Clustering of Missense Mutations in the C-Terminal Region of Factor H in Atypical Hemolytic Uremic Syndrome (original) (raw)
Abstract
Hemolytic-uremic syndrome (HUS) is a microvasculature disorder leading to microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure. Most cases of HUS are associated with epidemics of diarrhea caused by verocytotoxin-producing bacteria, but atypical cases of HUS not associated with diarrhea (aHUS) also occur. Early studies describing the association of aHUS with deficiencies of factor H suggested a role for this complement regulator in aHUS. Molecular evidence of factor H involvement in aHUS was first provided by Warwicker et al., who demonstrated that aHUS segregated with the chromosome 1q region containing the factor H gene (HF1) and who identified a mutation in HF1 in a case of familial aHUS with normal levels of factor H. We have performed the mutational screening of the HF1 gene in a novel series of 13 Spanish patients with aHUS who present normal complement profiles and whose plasma levels of factor H are, with one exception, within the normal range. These studies have resulted in the identification of five novel HF1 mutations in four of the patients. Allele HF1Δexon2, a genomic deletion of exon 2, produces a null HF1 allele and results in plasma levels of factor H that are 50% of normal. T956M, W1183L, L1189R, and V1197A are missense mutations that alter amino acid residues in the C-terminal portion of factor H, within a region—SCR16–SCR20—that is involved in the binding to solid-phase C3b and to negatively charged cellular structures. This remarkable clustering of mutations in HF1 suggests that a specific dysfunction in the protection of cellular surfaces by factor H is a major pathogenic condition underlying aHUS.
Hemolytic uremic syndrome (HUS) is a common cause of acute renal failure in children, leading to substantial morbidity and mortality (reviewed in Ruggenenti and Remuzzi 1998). HUS is characterized by a triad of symptoms—microangiopathic hemolytic anemia, thrombocytopenia and acute renal failure. Typical, epidemic or diarrhea-associated HUS is most common in infants and young children. In the majority of the cases, it is associated with the 0157:H7 strain of Escherichia coli, which produces a powerful exotoxin (either verocytotoxin or verotoxin). Other possible triggers of the disease are viruses and neuraminidase-producing microorganisms. The outcome of these patients is usually good, with a complete recovery of renal function within 2 or 3 wk. Atypical, non–diarrhea-associated HUS (aHUS) is most common in older children and adults. The prognosis is poorer than that in typical HUS, with a high (10%–30%) mortality. Renal involvement is a constant feature, and up to 50% of patients may need dialysis. Neurological symptoms are also common, and sequelae may persist for several years. Most cases of aHUS are idiopathic, but predisposing factors such as anticancer drugs, immunosuppressive agents, or oral contraceptives have been reported. aHUS can also develop during either the third trimester of pregnancy or the postpartum period, as well as in association with anti-endothelial antibodies. Although most cases are sporadic, familial cases of aHUS have been described. In these cases, both autosomal dominant (MIM 134370) and recessive (MIM 235400) modes of inheritance have been reported.
A number of observations, including the recurrence of the disease in transplanted individuals and the positive response to plasma exchange in some patients, have pointed to the involvement of a plasma factor in the etiology of aHUS. These facts, together with early reports describing the association of aHUS with deficiencies of the plasma protein factor H (Thompson and Winterborn 1981; Pichette et al. 1994; Ohali et al. 1998; Rougier et al. 1998), led to the decisive linkage studies by Warwicker et al. 1998, later confirmed by Ying et al. 1999, revealing that the disease segregates with a chromosome 1q32 region that includes the gene encoding factor H (HF1). Furthermore, in one of the families with aHUS that are included in these linkage studies, a missense mutation that had no effect on the levels of factor H was found in the HF1 gene, strongly supporting a role for factor H in the pathogenesis of aHUS.
