cholesterol in cells (Morris and Carstea 1998; Patterson et al. 2001). However, although >80% of NPC cases can be distinguished by cholesterol esterification rates of <10% of normal cells and obvious filipin staining, a minority of patients who present with classical clinical symptoms of NPC (e.g., supranuclear vertical gaze palsy, progressive neurological decline) exhibit a “variant” biochemical phenotype, with intermediate-to-normal cholesterol-esterification rates and less-definitive filipin staining (Vanier et al. 1991; Vanier et al. 1996). Thus, the diagnosis of NPC in these patients is difficult to confirm by existing methods, and the clinician must rely on a combination of clinical and ultrastructural data.
Throughout the length of the NPC1 protein in patients with NPC, >90 different deleterious mutations have been reported to occur, including point mutations and frameshifts leading to truncation of the protein (Carstea et al. 1997; Greer et al. 1999; Millat et al. 1999; Yamamoto et al. 1999; Ioannou 2000; Snow et al. 2000). The most commonly found NPC1 mutation, I1061T, has been shown to occur with relatively high frequency in patients of western European descent, and it generally correlates with a juvenile clinical phenotype with classical low cholesterol-esterification rates (Millat et al. 1999). A second mutation, G992W, is responsible for the Niemann Pick D (NPD) subtype of NPC disease in patients of common ancestry from southwestern Nova Scotia (Greer et al. 1998). Patients with NPD usually first present with clinical symptoms during the juvenile years, but their cholesterol-esterification rates are intermediate between those of classical NPC cells and normal fibroblasts (Sidhu et al. 1993; Jan and Camfield 1998). Other than these data, no significant correlations have been reported between the clinical presentation of the disease and either biochemical analyses or the occurrence of different NPC1 mutations among patients with NPC. In particular, nothing is known about whether the variant phenotype is dictated by specific NPC1 mutations or is due to offsetting effects of other aspects of the patient’s genetic background.
We previously described a broad screening assay for detection of multiple sphingolipid storage diseases (SLSDs), including NPC (Chen et al. 1999). This assay was based on the observation that a fluorescent sphingolipid analog (BODIPY-lactosylceramide [LacCer]) is transported to the Golgi apparatus in normal fibroblasts after endocytosis but is targeted to endosomes/lysosomes in cell samples from patients with an SLSD. This mistargeting of BODIPY-LacCer was found to be directly related to accumulation of cellular free cholesterol (Puri et al. 1999). Here, we have modified this technique so that it will detect NPC variants, in addition to patients with classical NPC. In addition, we performed mutational analysis of the NPC1 protein in variant and classical NPC samples and found that most mutations identified in variant samples were point mutations within the “cysteine-rich region” (Greer et al. 1999; Watari et al. 2000) of NPC1.
Patient Samples and Methods
Cells and Culture Conditions
All normal and one NPC (GM3123) human skin fibroblasts were obtained from the Coriell Institute for Medical Research. Other NPC human skin fibroblasts were from the NPC Cell Repository of the Division of Laboratory Genetics at the Mayo Clinic in Rochester, MN. Many of these samples were originally collected at the Developmental and Metabolic Neurology Branch, National Institute of Neurological Disorders and Stroke (NINDS). Patient samples were used anonymously and with consent for research purposes, under a protocol approved by the Mayo Clinic internal review board. NPC patient samples with cholesterol esterification rates >800 pmol/mg protein/6 h and nondefinitive filipin staining were defined as “variants.” Additional NPC samples with cholesterol esterification rates of <500 pmol/mg protein/6 h were defined as “classical” NPC samples. These definitions are based on those originally used by Vanier et al. (1991).
Cells were typically grown in Eagle’s minimum essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) (referred to hereafter as basal medium) as described (Martin and Pagano 1994). For some experiments, cells were incubated in medium supplemented with up to 150 μg/ml LDL (Sigma) for 24 h before incubation with BODIPY-LacCer or fixation for filipin and NPC1 immunostaining. All microscopy experiments were performed on cells grown to 30%–60% confluence on acid-etched 25-mm glass cover slips.
Esterification of LDL-Derived Cholesterol
Cholesterol esterification was performed essentially as described by Vanier et al. (1988). Briefly, cells seeded at a density of 5–10×105 cells in a 75 mm2 flask were maintained in McCoy’s 5A medium supplemented with 10% FBS and then were incubated at 37°C for 4 d in fresh medium supplemented with 10% lipoprotein-deficient serum (LPDS). Cholesterol esterification was initiated by the addition of fresh medium containing [3H]oleate (100 μmol/l in BSA solution), ±50 μg/ml of LDL. After 6 h, lipids were extracted and were separated by thin-layer chromatography, and the radioactivity in cholesterol ester was determined by scintillation counting.
Fluorescence Microscopy and BODIPY-LacCer Labeling
BODIPY-LacCer was custom synthesized as described (Martin and Pagano 1994). Cells were incubated with 5 μM BODIPY-LacCer/BSA for 45 min at 37°C in culture medium containing 1% FBS, were washed, were incubated in medium with 1% FBS for an additional 1 h at 37°C, and then were back exchanged with 5% BSA to remove cell-surface fluorescence (Martin and Pagano 1994; Chen et al. 1999). Fluorescence microscopy was performed using an Olympus IX70 microscope. Fluorescence micrographs for each experiment were exposed and printed identically and are representative of ⩾80% of the cells in a given experiment. Quantitation of BODIPY-LacCer in Cascade blue dextran-(Sigma) positive lysosomes of individual cells was determined by ratio imaging (Pagano et al. 2000). Filipin staining and NPC1 immunostaining of formaldehyde-fixed cells were performed as described elsewhere (Neufeld et al. 1999).
