Genomewide Multipoint Linkage Analysis of Seven Extended Palauan Pedigrees with Schizophrenia, by a Markov-Chain Monte Carlo Method (original) (raw)
Abstract
Palauans are an isolated population in Micronesia with lifetime prevalence of schizophrenia (SCZD) of 2%, compared to the world rate of ∼1%. The possible enrichment for SCZD genes, in conjunction with the potential for reduced etiological heterogeneity and the opportunity to ascertain statistically powerful extended pedigrees, makes Palauans a population of choice for the mapping of SCZD genes. We have used a Markov-chain Monte Carlo method to perform a genomewide multipoint analysis in seven extended pedigrees from Palau. Robust multipoint parametric and nonparametric linkage (NPL) analyses were performed under three nested diagnostic classifications—core, spectrum, and broad. We observed four regions of interest across the genome. Two of these regions—on chromosomes 2p13-14 (for which, under core diagnostic classification, _NPL_=6.5 and parametric _LOD_=4.8) and 13q12-22 (for which, under broad diagnostic classification, parametric _LOD_=3.6, and, under spectrum diagnostic classification, parametric _LOD_=3.5)—had evidence for linkage with genomewide significance, after correction for multiple testing; with the current pedigree resource and genotyping, these regions are estimated to be 4.3 cM and 19.75 cM in size, respectively. A third region, with intermediate evidence for linkage, was identified on chromosome 5q22-qter (for which, under broad diagnostic classification, parametric _LOD_=2.5). The fourth region of interest had only borderline suggestive evidence for linkage (on 3q24-28; for this region, under broad diagnostic classification, parametric _LOD_=2.0). All regions exhibited evidence for genetic heterogeneity. Our findings provide significant evidence for susceptibility loci on chromosomes 2p13-14 and 13q12-22 and support both a model of genetic heterogeneity and the utility of a broader set of diagnostic classifications in the population from Palau.
Introduction
Schizophrenia (SCZD [MIM 181500]) is a severe and debilitating psychiatric disorder that affects ∼1% of the world's population. Family, twin, and adoption studies have indicated substantial heritability (Gottesman and Shields 1982; Kendler and Diehl 1993). Segregation patterns indicate that a single-gene theory is not viable (Elston et al. 1978; O'Rourke et al. 1982; Tsuang et al. 1982_a;_ Risch and Baron 1984) and that, more likely, several genes of modest effect exist (Suarez et al. 1994). Reduced penetrance is evident on the basis of twin studies (Kendler 1983; Gottesman 1991), and phenocopies, of both environmental (Davison 1983) and drug-related types (Tsuang et al. 1982_b_), have been documented. Furthermore, the lack of consensus of existing results from linkage analyses suggests that substantial genetic heterogeneity may exist.
For mapping of genes for complex diseases, isolated populations have been suggested as a resource of choice (Wright et al. 1999). The expectation is of simplified genetic etiology and, thus, of diminished genetic heterogeneity, making the complex disease easier to dissect. Furthermore, for gene localization, extended pedigrees may be more statistically powerful than smaller family units, unless the disease genes are very frequent and have low risk (Durner et al. 1999). However, for SCZD, few extended pedigrees exist, primarily owing to the low marriage and reproduction rates of individuals who suffer from the disease (Böök et al. 1978).
The Republic of Palau, also known as “Belau,” is the westernmost archipelago in Micronesia. Blood-group clustering, linguistic analysis, and ethnographic studies indicate that the Palauan population has developed in relative isolation, even from other Micronesian populations, since the original settlement of Palau 2,000 years ago (Simmons et al. 1965). The basic unit of social organization in Palau is the clan, a blood-based, landholding group of families that share a common female founder; hence, extended pedigrees can be ascertained and studied. The lifetime prevalence of SCZD for individuals in Palau has been estimated, by complete ascertainment, to be 2%, compared to the worldwide rate of 0.8%–1% (Myles-Worsley 1999). For these reasons, the Palauan extended pedigrees with SCZD may represent an extremely important resource for gene localization, for SCZD.
In this study, we have chosen to analyze three nested diagnostic classifications—core, spectrum, and broad (see the “Psychiatric Assessment” subsection, below). Core and spectrum diagnostic classifications are considered under the assumption of a continuum for which common genetic mechanisms may exist (also see Wang et al. 1995; Brzustowicz et al. 2000; Gurling et al. 2001). A broad classification, including affective psychotic disorders, is a more controversial classification, although one that has also been suggested elsewhere (Straub et al. 1995; Kendler et al. 1996). It has been shown that SCZD and bipolar disorder do not significantly coaggregate (Baron et al. 1982; Maier et al. 1993). However, there still remains evidence for overlap; for example, families containing both individuals with SCZD and individuals with bipolar disorder are at increased risk for schizoaffective disorder, and several similarities, both clinical and epidemiological, exist between the two disorders. There is increasing evidence for shared chromosomal regions (Berrettini 2000; Baron 2001), and, for such loci, a broad classification has increased power for linkage detection (Kendler et al. 1996). Our approach, to consider analysis under all three classifications, allows for a more comprehensive set of hypotheses to be tested.
A two-point analysis has previously been performed using the core diagnostic phenotype in a single large pedigree from Palau (Coon et al. 1998). A single suggestive region on chromosome 2p13-14 was indicated. Multipoint analyses were not possible at that time (owing to lack of appropriate software) and hence were not performed. In the analyses presented in the present study, a genome search with seven extended pedigrees from Palau—including the same single large pedigree (15831) that was examined previously, analyzed under a more comprehensive set of diagnostic classifications—was performed. We performed multipoint analyses across the genome (22 autosomes) by Markov-chain Monte Carlo methodology and calculated both model-based and nonparametric statistics. The larger resource size that we considered, in addition to the more sophisticated analyses that we performed, results in a substantially more powerful study.