Factor H is a plasma protein (molecular mass 155,000 daltons) composed of 20 repetitive units of 60 amino acids called “short consensus repeats” (SCR) (Ripoche et al. 1988), also known as “complement control protein” (also known as “CCP”) modules. Factor H is encoded by a single gene located on human chromosome 1q32, within the RCA (regulators of complement activation) gene cluster (Rodríguez de Córdoba et al. 1985; Weis et al. 1987). Five additional factor H–related human plasma proteins have been identified and shown to be encoded by five genes, FHR1_–_FHR5, closely linked to the HF1 gene (Zipfel and Skerka 1994; Zipfel et al. 1999; Rodríguez de Córdoba et al. 1999; McRae et al. 2000). Factor H controls activation of the alternative pathway of complement in fluid phase and on cellular surfaces. It binds to C3b, accelerates the decay of the alternative-pathway C3-convertase and acts as a cofactor for the factor I–mediated proteolytic inactivation of C3b (Weiler et al. 1976; Whaley and Ruddy 1976; Pangburn et al. 1977). Factor H can also interact with polyanionic molecules (sialic acids or glycosaminoglycans) on certain cellular surfaces, conferring to them resistance to damage as a consequence of complement activation through the alternative pathway (Meri and Pangburn 1990; Pangburn et al. 1991). Complete factor H deficiency leads to a situation of hypocomplementemia that increases the risk of infection by pathogens and that is usually associated with recurrent infections by pyogenic microorganisms. In addition, deficiency of factor H has been associated with systemic lupus ertythematosis, type II membrane-proliferative glomerulonephritis, collagen type III glomerulopathy, and, as indicated above, familial HUS (reviewed in Ault 2000).
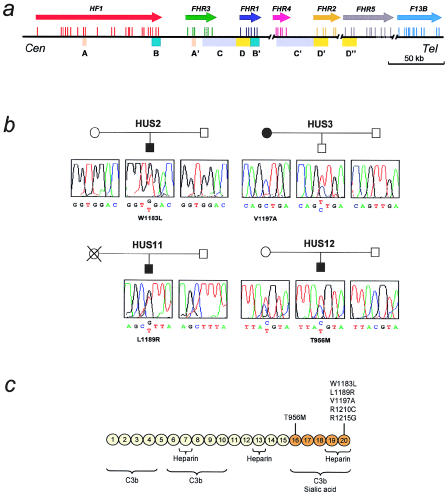
To get further insight into the genetic relationship between aHUS and factor H, we have performed mutational screening of HF1 in a novel series of patients with aHUS. These studies have been facilitated by previous work in our laboratory, related both to the organization of the HF1/FHR1–FHR5 region of the RCA gene cluster and to the structure of the human HF1 gene. Figure 1_a_ summarizes the organization of the 1q32 genomic region containing the human HF1 gene and FHR1–FHR5 genes (for genomic sequences of HF1 and of FHR1–FHR5, see the Entrez Nucleotide Sequence Search Web site). Besides showing the exon/intron organization of the genes within this region, figure 1_a_ also illustrates the existence of a number of large genomic duplications (1.2–38 kb long; 85%–97% identity) including different exons of the HF1/FHR1–FHR5 genes. Since two of these genomic duplications include exons 8/9 and exons 21–23, respectively, of the HFI gene, we took special care in the design of the PCR primers used to amplify these HF1 exons, so that homologous exons in either the FHR1 gene or the FHR3 gene would not be amplified. Table 1 shows the primer sequences and PCR conditions that we have used to amplify each of the exons of the HF1 gene from genomic DNA.
Figure 1.