Cholesterol Challenge and Blinded Study
Normal, classical NPC, and NPC variant fibroblasts were grown on glass cover slips, as described above. The cells were incubated for 24 h with 0–150 μg/ml of human LDL and then were pulse-labeled with BODIPY-LacCer, as described above. For the blinded study, coded samples of normal, variant, and classical NPC cells preincubated in basal medium with or without LDL were pulse-labeled with BODIPY-LacCer. Observers were then asked to score, in a blinded study, paired coded samples (the same cell sample with or without LDL) as either normal (Golgi labeling) or NPC (punctate labeling), on the basis of LacCer targeting patterns observed by fluorescence microscopy.
Complementation Analysis
Complementation analysis was performed on cell lines to determine whether the underlying genetic defect was in the NPC1 gene or NPC2 gene. Testing was performed as previously described (Vanier et al. 1996), using filipin staining to assess whether biochemical correction was present in fused cells. Control NPC1 and NPC2 cell lines had previously been determined to be in these complementation groups by analysis performed at the Developmental and Metabolic Neurology Branch at NINDS (Vanier et al. 1996).
NPC1 Mutation Detection
Genomic DNA was isolated from fibroblast cell lines, using the Puregene extraction kit (Gentra Systems). DNA samples were screened for mutations in NPC1 by conformation-sensitive gel electrophoresis (CSGE) and DNA sequencing. CSGE was utilized for initial screening of NPC1 exons and adjacent intronic sequences (Korkko et al. 1998; Markoff et al. 1998). To increase efficiency of the CSGE analysis, multiplex PCR (with primers to three different exons per multiplex) was used to generate products for CSGE (Snow et al. 2000). Homozygous alterations were detected by mixing PCR products from cell lines with PCR products from a normal control prior to heteroduplex formation and electrophoresis. Of the 25 NPC1 exons, 4 (1, 12, 14, and 17), which could not be analyzed by CSGE because repetitive sequences within the PCR products caused the formation of complex banding patterns, were analyzed by DNA sequencing. DNA sequencing was also used to analyze all samples with novel CSGE banding patterns and other samples and exons as indicated in the text. Cycle sequencing of PCR products utilized ABI Prism BigDye Terminator chemistry (PE Biosystems), an ABI Prism 377 Sequencer (PE Biosystems), and SEQUENCHER software (Gene Codes) for data analysis.
Southern blot assays using genomic DNA digested with _Bgl_II and _Hin_dIII (in separate reactions) were used to test for large deletions in some samples that were in the NPC1 complementation group but did not have mutations detected by the preceding methods. Probes used in Southern blot hybridization were generated by PCR of each of the NPC1 exons and were labeled with [32P]-dCTP using the HighPrime random prime labeling kit (Roche Molecular Biochemicals).
Results
Cholesterol Esterification Rates in NPC Fibroblast Samples
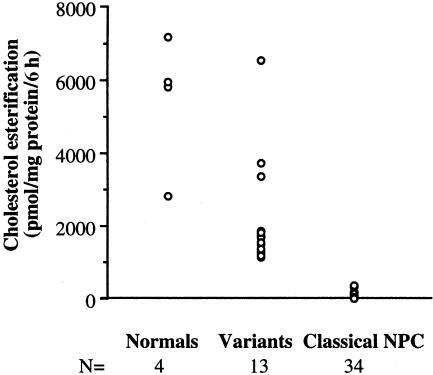
We used a subset of NPC patient fibroblast samples from the Mayo Clinic NPC cell repository. All samples were obtained from patients with clinical symptoms consistent with NPC disease. Among >200 fibroblast samples derived from patients with NPC, we identified 13 samples (referred to henceforth as “variants” and including one pair of siblings) with cholesterol esterification rates >800 pmol/mg of protein/6 h and nondefinitive filipin staining (see fig. 4_A_). An additional 34 samples, including three pairs of siblings, with cholesterol esterification rates of <500 pmol/mg of protein/6 h, were chosen from the cell repository as representative samples of classical NPC. A scattergram of cholesterol-esterification values of all variant, classical NPC, and normal fibroblasts used for our study is shown in fig. 1. This figure illustrates how cholesterol-esterification values of variants (range 1,161–6,561 pmol cholesterol ester/mg of protein/6 h; mean ± standard error [SE] = 2,235 ± 420) overlap with the range for normal cells (range 2,803–7,200; mean ± SE = 5,448 ± 934), making cholesterol-esterification assays insufficient for confirming a diagnosis of NPC in these patients.
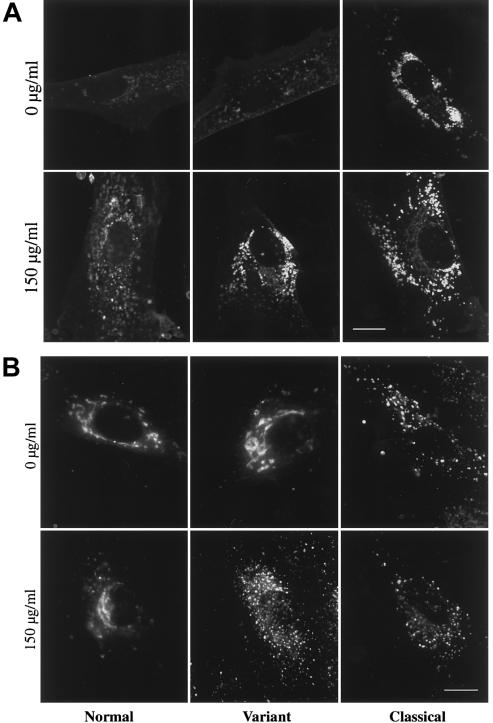
Figure 4.