Subjects and Methods
Psychiatric Assessment
After informed consent was obtained, subjects were interviewed by a board-certified psychiatrist (Dr. William Byerley), by a modified version of the semistructured interview called “the Schedule for Affective Disorders and Schizophrenia—Lifetime Version” (Endicott and Spitzer 1978). All interviews were conducted in the presence of a Belau National Hospital behavioral-health professional, and, when subjects preferred to be interviewed in their native language, the necessary translation was performed. Medical records of any individual who received hospital inpatient or outpatient psychiatric care were obtained. All available information was rendered anonymous and was reviewed separately and blind to genotypic data, by two board-certified psychiatrists (Drs. Fred Reimherr and Paul W. Wender). Research Diagnostic Criteria (Spitzer et al. 1978) were used to arrive at a consensual diagnosis. For any diagnostic disagreements, further information was requested. For diagnostic disparity that could not be resolved, the individual was coded as unknown.
Three nested diagnostic classifications were used in this study—core, spectrum, and broad. The core diagnostic classification was the narrowest definition for affected status and included diagnoses of chronic SCZD (symptoms >2 years duration), subchronic SCZD (symptoms >1 year but <2 years duration), and schizoaffective disorder (mainly schizophrenic course). The spectrum diagnostic classification included those classified as affected under the core definition, with, in addition, the nonaffective psychotic diagnoses of acute SCZD (symptoms <6 mo duration) and subacute SCZD (symptoms >6 mo but <1 year duration). In our sample, there were no schizotypal diagnoses, which would otherwise also fit in this classification. The broad diagnostic classification was the most inclusive and included all psychotic disorders, including the affective psychotic diagnoses of schizoaffective disorder (mainly affective course), bipolar disorder with psychotic symptoms, and psychotic depression.
Pedigree Sample
Medical records at Belau National Hospital were used to identify individuals who met diagnostic criteria for entry in the study. Consenting individuals were interviewed for family history and genealogical information, to ascertain other potentially affected individuals and to reveal genealogical ties to other previously ascertained pedigrees. The initial framework for the pedigree ascertainment was provided by Dr. Anthony Polloi. Since Palau is an island nation with a finite population, many complex marriage connections exist within and between the seven defined pedigrees. For these analyses, the pedigrees were constructed blind to genotypic data and with clusters of affected individuals grouped under common founders. There were no duplications of individuals in multiple pedigrees. Seven extended pedigrees from Palau that had comparable genotyping data were considered in this study. Each pedigree contained 3–10 genotyped individuals coded as affected under the core diagnostic classification (see the “Psychiatric Assessment” subsection and table 1). The pedigrees comprised 15–57 individuals (8–40 individuals with genotypic data) and 4–6 generations. Table 1 summarizes the number of individuals coded as affected under the core, spectrum, and broad diagnostic classifications, in the seven pedigrees. Additional individuals were genotyped (up to 3, 5, and 8 affected individuals for the core, spectrum, and broad diagnostic classifications, respectively) for a chromosome 2 region where previous interesting two-point evidence had been found in a single pedigree (Coon et al. 1998, pedigree 15831).
Table 1.
Characteristics of the Seven Pedigrees in the Linkage Analysis
| No. of Individuals Affecteda | ||||
|---|---|---|---|---|
| Pedigree | Core | Spectrum | Broad | Total |
| 15831 | 10 (10) | 10 (10) | 13 (12) | 57 (40) |
| 15841 | 9 (8) | 13 (12) | 14 (13) | 51 (29) |
| 20241 | 6 (6) | 6 (6) | 6 (6) | 15 (8) |
| 22381 | 3 (3) | 3 (3) | 3 (3) | 21 (14) |
| 24401 | 5 (4) | 5 (4) | 5 (4) | 31 (12) |
| 26491 | 4 (4) | 5 (5) | 5 (5) | 30 (14) |
| 26492 | 5 (5) | 5 (5) | 5 (5) | 24 (10) |
| Total | 42 (40) | 47 (45) | 51 (48) | 229 (127) |
The average of coefficient of kinship (ψ) between affected pairs was used to measure the level of clustering of the affected individuals in each pedigree. On average, across all seven pedigrees, ψ was found to range from .05 to .06, depending on the diagnostic classification used. The ψ value indicates the probability that a gene picked at random from two individuals is identical by descent (IBD); for example, ψ=1/4 for sibs, ψ=1/8 for uncle-nephew pairs, ψ=1/64 for second cousins, and ψ=0 for two individuals with no blood relation. The value ψ=.05 (or ψ=.06) indicates that, on average, the relatedness of affecteds within pedigrees is approximately the same as that of first cousins. This lack of dense clustering, although not unexpected, indicated that maintaining the full-pedigree structures for analysis would maximize the power of the resource (see the “Linkage Analysis” subsection, below).
Genotyping
Blood samples were obtained from consenting individuals, and high-molecular-weight DNA was extracted by standard techniques (Bell et al. 1981). A set of 391 di-, tri-, and tetranucleotide repeat markers from The Cooperative Human Linkage Center (Murray et al. 1994) screening set 6.0 were used to screen the genome (autosomes only). In addition, supplemental markers were genotyped, to cover telomeric regions thought to be gene rich (Saccone et al. 1992). The marker map was generated primarily on the basis of data from the Marshfield marker database (Center for Medical Genetics, Marshfield Medical Research Foundation) but also on the basis of data from The Cooperative Human Linkage Center and from the Genome Database (see the “Electronic-Database Information” section). Genotyping was performed by standard techniques (details in Coon et al. 1994). The average resolution for the genomic screen markers was 7–8 cM. For the region of interest on chromosome 2, additional markers were genotyped, and the resolution for this chromosome was higher (on average, 1 cM). Inconsistencies and incompatibilities were detected by a modified version of the UNKNOWN program from the LINKAGE analysis software suite (Lathrop et al. 1984). In addition, in candidate regions, improbable double recombinants were identified by comparison of observed and expected recombination rates. Clearly erroneous genotypes were deleted, and, in cases in which the discrepancy was not clearly apparent and the problem could not be resolved, genotypic data for the problematic marker were deleted for all individuals in the pedigree. Marker-allele frequencies were found to vary widely between the Palauan population and populations of northern-European descent. All marker-allele frequencies and haplotype frequencies used in the analyses were calculated on the basis of the pedigree sample.