Mutations in factor H in patients with aHUS. a, Organization of the 1q32 genomic region containing the human HF1 gene. Colored arrows indicate the location and transcriptional orientation of the HF1 gene and the FHR1–FHR5 genes. For each gene, the exon/intron organization is indicated by vertical bars. Genomic duplications within the region are indicated by colored boxes. These duplications are 1.2–38 kb in size and present a pairwise nucleotide identity of 85%–97%. The exon/intron organization of the HF1 gene, as well as the nucleotide sequences of the intronic flanking regions, was determined on the basis of data published by Male et al. (2000) and of data generated in our laboratory by PCR analysis and sequencing of PAC clones RPCIP704A14355, RPCIP704L20665, RPCIP704M03650, RPCIP704O14608 (authors' unpublished data). These data and sequence data generated by the Human Genome Project (clones AL049741, AL049744, AL139418, and AL353809) provided the genomic sequence for the HF1 gene and the FHR1–FHR5 genes. HF1 comprises 23 exons and spans >94 kb of genomic DNA. Exon 10 does not contribute to the factor H transcript; it is utilized only in the alternative HF1 transcript that codes for the factor H–like 1 molecule (also known as “FHL-1”) (Estaller et al. 1991). To amplify each of the HF1 exons present in the factor H transcript, we designed PCR primers to the intronic flanking sequences (table 1). Primers for exons 8/9 and 21–23 were designed to avoid amplification of homologous exons in the FHR3 gene and the FHR1 gene, respectively. b, Identification of mutations in the factor H gene in four patients with aHUS. The pedigrees of patients HUS2, HUS3, HUS11, and HUS12 are shown, as are the chromatograms corresponding to the DNA sequence surrounding the mutated nucleotide for each patient and family members studied. The positions of nucleotide changes are according to the cDNA sequence reported by Ripoche et al. (1988). The ATG initiation codon is located at nucleotide position c.74. Amino acid numbering includes the signal peptide. c, Functional domains and mutations in the factor H molecule. A diagram of the structure of human factor H with the 20 SCR repeats is shown. Functional domains are indicated schematically. The location of the six missense mutations thus far characterized in patients with aHUS (four of whom reported here) is indicated to illustrate that they are clustered within a specific region of factor H, a region that has been reported to be important for the control of C3b deposited on surfaces.
Table 1.
PCR Primers for Mutational Analysis of HF1
| Primer(5′→3′) | ||||
|---|---|---|---|---|
| Exon | Protein Region | Forward | Reverse | Fragment Sizea(bp) |
| 1 | Signal peptide | GACGTTGTGAACAGAGTTAGCTG | ACTCCTGTGAAAAGCATCATTAG | 181 |
| 2 | SCR1 | GTACATTTAAATAGACACTTTATGC | TACACCTAGTTTTCATAAATTTCAC | 281 |
| 3 | SCR2A | CCCACTCCTACATAAAATATATTCC | CCTATTTACTATCTTAATTATAAACC | 171 |
| 4 | SCR2B | TAAACACACATTATGTCAACGTTC | GAGAC TTTAAGATATTTTAATGTAAG | 206 |
| 5 | SCR3 | TACATACACATATTTTTCACAATAAAC | GCAAAAATACTAAAACAGTAAGTG | 294 |
| 6 | SCR4 | CCTTTAATTTGCAATAAACATTTTGG | TATGTGATAAATTTATAAAGATCCAG | 263 |
| 7 | SCR5 | CGGATACTTATTTCTGCATTATCC | AAATTTCAGAATTAAGAAATGGGTC | 264 |
| 8 | SCR6 | GTTTATTACAGTAAAATTTCTTTATAC | CTTCGATCTTTGAAAGTTTTATAC | 300 |
| 9 | SCR7 | TGAGCAAATTTATGTTTCTCATTTAC | TTAGAAAGACATGAACATGCTAGG | 279 |
| 11 | SCR8 | AAAATGTTATTGATCATATGCTTGTC | ACTTTTGTGTATCATCTGGATAATC | 284 |
| 12 | SCR9 | GTTTATTAGATGACATTAGAAATGAC | GGAAAACAGATTTATTTTCATTTTG | 280 |