LDL-challenge alteration of intracellular cholesterol distribution and BODIPY-LacCer transport in NPC variant fibroblasts. Normal, NPC variant, and classical NPC fibroblasts were incubated in culture medium ± 150 μg/ml of LDL for 24 h before filipin staining or incubation with BODIPY-LacCer. A, After fixation, cells were labeled with 50 μg/ml filipin (Neufeld et al. 1999), a polyene antibiotic which permeabilizes cells and binds to free cholesterol. Note that LDL treatment resulted in a greater alteration in intracellular cholesterol distribution in NPC variant cells than in normal cells. B, After treatment with and without LDL, cells were pulse labeled with BODIPY-LacCer, as in figure 2. LDL treatment shifted the BODIPY-LacCer targeting in variant cells from the Golgi apparatus to punctate structures but had little effect in normal or classical NPC cells. For each experiment, all micrographs were exposed and printed identically. Bar = 10 μm.
Figure 1.
LDL-derived cholesterol-esterification rates in the NPC subtypes. Esterification rates of LDL-derived cholesterol in normal, NPC variant, and classical NPC fibroblasts grown in lipoprotein-deficient serum were measured using [3H]oleate (see Patient Samples and Methods). Values are means of triplicate measurements of individual samples.
Detection of NPC Variants by a Sphingolipid-Targeting Assay
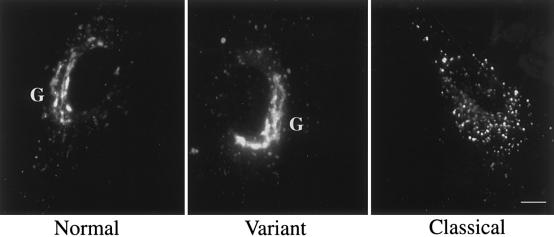
When we applied our original protocol for detection of SLSD cells, BODIPY-LacCer was targeted to the Golgi complex in normal human skin fibroblasts but was transported to endosome/lysosomes in classical NPC cell samples (fig. 2), which is consistent with our recent studies (Chen et al. 1999; Puri et al. 1999). All variant cell samples, however, displayed Golgi targeting of BODIPY-LacCer that was indistinguishable from that in normal cells (fig. 2 and data not shown).
Figure 2.
BODIPY-LacCer targeting in normal, variant, and classical NPC cells. Living NPC fibroblasts on glass cover slips were pulse-labeled with BODIPY-LacCer for 45 min at 37°C and were observed by fluorescence microscopy (see Patient Samples and Methods). Images are representative of >80% of cells observed for each experiment. Note that BODIPY-LacCer was targeted to the Golgi apparatus in both normal and NPC variant cells but was concentrated in punctate structures in classical NPC cells. Bar = 10 μm. G = Golgi complex.
We recently found that altered BODIPY-LacCer trafficking in SLSD cells is caused by elevated free cholesterol in these cells (Puri et al. 1999). The lack of lysosomal accumulation of BODIPY-LacCer in variant cells suggests that, under basal cell culture conditions (growth in media with 10% fetal bovine serum [FBS]), cholesterol levels in variants are similar to those in normal fibroblasts. Indeed, filipin staining in NPC variants is often similar to that in normal cells (Vanier et al. 1991; Patterson et al. 2001). We hypothesized (i) that NPC variant cells challenged by growth medium with excess LDL cholesterol may exhibit altered distribution or elevated levels of cholesterol compared with those in normal cells and (ii) that this would be reflected in altered BODIPY-LacCer targeting.
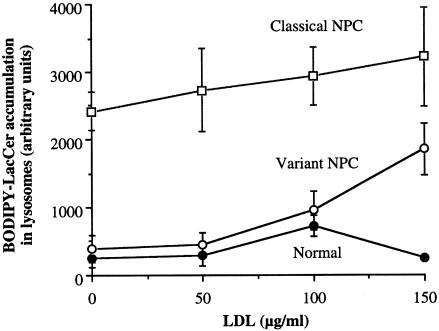
We first examined the dose-dependent effects of LDL on BODIPY-LacCer distribution in variant, classical NPC, and normal fibroblasts, using a quantitative approach. Cell samples were incubated for 24 h with 0–150 μg/ml of LDL (without prior lipid depletion) and with 2 mg/ml fluorescent blue dextran, a probe that accumulates in lysosomes after endocytosis (Koval and Pagano 1990). The cells were then pulse labeled with BODIPY-LacCer, and the relative concentration of BODIPY-LacCer in dextran-positive vesicles was calculated by red/green ratio imaging (Chen et al. 1997, 1999; Pagano et al. 2000). At 150 μg/ml, LDL dramatically increased the extent of colocalization of BODIPY-LacCer with dextran-labeled lysosomes in NPC variant fibroblasts but had little effect on normal or classical NPC cells (fig. 3).
Figure 3.
Selective dose-dependent LDL-induced lysosomal targeting of BODIPY-LacCer in NPC variants. Normal, variant, and classical NPC fibroblasts were preincubated overnight in culture medium with 10% FBS plus 0–150 μg/ml of LDL, along with 2 mg/ml Cascade blue dextran, a lysosomal marker. Cells were then pulse labeled with BODIPY-LacCer, as in figure 2. Blue, green, and red images of the same cells were acquired under the fluorescence microscope, and the amounts of BODIPY-LacCer in dextran positive-lysosomes were determined by red/green ratio imaging (Puri et al. 1999; Pagano et al. 2000). Each data point was obtained using multiple cultures of a single representative patient cell sample. Values are the mean ± SE for LacCer accumulation in the lysosomes of four or more cells (>100 lysosomes analyzed per cell). LDL added to culture media at 0, 50, 100, and 150 μg LDL protein/ml corresponded to ∼0.04, ∼0.40, ∼0.77, and ∼1.14 mg total cholesterol per ml of media, respectively.