Linkage Analysis
_Estimation of multipoint IBD vectors.—_The lack of dense clustering of affecteds in the pedigrees suggested that a methodology that allowed analysis of the pedigrees in their full structure would be advantageous, since extensive amounts of information on segregation and on sharing are lost when the pedigrees are split. In addition, multipoint analysis was considered essential—since marker heterozygosity was generally lower in the Palauan population compared to northern Europeans and since multipoint analysis would greatly reduce the effect of this poorer heterozygosity. The size and structure of the pedigrees prohibited the use of more-standard software—such as the GENEHUNTER suite (Kruglyak et al. 1996)—to calculate the multipoint inheritance vectors (haplotype frequencies) necessary to calculate multipoint linkage statistics. We have used the Markov-chain Monte Carlo method MCLINK (Thomas et al. 2000), which provides haplotype reconstructions by blocked Gibbs sampling (Jensen et al. 1995). In the Markov chain, these haplotype reconstructions appear in proportion to their true frequencies, and, hence, by sampling from the chain, a Monte Carlo estimate for the linkage statistics can be found.
_Linkage statistics.—_Multipoint LOD score and nonparametric linkage (NPL) analyses were performed. The TLOD statistic (Abkevich et al., in press) was used for the model-based LOD-score analysis. This statistic was first introduced by Göring and Terwilliger (2000). It is a combination of multipoint and two-point procedures, exploiting the advantages and discarding the disadvantages of both. Multipoint inheritance information is calculated. This is then used to determine the IBD probabilities for a specific marker, and the LOD score is calculated under a two-point paradigm. The use of multipoint data to determine the IBD vector for a specific marker reduces false-positive results, to which standard two-point analyses are prone, by better distinguishing between alleles that are IBD and those that are identical by state. The calculation of the TLOD statistic under the two-point paradigm allows the inclusion of the recombination fraction (θ) as a parameter, and, hence, the statistic is as robust to model misspecification as a standard two-point statistic. Göring and Terwilliger (2000, pp. 1097–1098) describe θ as “complex-valued,” indicating both “the true probability of recombination” and “modeling errors.” In other words, modeling errors can be accounted for by the inclusion of this parameter. In contrast, standard multipoint linkage analysis does not include θ as a free parameter (it is fixed at 0) and tends not to be robust to errors in the specified model (Risch and Giuffra 1992). As in standard two-point analyses, HOMOG (Ott 1986) was used to calculate heterogeneity TLOD (het-TLOD) values.
For the nonparametric analyses, a slightly different variation on the NPL as described by Kruglyak et al. (1996) was used (for full details, see Camp et al., in press). For ease of comparison between the het-TLOD scores and NPL scores, we transformed each “raw NPL” score >0 to an NPL score that could be interpreted on a het-TLOD scale. This transformation was achieved by mapping the empirical P value for the raw NPL score, calculated from simulations under the null hypothesis of no linkage, onto the distribution for het-TLOD scores calculated under the same null simulations. The het-TLOD score that corresponded to the empirical P value for the raw NPL was considered to be the NPL score, and these are the values that are reported throughout this study.
_Genetic models.—_For the model-based analyses, genetic models based on those derived by Kendler et al. (1996) were used. In their study, Kendler et al. describe models for three diagnostic definitions—narrow, intermediate, and broad—which correspond to the classifications used herein—core, spectrum, and broad, respectively. Lifetime risks under the three definitions were assumed to be 0.6%, 3.0%, and 4.0%, and gene frequencies were assigned for phenocopy rates of 10%, 20%, and 25%, for definitions of core (i.e., narrow), spectrum (i.e., intermediate), and broad, respectively. In Palau, a 2:1 male:female ratio was observed among all known individuals with SCZD (Myles-Worsley et al. 1999); hence, the penetrance for female gene carriers was set to half that for the males. The genetic models used in the model-based analyses are shown in table 2. The parameters in these models will not necessarily represent the true values for our population, particularly because a single-gene model itself is not viable for SCZD. However, several researchers (e.g., Clerget-Darpoux et al. [1986], Risch et al. [1989], Vieland et al. [1993], and Greenberg et al. [1998]) have shown that parametric analysis, under a single-gene model and with arbitrary genetic parameters, is a powerful method for detection linkage evidence, provided that both dominant and recessive models are considered. Hence, we adopted the previously established models described above, for use in our parametric analyses.
Table 2.
Genetic Models Used in the Model-Based Linkage Analyses
| Dominant Gene | Recessive Gene | |||||||
|---|---|---|---|---|---|---|---|---|
| P Valuea | P Valuea | |||||||
| DiagnosticClassification | Frequency | P(AA) | P(AB) | P(BB) | Frequency | P(AA) | P(AB) | P(BB) |
| Core: | ||||||||
| Male | .00492 | .0006 | .55 | .55 | .09909 | .0006 | .0006 | .55 |
| Female | .0006 | .275 | .275 | .0006 | .0006 | .275 | ||
| Spectrum: | ||||||||
| Male | .01613 | .0062 | .75 | .75 | .178885 | .0062 | .0062 | .75 |
| Female | .0062 | .375 | .375 | .0062 | .0062 | .375 | ||
| Broad: | ||||||||
| Male | .0178 | .0104 | .85 | .85 | .1882 | .0104 | .0104 | .85 |
| Female | .0104 | .425 | .425 | .0104 | .0104 | .425 |
_Multiple testing.—_The guidelines suggested by Lander and Kruglyak (1995) for the reporting of linkage findings from a single genomewide scan have been widely adopted; a genome with infinitely dense markers is considered in their calculations, to allow for the addition of hint markers without inflation of type I errors. However, these guidelines fail to address the issue of multiple testing, which arises from the analysis of multiple models and phenotype classifications. Here, we have analyzed six phenotype/model combinations—three nested diagnostic classifications for two genetic models (dominant and recessive). To address our multiple-testing problem, we have calculated the number (N) of effectively independent analyses of these six models, by considering the linear relationship between the analysis results (Camp and Farnham, in press). This value is used in conjunction with the theoretical rationale provided by Lander and Kruglyak (1995) to calculate thresholds for significant, intermediate, and suggestive evidence for linkage, which account for the multiple testing. For this study, _N_=1.7, and the thresholds for significant, intermediate, and suggestive were found to be 3.5, 2.4, and 2.1, representing false-positive rates, per genome, of .05, .5, and 1, respectively.