| 13 | SCR10 | TTGGCAATGATTAATTATATATTCTC | TCAAAGTTCTAATTCTTATTTCAGC | 274 |
| 14 | SCR11 | TATATTGTAAAACAGACAATTTAACC | ATACAAAATACAAAAGTTTTGACAAG | 289 |
| 15 | SCR12 | AAAACACATACATCATGTTTTCAC | GTTGTTACAATAAAAATATTAAACTTTG | 292 |
| 16 | SCR13 | AGTTGGTTTGATTCCTATCATTTG | ACACACATACCTATTACTTTTCC | 277 |
| 17 | SCR14 | ATATTTTTATTTTTTATTTTTTATTATAAC | TATTAACCTCATTTGAAAGAATTATG | 290b |
| 18 | SCR15 | GTATTTTATTTGTTTTTAACCCTTTG | ATGAATTCTACTATAAACAGAAATTG | 277 |
| 19 | SCR16 | TAAATTTATGAGTTAGTGAAACCTG | TGGTACCACTTACACTTTGAATG | 272 |
| 20 | SCR17 | TTTTAAAGATTTGCGGAACAAATAC | CCCACACATTATATAAATAAATTTTG | 263 |
| 21 | SCR18 | TTGCTACTCAAAATGAACACTAGG | CCTGCTATACTCCCCCAAAATG | 274 |
| 22 | SCR19 | TTGTATTTTGATTTGCTCTCACAAC | GTGAAATATCAGACTCATCACAGA | 296 |
| 23 | SCR20 | ATTTGCATACTACTTAATGTTTTATG | AGTTCTGAATAAAGGTGTGCAC | 284c |
Patients included in these studies were selected on the basis of a single criterion: presenting a clinical history of HUS of non–diarrhea-associated origin. Chronic renal failure and/or unsuccessful renal transplantation led most of them to hemodialysis. Twelve of the patients have no family history of HUS. In only one of the probands, HUS11, the death of the mother, as a consequence of postpartum aHUS, was reported. Our patients have a normal complement profile, and, with the only exception of HUS3, they all present with normal plasma levels of factor H, as determined by ELISA and semiquantitative western blot analyses (table 2). The mutational analysis of the HF1 gene was performed in genomic DNA from these patients and their available relatives, by both SSCP analysis and automatic DNA sequencing of the HF1 exons. In several of our patients, HF1 sequences were also obtained by reverse transcriptase–PCR using total RNA obtained from peripheral blood lymphocytes, as described elsewhere (Sánchez-Corral et al. 2000).
Table 2.
Complement Profiles and Factor H Levels in Patients with aHUS
| Patient(Age [years]) | Clinical History | CH50a | AP50b | C3c(mg/100 ml) | C4c(mg/100 ml) | Factor Hd(μg/100 ml) |
|---|---|---|---|---|---|---|
| HUS2 (23) | Chronic renal failure, transplanted twice | Normal | Normal | 71.2 | 30 | 1,158 |
| HUS3 (53) | Chronic renal failure, hemodialysis | Normal | Low | 59.2 | 26.3 | 109e |
| HUS5 (4) | Three HUS episodes, neurological symptoms | Normal | Normal | 112 | 15 | 350 |
| HUS8 (12) | Neurological symptoms | Normal | Normal | 96.5 | 30.3 | 501 |
| HUS9 (10) | Two HUS episodes, neurological symptoms | Normal | Normal | 113 | 23.5 | 286 |
| HUS10 (10) | One HUS episode | Normal | Normal | 94.1 | 17 | 484 |
| HUS11 (12) | Transplanted twice, deceased | Normal | Normal | 167 | 53 | 1,225 |
| HUS12 (13) | One HUS episode | Normal | Normal | 135 | 27.2 | 821 |
| HUS13 (10) | Transplanted, hemodialysis | Normal | Normal | 86.5 | 10.6 | 546 |
| HUS14 (21) | Cyclosporin related, chronic renal failure, transplanted | Normal | Normal | 84 | 24.4 | 195 |
| HUS15 (17) | Cyclosporin related, chronic renal failure, transplanted | Normal | Normal | 96.2 | 19.5 | 483 |
| HUS16 (2) | One HUS episode | Normal | Normal | 90.8 | 15.5 | 1,052 |
| HUS18 (10) | One HUS episode | Normal | Normal | 109 | 15.7 | 320 |