After treatment with 150 μg/ml of LDL, filipin staining showed an alteration, in the distribution of the intracellular free-cholesterol level in variant cells, to a pattern similar to that in classical NPC cells (fig. 4_A_); LDL treatment had little effect on filipin staining in normal or classical NPC cells (fig. 4_A_). The effect of LDL treatment on BODIPY-LacCer localization in variant cells was more striking; excess LDL shifted the distribution of BODIPY-LacCer from the Golgi to punctate structures in variant cells but had little effect on the Golgi labeling of normal cells or the punctate labeling of classical NPC cells (fig. 4_B_). This cholesterol-induced alteration in BODIPY-LacCer targeting was observed in all 13 variant cell samples studied (data not shown); the cholesterol challenge had no effect on the Golgi targeting of BODIPY-LacCer in all normal fibroblast samples examined (_n_=6).
Blinded Study of Sphingolipid Trafficking Assay
We conducted a blinded study to validate the usefulness of the modified BODIPY-LacCer targeting assay to detect NPC variants. Trained observers were asked to examine the distribution of BODIPY-LacCer by fluorescence microscopy in paired samples (the same cell sample preincubated with or without LDL) of normal, classical NPC, and NPC variant cells and to score the coded samples as either normal or NPC on the basis of the intracellular distribution of BODIPY-LacCer. As shown in table 1, the variant samples were scored as NPC samples in only 4 of 23 trials without LDL treatment, but were successfully detected as NPC in 22 of 23 trials with LDL. The frequency of successful detection of classical NPC cells was also increased with LDL, from 23 to 25 of 26 trials (table 1). When the modified BODIPY-LacCer assay with LDL was used, there were no false positives and only 2 false negatives in 47 trials. Thus, the technique should be useful for detection of NPC variants and may also increase the probability of detection of classical NPC in patient samples.
Table 1.
Blinded Study of LacCer Trafficking Screening Assay Performed with or without LDL[Note]
| No. of Trials Scored Correctly | |||
|---|---|---|---|
| SampleCategory(No. of UniqueSamples) | TotalTrials | Without LDL | With LDL |
| Normal (5) | 23 | 23 | 23 |
| NPC: | |||
| Variant (6) | 23 | 4 | 22 |
| Classical (6) | 26 | 23 | 25 |
NPC1 Mutation Analysis of NPC Subtypes
The results of NPC1 mutation analysis of individual NPC cell lines are shown in table 2 along with cholesterol-esterification rates for these samples. In 12 variant cell lines (NPC patient samples with cholesterol-esterification rates of >800 pmol cholesterol/mg protein/6 h) from unrelated patients, two NPC1 mutations were found in 5 fibroblast samples, and single mutations were identified in 2 additional samples. For the five fibroblast samples in which two mutant alleles were identified, all samples were compound heterozygotes, and 9 of 10 mutations were missense point mutations. For the two additional fibroblast samples in which only one mutation was found, one was a missense mutation and the other was a frameshift mutation (table 2). Of 12 mutant alleles found among variant samples, 7 (58%) occurred between amino acids 940 and 1007 (table 2) in the cysteine-rich region of NPC1 (amino acids 854–1014 [Greer et al. 1999; Watari et al. 2000]). In five additional variant samples, no NPC1 mutations were found, either by CSGE screening or by DNA sequencing of all 25 NPC1 exons. The identification of such a high percentage of non-NPC1 cases among variants was surprising, because, in general, ∼90% of patients with NPC are reported to possess NPC1 mutations (Patterson et al. 2001). In addition, mutations have been found in >90% of NPC1 alleles from ∼200 NPC cell lines in the Mayo NPC cell repository (K.S., unpublished data). One of the variant cell samples in which no NPC1 mutations were detected (sample V9 in table 2) was determined to be in the NPC2 complementation group. Complementation studies (based on correction of high filipin staining in heterokaryons) could not be interpreted with confidence by use of the other variant samples lacking in NPC1 mutations, because the normal-like filipin staining in these cells made assessments of correction in fused cells nondefinitive.
Table 2.