Results
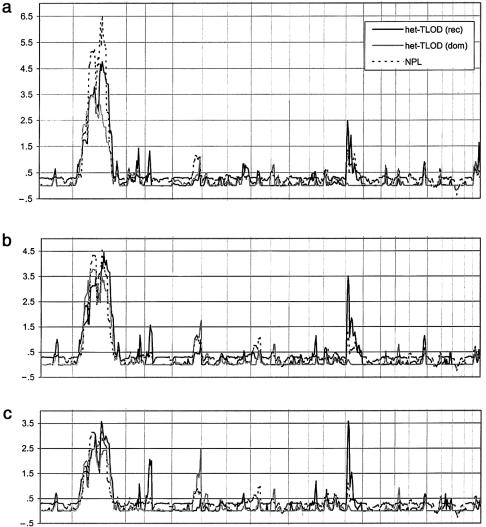
Figure 1 illustrates the multipoint-linkage results for the three diagnostic classifications—core, spectrum, and broad. For each classification, three linkage statistics were calculated—dominant and recessive parametric het-TLOD scores and an NPL score. Four regions of interest were identified, on chromosomes 2, 3, 5, and 13. Table 3 shows a summary of the maximum het-TLOD and NPL scores found for each of these regions.
Figure 1.
Multipoint parametric and nonparametric linkage scores across the genome (22 autosomes in chromosomal order) for core (a), spectrum (b), and broad (c) diagnostic classifications.
Table 3.
Maximum het-TLOD and NPL Scores for Regions on Chromosomes 2, 3, 5, and 13
| Chromosome 2a | Chromosome 3b | Chromosome 5b | Chromosome 13a | |||||
|---|---|---|---|---|---|---|---|---|
| DiagnosticClassification | het-TLODc | NPL | het-TLODc | NPL | het-TLODd | NPL | het-TLODc | NPL |
| Core | 4.8 R | 6.5 | 1.3 R | .5 | 1.0 D | 1.1 | 2.5 R | 1.4 |
| Spectrum | 4.4 R | 4.5 | 1.3 R | .5 | 1.7 D | 1.0 | 3.5 R | 1.3 |
| Broad | 3.6 R | 3.4 | 2.0 R | .5 | 2.5 D | .9 | 3.6 R | 1.1 |
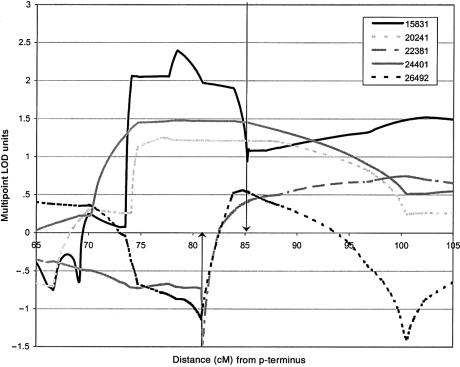
A genomewide significant result was found on chromosome 2p13-14, under the core diagnostic classification. The maximum NPL score of 6.5 and the maximum het-TLOD (4.8 under a recessive mode of inheritance) were observed at marker D2S358, ∼85 cM from the p-terminus. For both statistics, the score in the region was above the suggestive threshold (i.e., 2.1) from D2S378 to D2S1394 (∼14 cM) and above 1.0 from D2S2240 to D2S410 (∼56 cM). For marker D2S358, θ was estimated to be .02, and the proportion of families linked (α) was estimated to be 1.0, suggesting homogeneity. The value for θ remained low throughout the region (maximum θ=.05), with some evidence for heterogeneity apparent for markers toward the p-terminal end of the region (minimum α=.6). For further localization, we calculated standard multipoint by-pedigree LOD scores (θ fixed at 0). Table 4 shows the maximum by-pedigree recessive multipoint LOD scores across the region of interest on chromosome 2p13-14, under the core diagnostic classification. Dominant scores are also shown, because the dominant resourcewide het-TLOD score was borderline significant (_het_-_TLOD_=3.4). However, since the recessive model performs better on the whole, we have concentrated on this model. We have chosen a by-pedigree threshold of 0.6, to signify probable linkage to the region, since a multipoint LOD score of 0.6 corresponds to a pointwise (uncorrected) P value of .05. It can be seen that five of the seven pedigrees score ⩾0.6 under the recessive model, indicating that some genetic heterogeneity for this locus may exist. Figure 2 shows the recessive multipoint LOD traces for the five linked pedigrees. These traces may be used to estimate boundaries that delimit the region of interest. A large (>0.5) decrease in LOD score and a sharp change in gradient is indicative of an informative recombinant event in a pedigree. The base denotes the most outward point of the recombination event. A more gradual change in gradient indicates that the markers (or the string of markers) between which the recombinant event takes place are not fully informative. In this latter case, the addition of more markers could lead to better definition of the region. By this method, pedigrees 15831, 26492, and 22381 define recombinant boundaries encompassing a 4.3-cM region. Pedigree 15831 defines the q-boundary (toward the q-terminus) as being at 85.19 cM, and pedigrees 26492 and 22381 define the p-boundaries (toward the p-terminus) as being at 80.82 and 80.92 cM, respectively. The markers delimiting these inferred recombinant events are D2S136/D2S134 and D2S327.
Table 4.