In 4 of the 13 patients with aHUS, we identified HF1 mutations that were not found in a sample of >100 population-matched HF1 control chromosomes (fig. 1_b_). Patient HUS2 is heterozygous for a G→T substitution at HF1 nucleotide position c.3621 in exon 23. This mutation results in a tryptophan-to-leucine change, W1183L, in SCR20. No mutations were found in the other HF1 chromosome. The W1183L mutation was identified both in genomic DNA and in RNA obtained from the lymphocytes of the patient, but not in either the DNA or the mRNA obtained from his parents, indicating that it is a de novo mutation. Patient HUS3 presented a T→C substitution at nucleotide position c.3663 in exon 23, apparently in homozygosis. This mutation changes the amino acid valine 1197 to alanine in factor H SCR20. No additional mutations to V1197A were found in HUS3. Since levels of factor H in HUS3 were half of normal levels, and since she was also found to be homozygous at c. 257, c.994, c.1277, c.1492, c.1551, c.2089, c.2881, and c.3705 HF1 polymorphic sites (to be described elsewhere), we set up experiments to determine whether she carries a genomic deletion in HF1 in heterozygosis. Segregation analysis of HF1 polymorphisms in her son demonstrated that she carries a null allele at the c.257 polymorphic site in exon 2 (coding for SCR1), indicating a partial deletion of the HF1 gene in one of her chromosomes. Although the precise length of the deletion has not been established, we conclude that this deletion results in a null HF1 allele (HF1Δexon2) that is not transcribed into mRNA. Patient HUS3 was, therefore, interpreted to be an HF1 hemizygote who, in addition, carries the V1197A mutation in her only functional HF1 allele. Since the nucleotide change involved in the V1197A mutation is one of the two nucleotide differences that distinguish HF1 exon 23 from FHR1 exon 6, within the large genomic duplication involving the 3′ end of these two genes (fig. 1_a_), we suggest that the V1197A mutation could have originated via a gene-conversion event between HF1 and FHR1.
DNA sequence analysis of HF1 demonstrated that a third patient, HUS11, is heterozygous for a T→G substitution at HF1 nucleotide position c.3639 in exon 23. This mutation results in a leucine-to-arginine change, L1189R, in SCR20. The mutation was not found in the father of the patient, suggesting a maternal origin. However, DNA from the mother, who had died of postpartum HUS, was not available. Finally, in a fourth patient, HUS12, HF1 mutational analysis demonstrated a C→T substitution in heterozygosis at nucleotide position c.2940 in exon 19. This change results in a threonine-to-methionine change, T956M, in SCR16. Analysis of the DNA from the parents of HUS12 revealed that the mother also carries the T956M mutation in heterozygosis. No mutations in HF1 were found in the remaining nine patients with aHUS, by either SSCP or DNA sequencing analyses.
Different strategies, involving the use of monoclonal antibodies, enzymatic digestion, or deletion mutagenesis, have allowed the identification and characterization of distinct functional domains in the factor H molecule, although it has not been exactly determined which amino acid residues are responsible for these functions (Alsenz et al. 1985; Gordon et al. 1995; Kühn et al. 1995; Jokiranta et al. 1996; Prodinger et al. 1998). Figure 1_c_ depicts schematically some of these functional domains. Factor H has three binding sites for C3b—in SCR1–SCR4, SCR6–SCR10, and SCR16–SCR20. The C3b-binding site in SCR1–SCR4 is the only site essential for the cofactor activity with factor I, but deletion of any of the C3b-binding sites significantly decreases factor H binding to C3b deposited on cellular surfaces (Sharma and Pangburn 1996). In addition, figure 1_c_ illustrates that SCR7, SCR13, and SCR16–SCR20 have been found to contain heparin- and sialic acid–binding sites (Pangburn et al. 1991; Blackmore et al. 1996, 1998; Ram et al. 1998).