NPC1 Mutations Found for Cell Lines with Variant and Classical Biochemical Phenotypes[Note]
| Mutations | |||
|---|---|---|---|
| Cell Line | Allele 1 | Allele 2 | Cholesterol Esterification(pmol cholesterol/mg protein/6 h) |
| Variant: | |||
| V1 | T137M | D948N | 1,830 |
| V2 | P237S | S1004L | 1,498 |
| V3 | S940L | R978C | 1,161 |
| V4a | R958Q | P1007A | 1,726; 3,716 |
| V5 | P1007A | IVS21(-10)delTCC | 1,867 |
| V6 | P237S | None found | 1,498 |
| V7 | 2336insT | None found | 1,692 |
| V8 | None found | None found | 1,532 |
| V9b | None found | None found | 1,263 |
| V10 | None found | None found | 3,352 |
| V11 | None found | None found | 6,561 |
| V12 | None found | None found | 1,370 |
| Classical: | |||
| C1 | Q92R | 394delC | 0 |
| C2 | P237S | I1061T | 159 |
| C3 | D242N | S940L | 0 |
| C4 | G248V | M1142T | 48 |
| C5 | P401T | I1061T | 145 |
| C6 | R404Q | D874V | 16 |
| C7 | R404Q | R404Q or deletionc | 41 |
| C8 | 1628delC | 1628delC or deletionc | 0 |
| C9 | E612D | 1628delC | 0 |
| C10 | S652W | I1061T | 2 |
| C11 | S652W | IVS9(+1)G→C | 30 |
| C12 | R789C | V1023G | 187 |
| C13 | Y825C | 451-452delAG | 10 |
| C14a | D874V | Y890X | 44; 110 |
| C15 | D874V | del1271 | 25 |
| C16 | P888S | R1186H | 30 |
| C17 | Q921X | I1061T | 24 |
| C18 | L929P | 3322insG | 198 |
| C19a | S940L | 688del6bp | 131; 303 |
| C20 | D944N | P237S, IVS23(+3)G→C | 46 |
| C21 | R958X | R958X or deletionc | 145 |
| C22 | C976R | E1089K | 71 |
| C23 | P1007A | Deletiond | 365 |
| C24 | I1061T | 636delT | 0 |
| C25 | I1061T | I1061T | 29 |
| C26 | I1061T | I1061T | 43 |
| C27a | I1061T | NII50K | 0; 0 |
| C28 | I1061T | V1165M | 300 |
| C29 | N1156S | R1186H | 127 |
| C30 | R1186H | R1186H or deletionc | 0 |
| C31 | E1189G | 3135insG | 32 |
Among 31 unrelated classical NPC samples, 38 different DNA alterations were found, including missense, frameshift, and splicing mutations that occurred throughout much of the NPC1 gene (table 2). The I1061T substitution was found to be the most common mutation among classical NPC cases, occurring in 11 of 62 unrelated alleles in nine unrelated patients. In contrast, this mutation was not identified in any of the variant samples. The high occurrence of I1061T among classical NPC patient samples is similar to previously published findings and is consistent with the idea that this mutation leads to a classical NPC phenotype (Millat et al. 1999). In contrast with the findings for the variant samples, only 10 of 62 NPC1 mutations in classical NPC samples occurred within the cysteine-rich region. Four mutations in this region that occurred among variant patients (D948N, R958Q, R978C, and S1004L) were not present in any classical patient samples (table 2). Mutations in the cysteine-rich region found in both variant and classical samples (S940L and P1007A) occurred in combinations with different mutations between the two phenotypic types, suggesting that interactions between both alleles play a role in determining the classical versus variant phenotype. From these few samples, it cannot be determined with certainty if the variant and classical phenotypes are determined entirely by specific combinations of NPC1 mutations or if other individual genetic differences also play a role. However, for four pairs of siblings, identical mutations resulted in the same NPC subtype classification (table 2), suggesting a strong influence of specific mutations on NPC phenotypes.
Discussion
We studied the feasibility of using BODIPY-LacCer trafficking in human skin fibroblasts to detect NPC variants. We found that, under basal conditions (cells cultured in 10% FBS), NPC variant samples were indistinguishable from normal fibroblasts in their ability to target BODIPY-LacCer to the Golgi apparatus. However, overnight treatment with LDL shifted the distribution of BODIPY-LacCer in NPC variant samples from the Golgi to punctate, endosomal structures typically seen in classical NPC and many other SLSDs (Chen et al. 1999) but did not affect the Golgi targeting of BODIPY-LacCer in normal fibroblasts. We validated the reproducibility of this modified assay in a blinded study which demonstrated that BODIPY-LacCer trafficking (with the LDL modification) detected variant samples as NPC in >95% of total trials. Finally, we performed NPC1 mutational analysis in variant and classical samples and found that specific mutations within the cysteine-rich region of NPC1 are correlated with the variant phenotype.
We have demonstrated elsewhere that BODIPY-LacCer trafficking is altered in many SLSDs (NPC, Niemann Pick A and B, prosaposin deficiency, Fabry disease, metachromatic leucodystropy, GM1 and GM2 gangliosidoses, and mucolipidosis type IV), and we have proposed this technique as a broad initial screening assay for detection of SLSDs (Chen et al. 1999). We subsequently determined that the altered trafficking of BODIPY-LacCer in SLSD cells results from an accumulation of excess intracellular free cholesterol, either due to a primary defect (e.g., NPC) or secondary to the accumulation of sphingolipids in cells deficient in specific lysosomal hydrolase activities (e.g., Niemann-Pick A and gangliosidoses) (Puri et al. 1999).
Here, we found that, unlike classical NPC fibroblasts, all of the NPC variant cells that we tested exhibited no defect in BODIPY-LacCer targeting when cultured in basal medium. These results suggest that, under these conditions, variant cells have little intracellular accumulation of LDL-derived cholesterol, which is consistent with low-to-intermediate filipin staining in these cells (e.g., see fig. 4_A_). However, given the reported multiple defects in cholesterol homeostasis reported for NPC cells (Pentchev et al. 1987; Patterson et al. 2001), we hypothesized that challenging variant cells with excess exogenous LDL would induce an NPC-like phenotype without perturbing normal cells. We optimized the LDL treatment and found a dosage that selectively induced altered BODIPY-LacCer trafficking in variant (but not normal) cells. It should be noted that, of the cell types that we have studied, classical NPC cells and other storage-disease cells (noted above) all show abnormal BODIPY-LacCer trafficking, even when assayed in basal medium, whereas normal cells do not shift to a SLSD-like pattern of BODIPY-LacCer targeting, even when challenged overnight with 150 μg/ml of LDL. Thus, the shift from normal trafficking of BODIPY-LacCer to abnormal, SLSD-like trafficking, on challenge with cholesterol, appears to be specific to NPC variant cells (fig. 4_B_). We propose that the BODIPY-LacCer trafficking assay, modified by the cholesterol challenge, may be useful for the confirmation of a diagnosis of NPC in the case of variants. Our limited blinded study confirmed the potential usefulness of the method. However, more-extensive trials of this assay are required for full evaluation of its value as a diagnostic procedure for NPC variants. We also tested the modified BODIPY-LacCer–trafficking assay as a possible means of detection of carriers of NPC. However, trials with several obligate heterozygote parents of patients with classical NPC showed no perturbations in BODIPY-LacCer trafficking, before or after cholesterol treatment (data not shown), suggesting that this technique will not be useful for the detection of NPC heterozygotes.