Maximum Parametric By-Pedigree Multipoint LOD Scores Calculated under the Classification/Best Model Combination for Each Region
| Chromosome 2 | Chromosome 3 | Chromosome 5 | Chromosome 13 | |
|---|---|---|---|---|
| Region | D2S378–D2S1394 | D3S2398–qter | D5S2008–qter | D13S1493–D13S325 |
| Diagnostic classification/best model | Core/Recessive (Dominantb) | Broad/Recessive | Broad/Dominant | Broad (Spectrumc)/Recessive |
| Pedigree:a | ||||
| 15831 | 2.4 (2.8)d | .7 | 3.5 | −.2 (−.1) |
| 15841 | −.2 (−3.0) | .7 | −1.6 | 1.1 (.7) |
| 20241 | 1.2 (−.3) | .3 | −1.5 | .9 (.8) |
| 22381 | .6 (−1.1) | .3 | −1.2 | .6 (.8) |
| 24401 | 1.5 (.7) | .1 | 0 | .4 (.4) |
| 26491 | .4 (.6) | .5 | −.5 | .1 (.2) |
| 26492 | .6 (−.4) | .4 | 0 | 1.1 (1.1) |
Figure 2.
Recessive multipoint LOD traces for the five pedigrees with linkage to the region on chromosome 2
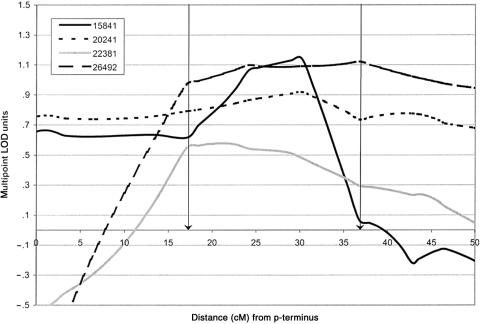
A second significant genomewide result was found for a region on chromosome 13q12-22, under a recessive mode of inheritance, for both the broad and spectrum diagnostic classifications. The maximum score was a recessive het-TLOD score of 3.6, under the broad classification, at marker D13S894, ∼24 cM from the p-terminus. An almost equivalent score was found for the same marker under the spectrum diagnostic classification (recessive _het_-_TLOD_=3.5). The recessive score was above the suggestive threshold between markers D13S1493 and D13S325 (∼13 cM) and was above 1.0 between markers D13S1493 and D13S317 (∼38 cM). The NPL scores in the region were much lower (1.1 and 1.3, under broad and spectrum diagnostic classifications, respectively) and were not significant. For both classifications, the estimates for θ and α were 0 and 1, respectively, at the peak marker; however, some evidence for heterogeneity was found to exist toward the p-terminal end of the region (α=.86). Table 4 shows the maximum by-pedigree recessive multipoint LOD scores across the region of interest on chromosome 13q12-22, under both the broad and spectrum diagnostic classifications. We focused on the broad classification since it performed marginally better; however, conclusions were equivalent under the spectrum classification. For this region, four of the seven pedigrees scored ⩾0.6, indicating genetic heterogeneity, which was not apparent from the het-TLOD analysis. In figure 3, the recessive multipoint LOD traces for the four linked pedigrees under the broad diagnostic classification are shown. There were no sharp changes in gradient. The estimated p- and q-boundaries for this region were both gradual gradient changes and were both from recombinants in pedigree 15841. The p-boundary was found at 17.03 cM, and the q-boundary was found at 36.78 cM, defining a 19.75-cM region between markers D13S1493 and D13S788. The gradual gradient changes for these estimated boundaries indicate that the exact positions of the recombinants are not well identified. The addition of informative and highly polymorphic markers around the boundary regions could allow better determination of the location of these boundary-defining recombinant events.
Figure 3.
Recessive multipoint LOD traces for the four pedigrees with linkage to the region on chromosome 13
A region on chromosome 5q22-qter, with genomewide intermediate evidence for linkage, was found under a dominant model for the broad diagnostic classification (dominant _het_-_TLOD_=2.5; θ=0; α=.2). The identified region (defined by a dominant het-TLOD score of ⩾1.0) lies between marker D5S2008 at 190 cM to the most telomeric marker, a distance of ∼15 cM. Table 4 shows the maximum by-pedigree dominant multipoint LOD scores across the region. These results are concordant with the resourcewide dominant het-TLOD results and suggest that much heterogeneity exists for this region. In fact, this intermediate hint seems to be pedigree specific (for pedigree 15831, by-pedigree multipoint _LOD_=3.5).
A borderline genomewide suggestive region on chromosome 3q24-28 was identified, under a recessive model and the broad diagnostic classification (recessive _het_-_TLOD_=2.0; θ=0; α=1). The score remained ⩾1.0 from marker D3S2398 at 204 cM to the most telomeric marker, ∼16 cM. Table 4 shows that, although all by-pedigree scores were positive, most linkage signals were weak, and the scores for only two pedigrees were at or above the by-pedigree threshold of 0.6 (pedigrees 15831 and 15841, which both scored 0.7).
Discussion
Our results suggest that the chromosomal regions of 2p13-14 and 13q12-22 contain genes involved in susceptibility to SCZD in Palau. Our analyses indicated that the locus on 2p13-14 is within a 4.3-cM region. Furthermore, the region could potentially be narrowed, by use of markers alone, to 0.8 cM. The strength of the linkage evidence (under core diagnostic classification, _NPL_=6.5 and _het_-_TLOD_=4.8), in addition to the small size of the region, makes the 2p13-14 region a strong candidate for gene isolation via positional cloning. There are several candidate genes in this region, including ASCT1, a neutral amino acid transporter, which was previously screened in some affecteds in pedigree 15831, with only polymorphisms not segregating with the disease found (Bennett et al. 2000). Two studies (DeLisi 1997; Faraone et al. 1998) have found nominal evidence in this region, although most studies have not found evidence here. Hence, the locus on 2p13-14 may be of smaller effect in other populations.