The T956M, W1183L, L1189R, and V1197A mutations identified in this study alter the amino acid sequence of the factor H molecule without apparently affecting the normal secretion of the protein into plasma (table 2) (for the factor H sequence, see Entrez Nucleotide Sequence Search). One of these mutations lies in SCR16, and the other three in SCR20, the most C-terminal domain of factor H. There are only two other cases of aHUS that have normal levels of factor H and in which mutations in HF1 have been previously identified, and, most interesting, in both cases the mutations (R1215G and R1210C) also lie in SCR20 (Warwicker et al. 1998; Caprioli et al. 2000). The clustering of missense mutations in the SCR16–SCR20 domains of factor H in patients with aHUS contrasts with the random distribution of (1) HF1 mutations in individuals with factor H deficiency (Ault et al. 1997; Warwicker et al. 1998; Buddles et al. 2000; Sánchez-Corral et al. 2000) and (2) the HF1 single-nucleotide polymorphisms thus far described (see above), strongly suggesting a genotype-phenotype correlation in aHUS, which points to the involvement, in the pathogenesis of this disease, of a specific dysfunction in the C-terminal region of factor H.
Although the relative contributions of SCR16 and SCR20 to the functional map of factor H have not been established, it is clear that both SCRs are part of a region that is involved in the binding to solid-phase C3b and to negatively charged cellular structures (such as sialic acid)—and that this region contributes significantly to the capacity of factor H to control activation of the complement system on certain cellular surfaces (Pangburn et al. 2000). Therefore, it is likely that the SCR16–SCR20 mutations found in patients with aHUS result in a defective protection of cellular surfaces by factor H. On the other hand, the T956M, W1183L, L1189R, and V1197A mutations should not alter the cofactor activity of factor H in the fluid phase, which is in agreement with the normal complement activity and C3 levels observed in our patients with aHUS. It is noticeable that in patients HUS2, HUS11, and HUS12 the levels of factor H are considerably elevated. Whether this situation results from an increased transcription of the HF1 gene or is a consequence of a low turnover of the mutated protein in plasma remains to be determined.
Thus far, two types of mutations in factor H have been observed in patients with aHUS: null HF1 alleles resulting in decreased levels of factor H and missense mutations in the SCR16–SCR20 region. Interestingly, one of our patients with aHUS (i.e., HUS3) carries both an HF1 null allele and the V1197A mutation in her only functional HF1 copy, which suggests that, in some cases, both decreased levels of factor H and mutations in SCR16–SCR20 may be necessary to produce pathologically significant dysfunction of factor H. In this regard, it should be interesting to reevaluate the previously described cases of aHUS that have decreased levels of factor H, in a search for additional HF1 missense mutations.
As indicated above, nine of our patients with aHUS have no mutations in HF1 and present normal levels of factor H in plasma. Experiments to further characterize the activity of factor H in the sera of these patients, including the search for potential plasma components that may interfere with the role of factor H in controlling the surface activation of the complement system, would be needed before involvement of factor H can be excluded from the pathogenesis of aHUS in these patients.
Our patients with aHUS who have missense mutations in the SCR16–SCR20 region are HF1 heterozygotes, which make difficult the purification and analysis of the mutant factor H proteins. Functional characterization of these mutations in recombinant factor H molecules would be needed to confirm that, in patients with aHUS, there is a defective control, by factor H, of the activation of the complement system on cellular surfaces. If this hypothesis is correct, administration of exogenous, functionally active factor H may become a successful therapeutic approach for a significant number of patients with aHUS.