Defective cholesterol transport is accepted as a hallmark of the cell-biological pathology in NPC disease (Patterson et al. 2001). The selective response by variants to LDL-cholesterol supports the concept that variants have diminished capacity for cholesterol transport and are part of the continuum of phenotypes in the NPC disease complex. In addition to an impairment in cholesterol trafficking, patients with NPC exhibit storage of sphingolipids in visceral organs and the brain, and NPC cells in culture also accumulate sphingolipids (Vanier 1983; Walkley 1995; Vanier 1999; Patterson et al. 2001). Thus, a possible direct role for the NPC1 protein in sphingolipid transport has been proposed (Neufeld et al. 1999; Vanier 1999). Indeed, in a recent study, anti-glycosphingolipid antibodies were used to demonstrate the presence of several endogenous glycosphingolipids (including LacCer) within the NPC1-containing endosomal compartment in normal fibroblasts, as well as the absence of these lipids from this compartment in NPC cells (Zhang et al. 2001). Although we demonstrated elsewhere that the regulation of targeting of BODIPY-LacCer between the Golgi or punctate endosomes in SLSD cells in general is closely linked to intracellular cholesterol levels and/or distribution (Puri et al. 1999), we cannot rule out the possibility that, in the case of classical or variant NPC fibroblasts, mutations in the NPC1 protein have a more direct effect on the intracellular transport of BODIPY-LacCer.
We found a total of 49 different NPC1 mutations among the variant and classical NPC samples that were analyzed. Genotype-phenotype correlations among these data are difficult, because of the large number of private mutations and because most of the samples were found to be heteroallelic. However, several observations of genotype-phenotype correlation among certain mutations may be significant. Among the variant samples for which we detected NPC1 mutations, we found a high incidence of point mutations between amino acids 940–1007 within the NPC1 cysteine-rich region (Greer et al. 1999). An association between missense mutations in this region and the NPC variant phenotype has also been recently noted elsewhere (Vanier and Millat 2000). The NPC1 cysteine-rich region contains eight cysteine residues that are conserved between mammals, Caenorhabditis elegans and yeast (Greer et al. 1999) and forms part of a hydrophilic loop that has a luminal orientation within the endosome (Davies and Ioannou 2000). Some of the conserved cysteines in this region have been shown to be critical for normal NPC1 function by transfection of cysteine→serine NPC1 mutant constructs into NPC1 deficient cells (Watari et al. 2000). In a study of NPC1 mutations in patients from 13 apparently unrelated families, Greer et al. (1999) noted a prevalence of missense mutations within the cysteine-rich region and interpreted this as evidence that this region is critical for NPC function. Our observations suggest that mutations in this region may be associated with either a variant phenotype (e.g., D948N, S1004L, R978C, or R958Q), a classical phenotype (e.g., D874V, P888S, L929P, or C976R), or either phenotype (e.g., S940L and P1007A). For those mutations that are found in association with both phenotypes, we propose that it is the second mutation that primarily determines the cell phenotype. Thus, we suggest that R978C, R958Q, and IVS21(-10)delTCC are biochemically mild mutations and that D242N and 688del6bp are biochemically severe mutations. From our study, the relative roles that specific NPC1 mutations, other genetic factors, and nongenetic factors play in determining phenotype cannot be determined with certainty. However, evidence for NPC1 genotype influence on phenotype is provided by the observation that four pairs of siblings (in which each sib pair showed identical mutations) were classified concordantly as having either the variant or the classical phenotype (table 2).
One recurrent mutation (P237S) reported here is worthy of note. This point mutation occurred in both variant and classical samples in the present study (table 2) and was reported elsewhere to be a deleterious mutation with no occurrence in 100 normal samples (Yamamoto et al. 2000). However, we have identified this substitution in 2 of 98 chromosomes in a screen of 49 anonymized samples from the general population (K.S., unpublished data). In addition, we have identified several NPC fibroblast samples that have two mutations, in addition to the P237S substitution (sample C20 in table 2 and data not shown). On the basis of these observations, we believe that the P237S substitution is more likely to be a benign polymorphism than a deleterious mutation, although we cannot rule out the possibility that it could have a modifying effect on NPC1 function. This uncertainty could be resolved by examination of the phenotype of NPC null cells transfected with a NPC1-P237S mutation construct.