The second locus, on chromosome 13q12-22, was narrowed to a 19.75-cM region. By the addition of markers alone, this region has the potential to be reduced to ∼6 cM in size. Hence, for this region, further Palauan pedigrees or an alternative pedigree resource will have to be utilized to progress to gene isolation. Other studies have identified evidence for linkage to 13q, although these have generally found it at points more distal than our defined region. Blouin et al. (1998) reported significant linkage at 13q32, and this was confirmed by Brzustowicz et al. (1999); in addition, Lin et al. (1995, 1997) and Shaw et al. (1998) have found linkage evidence for this more distal region, although others (e.g., Levinson et al. [2000]) have not. Furthermore, evidence for linkage to bipolar disorder has been found in this region (Kelsoe et al. 2001).
Two other regions in the genome, with lower evidence for linkage, were found, on chromosomes 5q22-qter and 3q24-28. There have been several reports of linkage to the 5q22-qter region, in different populations. The first report of linkage was in a set of Irish families (Straub et al. 1997), followed by confirmation in 13 British families (Gurling et al. 2001). Linkage has also been reported in Finnish families (Paunio et al. 2000) and German families (Schwab et al. 1997). In addition, Edenberg et al. (1997) found, in this region, evidence for linkage to bipolar disorder. In our analyses, the signal at 5q22-qter was the consequence of a single pedigree (pedigree 15831) with a multipoint LOD score of 3.5. However, the consistent evidence for linkage in the same region in other populations makes this region one of interest and worthy of further investigation. The region on chromosome 3q24-28 is our lowest hint region and has poor by-pedigree support. For SCZD phenotypes, it has not been identified by other groups, although Kelsoe et al. (2001) found evidence for linkage to bipolar disorder. This region is of only tentative interest, on the basis of our current results.
Full-chromosome multipoint analyses were performed in this study, which are advantageous for several reasons. Multipoint linkage analyses are more robust to false positives, which are associated with two-point analyses, and hence are more reliable. In addition, a positive result implies a segregating haplotype, rather than a single marker; hence, a positive result implies flanking support for the region. Furthermore, it increases the informativeness of the markers used in the analysis. This increased informativeness proved important in our current analyses, since several genomic-search markers were not as polymorphic in Palauans as in other populations. This, in addition to the ability to analyze the full-pedigree structures by Markov-chain Monte Carlo methods, enabled us to more fully exploit the power of the resource. Here, the utility of haplotype information in linkage analysis was illustrated by the region on chromosome 5q22-qter. This region was not previously identified in the two-point analyses performed on pedigree 15831 (Coon et al. 1998). In the present study, pedigree 15831 scored >2.0, both by TLOD and standard multipoint LOD analysis, under a core diagnostic classification and dominant model, as used by Coon et al. (1998). A similar observation was made recently by Garner et al. (2001), who identified a new region of interest for severe bipolar disorder, by utilizing Markov-chain Monte Carlo techniques to perform multipoint analyses in a single complex pedigree.
Three nested diagnostic classifications were used in the analyses presented here. For each classification, a different genetic model was implemented for the parametric analyses. The incremental changes in the number of individuals considered as affected under these three classifications was small (table 1). However, for some loci, large differences in linkage evidence, between classifications, were found. Caution must be exercised when conclusions are drawn on the basis of analyses performed both under dynamic parametric genetic models and under diagnostic classifications, since any conclusions regarding diagnostic classification and changes in genetic parameters are confounded. To attempt to distinguish between the effect of diagnostic classification and changes in genetic parameters, we examined both the by-pedigree parametric and the by-pedigree nonparametric results, the latter of which was not influenced by change in model parameters. For chromosome 2p13-14, the difference in linkage evidence observed, between core and spectrum, was due to the performance of unlinked pedigrees under the spectrum genetic model, with sharing (assessed nonparametrically) in linked pedigrees unchanged between the two classifications. However, there was evidence for reduced sharing in pedigree 15831 under the broad classification. This indicates that chromosome 2p13-14 could be influential in spectrum diagnoses, in addition to core diagnoses, but that it is not associated with affective psychoses. For chromosome 13q12-22, there was evidence for increased sharing in pedigree 15841 under the spectrum diagnostic classification versus the core diagnostic classification, but there was no additional sharing evident under the broad classification. All other pedigrees are consistent in sharing, over the three classifications, as would be expected (table 1). These results would suggest that the locus on 13q12-22 is a susceptibility locus for spectrum SCZD and that evidence of linkage to the broad phenotype may be due to a genetic model that better fitted the data, rather than to the additional sharing of individuals that was evident under the broad classification. For the single-pedigree region on chromosome 5q22-qter, both the broad classification and the change in model parameters influenced the linkage evidence, suggesting that this locus may also predispose to affective psychoses. Finally, for the region on chromosome 3q24-28, there was evidence that sharing increased under the spectrum diagnostic classification versus the core diagnostic classification and further increased under the broad classification, suggesting that this locus may also be involved in a more extensive set of psychiatric diagnoses.
Our results suggest that use of a range of parametric models and of phenotypes may be advantageous for gene localization. The cost:benefit ratio for multiple testing must, of course, be considered, since a correction for the multiple analyses performed must be made. However, the phenotypes are likely to be related, as are the genetic models considered, and the correction may be acceptable compared to the potential advantage of increased power to detect linkage. In this study, the phenotypes and genetic parameters were both nested. In addition, the incremental changes across the three diagnostic classifications were small. This led to a set of highly correlated analyses (_N_=1.7) and a relatively modest multiple-testing correction (linkage thresholds in LOD scores were raised by 0.2).
It is clear that SCZD is a disease with complex inheritance and with challenges both genetic and phenotypic. Our results support the use of a comprehensive set of diagnostic classifications for linkage analysis, the utility of haplotype information, and the use both of robust parametric linkage statistics (Göring and Terwilliger 2000; Abkevich et al., in press) and of nonparametric linkage statistics. Our findings provide significant evidence for susceptibility loci on chromosomes 2p13-14 and 13q12-22 and support a model of genetic heterogeneity. Furthermore, our findings suggest that loci identified in a Palauan population may be both distinct and shared with those in other populations.