Acknowledgments
We thank the families with aHUS and all the clinicians, particularly Drs. Ángel Alonso and Mª Auxiliadora Bajo, for their collaboration and donation of blood samples. We would also like to thank S. Vara de Rey, D. Beltrán Valero de Bernabé, L. Gulliksen, and the DNA Sequencing Laboratory at the Centro de Investigaciones Biológicas, for their contribution to this work. We also thank the reviewers of the manuscript, for their helpful comments. This research was supported by Spanish Fondo de Investigaciones Sanitarias grant FIS 98/0687, Comisión Interministerial de Ciencia y Tecnología grant SAF-99/0013-C02-01, and Comunidad de Madrid grants 08.1/0008/1998 and 08.6/0028/2000. D.P.-C. and P.S.-C. have been awarded grants from Glaxo Wellcome/CSIC and the Comunidad de Madrid, respectively.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Entrez Nucleotide Sequence Search, http://www.ncbi.nlm.nih.gov/Entrez/nucleotide.html (for factor H cDNA sequence [accession number Y00716] and genomic sequences of the HF1 gene and the FHR1–FHR5 genes [accession numbers AL049744, AL049741, AL139418, and AL353809])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for autosomal dominant [MIM 134370] and recessive [MIM 235400] HUS)
References
- Alsenz J, Schulz TF, Lambris JD, Sim R-B, Dierich MP (1985) Structural and functional analysis of the complement component factor H with the use of different enzymes and monoclonal antibodies to factor H. Biochem J 232:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault BH (2000) Factor H and the pathogenesis of renal diseases. Pediatr Nephrol 14:1045–1053 [DOI] [PubMed] [Google Scholar]
- Ault BH, Schmidt BZ, Fowler NL, Kashtan CE, Ahmed AE, Vogt BA, Colten HR (1997) Human factor H deficiency: mutations in framework cysteine residues and block in H protein secretion and intracellular catabolism. J Biol Chem 272:25168–25175 [DOI] [PubMed] [Google Scholar]
- Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL (1998) Identification of the second heparin-binding domain in human complement factor H. J Immunol 160:3342–3348 [PubMed] [Google Scholar]
- Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL (1996) Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J Immunol 157:5422–5427 [PubMed] [Google Scholar]
- Buddles MRH, Dome RL, Richards A, Goodship J, Goodship THJ (2000) Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am J Hum Genet 66:1721–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli J, Bettinaglio P, Zipfel PF, Vasile B, Gamba S, Amadei B, Orisio S, Remuzzi G, Noris M (2000) Mutations of factor H (HF) in familial hemolytic uremic syndrome (HUS)/thrombotic thrombocytopenic purpura (TTP). Immunopharmacology 49:13 [Google Scholar]
- Estaller C, Schwaeble W, Dierich M, Weiss EH (1991) Human complement factor H: two factor H proteins are derived from alternatively spliced transcripts. Eur J Immunol 21:799–802 [DOI] [PubMed] [Google Scholar]
- Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM (1995) Identification of complement regulatory domains in human factor H. J Immunol 155:348–356 [PubMed] [Google Scholar]
- Jokiranta TS, Zipfel PF, Hakulinen J, Kühn S, Pangburn MK, Tamerius JD, Meri S (1996) Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal antifactor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett 393:297–302 [DOI] [PubMed] [Google Scholar]
- Kühn S, Skerka C, Zipfel PF (1995) Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H. J Immunol 155:5663–5670 [PubMed] [Google Scholar]
- Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL (2000) Complement factor H: sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 37:41–52 [DOI] [PubMed] [Google Scholar]
- McRae JL, Cowan PJ, Power DA, Mitchelhill KI, Kemp BE, Morgan BP, Murphy BF (2000) Human factor H-related protein 5 (FHR-5): a new complement-associated protein. J Biol Chem (in press) [DOI] [PubMed] [Google Scholar]