All the cell samples studied here were from patients whose clinical symptoms were consistent with a diagnosis of NPC. We classified samples as NPC variants solely on the basis of relatively higher esterification rates and nondefinitive filipin staining. From the limited clinical data we have available for the NPC cell lines studied, we found cases of juvenile- and adult-onset disease among both variant and classical biochemical phenotypes. Postmortem histological analysis of individual cases also showed similar brain pathology for variant versus classical patients. Our data thus suggested no distinction on clinical grounds between the variant and classic biochemical phenotypes. We found that 5 of 12 of the unique variant patient samples possessed no detectable defect in the NPC1 protein, although we screened by CSGE and fully sequenced all NPC1 exons in these samples, and, clinically, the patients from which these cells were derived presented with typical NPC symptoms (e.g., vertical supranuclear gaze palsy, dementia and movement disorders). Preliminary immunofluorescence studies using a polyclonal antibody against the C-terminus of the NPC1 protein showed a level of NPC1 expression in all variant samples (both with and without NPC1 mutations) at least as high as that in normal fibroblasts (data not shown). Thus, we have no evidence for regulatory NPC1 mutations leading to a lower expression of NPC1 in the non-NPC1 variant patients. Interestingly, our immunofluorescence studies are consistent with a recent report which showed that a high level of NPC1 expression (determined by western blotting) was typical of patients in the NPC1 complementation group who had a less severe clinical phenotype (Yamamoto et al. 2000). Except in one case, where a cell line was identified as NPC2, complementation studies to classify these samples as either NPC1 or NPC2 were found to be impractical in these cell lines because of low levels of filipin staining. Future studies will be required to determine if any of these other cell samples lacking in mutations in NPC1 are defective in HE1, the newly identified NPC2 gene product (Naureckiene et al. 2000) or are the result of a different disorder that has overlapping clinical and biochemical features with NPC.
In conclusion, we have developed an assay, based on the altered intracellular trafficking of BODIPY-LacCer, that can be used to detect NPC variants. All variant NPC fibroblast samples tested responded to an LDL-cholesterol challenge, distinguishing them from normal fibroblasts and demonstrating that these patient samples do possess a reduced ability to handle free cholesterol, which is consistent with their inclusion in the NPC disease category. Finally, by NPC1 mutation analysis, we found that most variant samples could be placed in one of two categories: those having mutation(s) present within the cysteine-rich region of NPC1 and those in which no NPC1 mutation was detected. These data provide a new technique which may be useful for the diagnosis of NPC variants, and lend new insight into the function of the NPC1 protein.
Acknowledgments
This work was supported by National Institutes of Health grants GM22942 and GM60934 (to R.E.P.) and grants from the Ara Parseghian Medical Research Foundation. We thank Shutish Patel of the Neurobiology Research Laboratory of the Veterans Affairs Healthcare System in Newington, CT, for the gift of polyclonal antibodies against NPC1.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NPC [MIM 257220] and NPC2 [MIM 601015])
References
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, et al (1997) Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228–231 [DOI] [PubMed] [Google Scholar]
- Chen CS, Martin OC, Pagano RE (1997) Changes in the spectral properties of a plasma membrane lipid analog during the first seconds of endocytosis in living cells. Biophys J 72:37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Patterson MC, Wheatley CL, O’Brien JF, Pagano RE (1999) Broad screening test for sphingolipid storage diseases based on alterations in lipid trafficking. Lancet 354:901–905 [DOI] [PubMed] [Google Scholar]
- Cruz JC, Sugii S, Yu C, Chang TY (2000) Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J Biol Chem 275:4013–4021 [DOI] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA (2000) Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem 275:24367–24374 [DOI] [PubMed] [Google Scholar]
- Greer WL, Donson MJ, Girouard GS, Byers DM, Riddell DC, Neumann PE (1999) Mutations in NPC1 highlight a conserved NPC1-specific cysteine-rich domain. Am J Hum Genet 65:1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer WL, Riddell DC, Gillan TL, Girouard GS, Sparrow SM, Byers DM, Dobson MJ, Neumann PE (1998) The Nova Scotia (Type D) form of Niemann-Pick Disease is caused by a G3097ÆT transversion in NPC1. Am J Hum Genet 63:52–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou YA (2000) The structure and function of the Niemann-Pick C1 protein. Mol Genet Metab 71:175–181 [DOI] [PubMed] [Google Scholar]
- Jan MM, Camfield PR (1998) Nova Scotia Niemann-Pick (type D): clinical study of 20 cases. J Child Neurol 13:75–78 [DOI] [PubMed] [Google Scholar]
- Korkko J, Annunen S, Pihlajamaa T, Prockop DJ, Ala-Kokko L (1998) Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc Natl Acad Sci USA 95:1681–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval M, Pagano RE (1990) Sorting of an internalized plasma membrane lipid between recycling and degradative pathways in normal and Niemann-Pick, type A fibroblasts. J Cell Biol 111:429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Ye J, Rigney M, Steck T (2000) Cholesterol movement in Niemann-Pick type C cells and in cells treated with amphiphiles. J Biol Chem 275:17468–17475 [DOI] [PubMed] [Google Scholar]