Acknowledgments
We would like to thank William Byerley for his contribution to ascertainment, phenotyping, and genotyping. We would also like to thank Fred Reimherr and Paul Wender, for their role in establishing diagnostic reliability, and Mark Hoff, Jude Rosenthal, and Pamela Bennett, for their technical skills with regard to genotyping. This research was supported by National Institute of Mental Health research grants R01-MH54186 (to M.M.-W.) and R01-MH560908 (to William Byerley), a NARSAD Independent Investigator award (to M.M.-W.), and a NARSAD Senior Investigator award (to William Byerley). We thank Myriad Genetics, and Alun Thomas especially, for permission to use their suite of proprietary software, to perform the necessary multipoint analyses.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Cooperative Human Linkage Center, The, http://lpg.nci.nih.gov/CHLC/
- Genome Database, The, http://gdbwww.gdb.org/ (for information on genetic mapping)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCZD [MIM 181500])
References
- Abkevich V, Camp NJ, Gutin A, Farnham JM, Cannon-Albright L, Thomas A. A robust multipoint linkage statistic (TLOD) for mapping complex trait loci. Genet Epidemiol (in press) [DOI] [PubMed] [Google Scholar]
- Baron M (2001) Genetics of schizophrenia and the new millennium: progress and pitfalls. Am J Hum Genet 68:299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Gruen R, Asnis L, Kane J (1982) Schizoaffective illness, schizophrenia and affective disorders: morbidity risk and genetic transmission. Acta Psychiatr Scand 65:253–262 [DOI] [PubMed] [Google Scholar]
- Bell G, Karam J, Rutter W (1981) Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci USA 78:5759–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett PJ, Hoff M, Rosenthal J, Zhao M, Coon H, Myles-Worsley M, Byerley W (2000) Mutation screening of a neutral amino acid transporter, ASCT1, and its potential role in schizophrenia. Psychiatr Genet 10:79–82 [DOI] [PubMed] [Google Scholar]
- Berrettini WH (2000) Are schizophrenic and bipolar disorders related? A review of family and molecular studies. Biol Psychiatry 48:531–538 [DOI] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyneic PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 [DOI] [PubMed] [Google Scholar]
- Böök JA, Wetterberg L, Modrzeewska K (1978) Schizophrenia in a North Swedish geographical isolate, 1900–1977: epidemiology, genetics and biochemistry. Clin Genet 14:373–394 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EWC, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EWC, Little D, Hogen J, Hodgkinson K, Bassett A (1999) Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 65:1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp NJ, Farnham JM. Correcting for multiple analyses in genomewide linkage studies. Ann Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Camp NJ, Gutin A, Abkevich V, Farnham JM, Cannon-Albright L, Thomas A. A new non-parametric linkage (NPL) statistic for mapping both qualitative and quantitative trait loci. Genet Epidemiol (in press) [DOI] [PubMed] [Google Scholar]
- Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J (1986) Effects of misspecifying genetic parameters in lod score analysis. Biometrics 42:393–399 [PubMed] [Google Scholar]
- Coon H, Jensen S, Holik J, Hoff M, Myles-Worsley M, Reimherr F, Wender P, Waldo M, Freedman R, Leppert M, Byerley W (1994) Genomic scan for genes predisposing to schizophrenia. Am J Med Genet 54:59–71 [DOI] [PubMed] [Google Scholar]
- Coon H, Myles-Worsley M, Tiobech J, Hoff M, Rosenthal J, Bennett P, Reimherr F, Wender P, Dale P, Polloi A, Byerley W (1998) Evidence for a chromosome 2p13-14 schizophrenia susceptibility locus in families from Palau, Micronesia. Mol Psychiatry 3:521–527 [DOI] [PubMed] [Google Scholar]
- Davison K (1983) Schizophrenia-like psychoses associated with organic cerebral disorders: a review. Psychiatr Dev 1:1–33 [PubMed] [Google Scholar]
- DeLisi L (1997) A genome scan for schizophrenia. Paper presented at The 1997 Park City Molecular Psychiatry Conference. Park City, UT, February 2–5 [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, Conneally PM, Sorbel JJ, Carr K, Crose C, Willig C, Zhao J, Miller M, Bowman E, Mayeda A, Rau NL, Smiley C, Rice JP, Goate A, Reich T, Stine OC, McMahon F, DePaulo JR, Meyers D, Detera-Wadleigh SD, Goldin LR, Gershon ES, Blehar MC, Nurnberger JI (1997) Initial genomic scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 3, 5, 15, 16, 17, and 22. Am J Med Genet 74:238–246 [PubMed] [Google Scholar]
- Elston RC, Namboodri KK, Spence MA, Rainer JD (1978) A genetic study of schizophrenia pedigrees. II. One locus hypotheses. Neuropsychobiology 4:193–206 [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL (1978) A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry 35:837–844 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, Markel P, Lee H, Harkavry-Freidman J, Kaufmann C, Cloninger CR, Tsuang MT (1998) Genome scan of European-American schizophrenia pedigrees: results of the NIMH Genetics Initiative and Millenium Consortium. Am J Med Genet 81:290–295 [PubMed] [Google Scholar]
- Garner C, McInnes LA, Service SK, Spesny M, Fournier E, Leon P, Freimer NB (2001) Linkage analysis of a complex pedigree with severe bipolar disorder, using a Markov chain Monte Carlo method. Am J Hum Genet 68:1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HHH, Terwilliger JD (2000) Linkage analysis in the presence of errors. I. Complex-valued recombination fractions and complex phenotypes. Am J Hum Genet 66:1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II (1991) Schizophrenia genesis: the origins of madness. WH Freeman, New York [Google Scholar]
- Gottesman II, Shields J (1982) Schizophrenia: the epigenetic puzzle. Cambridge University Press, New York [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurling HMD, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CS, Kong A, Kjaerulff U (1995) Blocking-Gibbs sampling in very large probabilistic expert systems. Int J Hum Comput Stud 42:647–666 [Google Scholar]
- Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H (2001) A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA 98:585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS (1983) Overview: a current perspective on twin studies of schizophrenia. Am J Psychiatry 140:1413–1425 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Diehl SR (1993) The genetics of schizophrenia: a current genetic-epidemiologic perspective. Schizophr Bull 19:261–285 [DOI] [PubMed] [Google Scholar]
- Kendler KS, O'Neill FA, Burke J, Murphy B, Duke F, Straub RE, Shinkwin R, Nuallain MN, MacLean CJ, Walsh D (1996) Irish study on high-density schizophrenia families: field methods and power to detect linkage. Am J Med Genet 67:179–190 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Holmans P, Straub RE, Owen MJ, Wildenauer DB, Gejman PV, Pulver AE, Laurent C, Kendler KS, Walsh D, Norton N, Williams NM, Schwab SG, Lerer B, Mowry BJ, Sanders AR, Antonarakis SE, Blouin JL, DeLeuze JF, Mallet J (2000) Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: Schizophrenia Linkage Collaborative Group III. Am J Hum Genet 67:652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MW, Curtis D, Williams N, Arranz M, Nanko S, Collier D, McGuffin P, Murray R, Owen M, Gill M, Powell J (1995) Suggestive evidence for linkage of schizophrenia to markers on chromosome 13q14.1-q32. Psychiatr Genet 5:117–126 [DOI] [PubMed] [Google Scholar]
- Lin MW, Sham P, Hwu HG, Collier D, Murray R, Powell JF (1997) Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum Genet 99:417–420 [DOI] [PubMed] [Google Scholar]
- Maier W, Lichtermann D, Minges J, Heun R, Hallmayer J (1993) The impact of gender and age at onset on the familial aggregation of schizophrenia. Eur Arch Psychiatry Clin Neurosci 242:279–285 [DOI] [PubMed] [Google Scholar]
- Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Manion F, Quillen J, et al (1994) A comprehensive human linkage map with centimorgan density: Cooperative Human Linkage Center (CHLC). Science 265:2049–2054 [DOI] [PubMed] [Google Scholar]
- Myles-Worsley M, Coon H, Tiobech J, Collier J, Dale P, Wender P, Reimherr F, Polloi A, Byerley W (1999) Genetic epidemiological study of schizophrenia in Palau, Micronesia: prevalence and familiality. Am J Med Genet 88:4–10 [DOI] [PubMed] [Google Scholar]
- O'Rourke DH, Gottesman II, Suarez BK, Rice J, Reich T (1982) Refutation of the general single-locus model for the etiology of schizophrenia. Am J Hum Genet 34:630–649 [PMC free article] [PubMed] [Google Scholar]
- Ott J (1986) Linkage probability and its approximate confidence interval under possible heterogeneity. Genet Epidemiol Suppl 1:251–257 [DOI] [PubMed] [Google Scholar]
- Paunio T, Ekelund J, Hovatta I, Varilo T, Terwilliger JD, Meyer J, Parker A, Maruti S, Suvisaari J, Arajarvi R, Partonen T, Juvonen H, Lonnqvist J, Peltonen L (2000) Genomewide scan of an extended Finnish schizophrenia study sample. Am J Med Genet 96:460 [Google Scholar]
- Risch N, Baron M (1984) Segregation analysis of schizophrenia and related disorders. Am J Hum Genet 36:1039–1059 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Claus E, Giuffra L (1989) Linkage and mode of inheritance in complex traits. Prog Clin Biol Res 329:183–188 [PubMed] [Google Scholar]
- Risch N, Giuffra (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed] [Google Scholar]
- Saccone S, De Sario A, DellaValle G, Bernardi G (1992) The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes. Proc Natl Acad Sci USA 89:4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab S, Eckstein GN, Hallmayer J, Lerer B, Albus M, Borrmann M, Lichtermann D, Ertl MA, Maier W, Wildenauer DB (1997) Evidence suggestive of a locus on chromosome 5q31 contributing to susceptibility for schizophrenia in German and Israeli families by multipoint affected sib-pair linkage analysis. Mol Psychiatry 2:156–160 [DOI] [PubMed] [Google Scholar]
- Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, Laval SH, Vita A, De Hert M, Cardon LR, Crow TJ, Sherrington R, DeLisi LE (1998) A genomewide search for schizophrenia susceptibility genes. Am J Med Genet 81:364–376 [DOI] [PubMed] [Google Scholar]
- Simmons RT, Graydon JJ, Gajdusek DC, Brown P, Reisenberg SH (1965) Blood group genetic variations in natives of the Caroline Islands and in other parts of Micronesia. Oceania 36:132–170 [Google Scholar]
- Spitzer RL, Endicott J, Robins E (1978) Research diagnostic criteria: rationale and reliability. Arch Gen Psychiatry 35:773–782 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O'Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Webb BT, Zhang J, Walsh D, Kendler KS (1995) A potential vulnerability locus for schizophrenia on chromosome 6p24-22: evidence for genetic heterogeneity. Nat Genet 11:287–293 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O'Neill FA, Walsh D, Kendler KS (1997) Support for a possible schizophrenia vulnerability locus in region 5q22-31 in Irish families. Mol Psychiatry 2:148–155 [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, Van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR (eds) Genetic approaches to mental disorders. American Psychiatric Association, Washington, DC [Google Scholar]
- Thomas A, Gutin A, Abkevich V, Bansal A (2000) Multilocus linkage analysis by blocked Gibbs sampling. Stat Comput 10:259–269 [Google Scholar]
- Tsuang MT, Bucher KD, Fleming JA (1982_a_) Testing the monogenic theory of schizophrenia: an application of segregation analysis to blind family study data. Br J Psychiatry 140:595–599 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Simpson JC, Kronfol Z (1982_b_) Subtypes of drug abuse with psychosis: demographic characteristics, clinical features, and family history. Arch Gen Psychiatry 39:141–147 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Greenberg DA, Hodge SE (1993) Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered 43:329–336 [DOI] [PubMed] [Google Scholar]
- Wang S, Sun C, Walczak CA, Ziegle JS, Kipps BR, Goldin LR, Diehl SR (1995) Evidence for a susceptibility locus for schizophrenia on chromosome 6pter-p22. Nat Genet 10:41–46 [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]