- Meri S, Pangburn MK (1990) Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci USA 87:3982–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohali M, Shalev H, Schlesinger M, Katz Y, Kachko L, Carmi R, Sofer S, Landau D (1998) Hypocomplementemic autosomal recessive hemolytic uremic syndrome with decreased factor H. Pediatr Nephrol 12:619–624 [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Atkinson MAL, Meri S (1991) Localization of the heparin-binding site on complement factor H. J Biol Chem 266:16847–16853 [PubMed] [Google Scholar]
- Pangburn MK, Pangburn KLW, Koistinen V, Meri S, Sharma AK (2000) Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J Immunol 164:4742–4751 [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Schreiber RD, Müller-Eberhard HJ (1977) Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J Exp Med 146:257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichette V, Quérin S, Schürch W, Brun G, Lehner-Netsch G, Delâge JM (1994) Familial hemolytic-uremic syndrome and homozygous factor H deficiency. Am J Kidney Dis 24:936–941 [DOI] [PubMed] [Google Scholar]
- Prodinger WM, Hellwage J, Spruth M, Dierich MP, Zipfel PF (1998) The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochem J 331:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA (1998) A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med 187:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche J, Day AJ, Harris TJR, Sim RB (1988)The complete amino acid sequence of human complement factor H. Biochem J 249:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S, Díaz-Guillén MA, Heine-Suñer D (1999) An integrated map of the human regulator of complement activation (RCA) gene cluster on 1q32. Mol Immunol 36:803–808 [DOI] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S, Lublin DM, Rubinstein P, Atkinson JP (1985) Human genes for three complement components that regulate the activation of C3 are tightly linked. J Exp Med 161:1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Kazatchkine MD, Rougier JP, Fremeaux-Bacchi V, Blouin J, Deschenes G, Soto B, Baudouin V, Pautard B, Proesmans W, Weiss E, Weiss L (1998) Human complement factor H deficiency associated with hemolytic uremic syndrome. J Am Soc Nephrol 9:2318–2326 [DOI] [PubMed] [Google Scholar]
- Ruggenenti P, Remuzzi G (1998) Thrombotic microangiopathy. In: Suki WN, Massry SG (eds) Suki and Massrý's therapy of renal diseases and related disorders. Kluwer Academic, Boston, pp 513–527 [Google Scholar]
- Sánchez-Corral P, Bellavia D, Amico, L, Brai M, Rodríguez de Córdoba S (2000) Molecular basis for factor H and FHL-1 deficiency in an Italian family. Immunogenetics 51:366–369 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Pangburn MK (1996) Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc Natl Acad Sci USA 93:10996–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RA, Winterborn MH (1981) Hypocomplementemia due to a genetic deficiency of β1H globulin. Clin Exp Immunol 46:110–119 [PMC free article] [PubMed] [Google Scholar]
- Warwicker P, Goodship THJ, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA (1998) Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int 53:836–844 [DOI] [PubMed] [Google Scholar]
- Weiler JM, Daha MR, Austen KF, Fearon DT (1976) Control of the amplification convertase of complement by the plasma protein beta 1 H. Proc Natl Acad Sci USA 73:3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis JH, Morton CC, Bruns GAP, Weis JJ, Klickstein LB, Wong WW, Fearon DT (1987) A complement receptor locus: genes encoding C3b/C4b receptor and C3d/Epstein-Barr virus receptor map to 1q32. J Immunol 138:312–315 [PubMed] [Google Scholar]
- Whaley K, Ruddy S (1976) Modulation of C3b hemolytic activity by a plasma protein distinct from C3b inactivator. Science 193:1011–1013 [DOI] [PubMed] [Google Scholar]
- Ying L, Katz Y, Schlesinger M, Carmi R, Shalev H, Haider N, Beck G, Sheffield VC, Landau D (1999) Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am J Hum Genet 65:1538–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S (1999) The factor H protein family. Immunopharmacology 42:53–60 [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C (1994) Complement factor H and related proteins: an expanding family of complement-regulatory proteins? Immunol Today 15:121–126 [DOI] [PubMed] [Google Scholar]