- Markoff A, Sormbroen H, Bogdanova N, Preisler-Adams S, Ganev V, Dworniczak B, Horst J (1998) Comparison of conformation-sensitive gel electrophoresis and single-strand conformation polymorphism analysis for detection of mutations in the BRCA1 gene using optimized conformation analysis protocols. Eur J Hum Genet 6:145–150 [DOI] [PubMed] [Google Scholar]
- Martin OC, Pagano RE (1994) Internalization and sorting of a fluorescent analog of glucosylceramide to the Golgi apparatus of human skin fibroblasts: utilization of endocytic and nonendocytic transport mechanisms. J Cell Biol 125:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Marcais C, Rafi MA, Yamamoto T, Morris JA, Pentchev PG, Ohno K, Wenger DA, Vanier MT (1999) Niemann-Pick C1 disease: the 1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet 65:1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Carstea ED (1998) Niemann-Pick C disease: cholesterol handling gone awry. Mol Med Today 4:525–531 [DOI] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Rattiaux R, Jadot M, Lobel P (2000) Identification of HE1 as the second gene in Niemann-Pick C disease. Science 290:2298–2301 [DOI] [PubMed] [Google Scholar]
- Neufeld EB, Wastney M, Patel S, Suresh S, Cooney AM, Dwyer NK, Roff CF, Ohno K, Morris JA, Carstea ED, Incardona JP, Strauss JF III, Vanier MT, Patternson MC, Brady RO, Pentchev PG, Blanchette-Mackie EJ (1999) The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J Biol Chem 274:9627–9635 [DOI] [PubMed] [Google Scholar]
- Pagano RE, Watanabe R, Wheatley C, Dominguez M (2000) Novel applications of BODIPY™-sphingolipid analogs to study lipid traffic and metabolism in cells. Methods Enzymol 312:523–534 [DOI] [PubMed] [Google Scholar]
- Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea ED, Neufeld EB, Blanchette-Mackie EJ, Pentchev PG (2001) Niemann-Pick disease type C: a lipid trafficking disorder. In: Scriver C, Beaudet A, Sly W, Vale D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw Hill, New York, pp 3611–3634 [Google Scholar]
- Pentchev PG, Comly ME, Kruth HS, Tokoro T, Butler J, Sokol J, Filling-Katz M, Quirk JM, Marshall DC, Patel S, Vanier MT, Brady RO (1987) Group C Niemann-Pick disease: faulty regulation of low-density lipoportein uptake and cholesterol storage in cultured fibroblasts. FASEB J 1:40–45 [DOI] [PubMed] [Google Scholar]
- Puri V, Watanabe R, Dominguez M, Sun X, Wheatley CL, Marks DL, Pagano RE (1999) Cholesterol modulates membrane traffic along the endocytic pathway in sphingolipid storage diseases. Nat Cell Biol 1:386–388 [DOI] [PubMed] [Google Scholar]
- Sidhu HS, Rastogi SA, Byers DM, Guernsey DL, Cook HW, Palmer FB, Spence MS (1993) Regulation of low density lipoprotein receptor and 3-hydroxy-3-methyl glutaryl-CoA reductase activities are differentially affected in Niemann-Pick type C and type D fibroblasts. Biochem Cell Biol 71:467–474 [DOI] [PubMed] [Google Scholar]
- Snow K, Park WD, Lundquist PA, Vockley CW, Patterson MC, Karnes PS, O’Brien JF (2000) Genetic testing for Niemann-Pick Type C disease. Genet Med 2:107 [Google Scholar]
- Uc EY, Wenger DA, Jankovic J (2000) Niemann-Pick Disease Type C: two cases and an update. Mov Disord 15:1199–1203 [DOI] [PubMed] [Google Scholar]
- Vanier MT (1983) Biochemical studies in Niemann-Pick disease. I. Major sphingolipids of liver and spleen. Biochim Biophys Acta 750:178–184 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Lipid changes in Niemann-Pick disease type C brain: personal experience and review of the literature. Neurochem Res 24:481–489 [DOI] [PubMed] [Google Scholar]
- Vanier MT, Duthel S, Rodriquez-Lafrasse C, Pentchev P, Carstea ED (1996) Genetic heterogeneity in Niemann-Pick C disease: a study using somatic cell hybridization and linkage analysis. Am J Hum Genet 58:118–125 [PMC free article] [PubMed] [Google Scholar]
- Vanier MT, Millat G (2000) Niemann-Pick C disease: insight from studies on mutated NPC1 gene and protein. J Inherit Metab Dis Suppl 23:232 [Google Scholar]
- Vanier MT, Rodriguez-Lafrasse C, Rousson R, Gazzah N, Juge MC, Pentchev PG, Revol A, Lousot P (1991) Type C Niemann-Pick disease: spectrum of phenotypic variation in disruption of intracellular LDL-derived cholesterol processing. Biochim Biophys Acta 1096:328–337 [DOI] [PubMed] [Google Scholar]
- Vanier MT, Wenger DA, Comly ME, Rousson T, Brady RO, Pentchev PG (1988) Niemann-Pick disease group C: clinical variability and diagnosis based on defective cholesterol esterification. Clin Genet 33:331–348 [DOI] [PubMed] [Google Scholar]
- Walkley SU (1995) Pyramidal neurons with ectopic dendrites in storage diseases exhibit increased GM2 ganglioside immunoreactivity. Neuroscience 68:1027–1035 [DOI] [PubMed] [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Watari M, Burd CG, Patel S, Pentchev PG, Strauss JF (2000) Determinants of NPC1 expression and action: key promoter regions, posttranscriptional control, and the importance of a “cysteine-rich” loop. Exp Cell Res 259:247–256 [DOI] [PubMed] [Google Scholar]
- Winsor EJT, Welch JP (1978) Genetic and demographic aspects of Nova Scotia Niemann-Pick disease. Am J Hum Genet 30:530–538 [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nanba E, Ninomiya H, Higaki K, Taniguchi M, Zhang H, Takeshima T, Inui K, Okada S, Tanaka A, Sakuragawa N, Millat G, Vanier MT, Morris JA, Pentchev PG, Ohno K (1999) NPC1 gene mutations in Japanese patients with Niemann-Pick disease type C. Hum Genet 105:10–16 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Ninomiya H, Matsumoto M, Ohta Y, Nanba E, Tsutsumi Y, Yamakawa K, Millat G, Vanier MT, Pentchev PG, Ohno K (2000) Genotype-phenotype relationship of Niemann-Pick disease type C: a possible correlation between clinical onset and levels of NPC1 protein in isolated skin fibroblasts. J Med Genet 37:707–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Dwyer MK, Neufeld EB, Love DC, Cooney A, Comly M, Patel S, Watari H, Strauss III JF, Pentchev PG, Hanover JA, Blanchette-Mackie EJ (2001) Sterol-modulated glycolipid sorting occurs in Niemann-Pick C1 late endosomes. J Biol Chem 276:3417–3425 [DOI] [PubMed] [Google Scholar]