Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase (original) (raw)
Abstract
Selenoprotein R (SelR) is a mammalian selenocysteine-containing protein with no known function. Here we report that cysteine homologs of SelR are present in all organisms except certain parasites and hyperthermophiles, and this pattern of occurrence closely matches that of only one protein, peptide methionine sulfoxide reductase (MsrA). Moreover, in several genomes, SelR and MsrA genes are fused or clustered, and their expression patterns suggest a role of both proteins in protection against oxidative stress. Consistent with these computational screens, growth of_Saccharomyces cerevisiae_ SelR and MsrA mutant strains was inhibited, and the strain lacking both genes could not grow, in the presence of H2O2 and methionine sulfoxide. We found that the cysteine mutant of mouse SelR, as well as the_Drosophila_ SelR homolog, contained zinc and reduced methionine-_R_-sulfoxide, but not methionine-_S_-sulfoxide, in _in vitro_assays, a function that is both distinct and complementary to the stereo-specific activity of MsrA. These findings identify a function of the conserved SelR enzyme family, define a pathway of methionine sulfoxide reduction, reveal a case of convergent evolution of similar function in structurally distinct enzymes, and suggest a previously uncharacterized redox regulatory role of selenium in mammals.
Dietary selenium has many biological functions and biomedical applications. Its essential role in mammals is explained by the fact that it is present in several proteins as selenocysteine (Sec), the 21st amino acid in protein that is inserted cotranslationally in response to TGA codons (1). With the exception of Sec, which is typically located in enzyme active centers and is essential for catalytic activity, previously characterized selenoproteins lack any common amino acid motifs or patterns. However, selenoprotein genes contain a common mRNA stem–loop structure, designated the Sec insertion sequence (SECIS) element that is necessary for incorporation of Sec at TGA codons (2).
Bioinformatics methods have recently been developed that allow detection of SECIS elements in large nucleotide sequence databases (3–6). Identification of SECIS elements led to the discovery of several previously uncharacterized selenoproteins. Because of the lack of sequence homology to known proteins, further functional characterization of these selenoproteins has been difficult. However, recently developed bioinformatics techniques of domain fusion patterns (7, 8), phylogenetic profiles (correlated evolution), gene clustering, and expression profiling (9) offer new tools for prediction of protein function that are independent of homology analyses (10). In this report, we used these methods for functional characterization of mammalian selenoprotein R (SelR) (3), also called SelX (4). SelR is a small (≈12 kDa) Sec-containing protein. Homologs of this protein were previously identified in bacteria, archaea and eukaryotes, and with the exception of vertebrate SelR, all homologs contained cysteine in place of Sec.
Our in silico analyses initially suggested a functional linkage of SelR to a pathway of methionine sulfoxide reduction, as well as the role of SelR in protection against oxidative stress and/or redox regulation of cellular processes. The methionine sulfoxide reduction pathway has been previously characterized in considerable detail. Its principal component, peptide methionine sulfoxide reductase (MsrA), catalyzes thioredoxin-dependent methionine sulfoxide reduction (11, 12). However, although MsrA accounts for most of the cellular methionine sulfoxide reductase activity, the role of this protein as the sole peptide methionine sulfoxide reductant contrasts with experimental data. For example, bacterial, yeast, and mammalian MsrA gene knockout cells exhibit residual methionine sulfoxide reductase activity (13–15), and recent data indicate that MsrA is stereo-specific for methionine-_S_-sulfoxide derivatives (16, 17). Thus, even in the presence of fully active and abundant MsrA, the possibility remained for a potentially detrimental accumulation of methionine-_R_-sulfoxides in cellular proteins. Several possibilities have been raised to explain these puzzling observations, including the suggestion that a methionine-R,_S_-sulfoxide racemase might be present in organisms (17). However, no data have yet been reported that offer an explanation of this phenomenon.
We tested catalytic activities of mouse and _Drosophila_SelR that are relevant to the pathway of methionine sulfoxide reduction and found that these proteins can reduce methionine-R_-sulfoxide, but are not active with the_S stereoisomer. This report provides an example of how computational analyses of genome databases and subsequent experimental testing of functional predictions turned characterization of a mammalian selenoprotein into definition of an important metabolic pathway in all three major domains of life.
Experimental Procedures
Bioinformatics Methods.
Patterns of gene occurrence were initially analyzed in 44 (33 bacterial, 9 archaeal, and 2 eukaryotic) completely sequenced genomes by using Clusters of Orthologous Groups (COG) (http://www.ncbi.nlm.nih.gov/COG). SelR homologs formed aCOG02299 cluster that was represented by 33 sequences in 29 genomes. The COG Phylogenetic Patterns and Phylogenetic Patterns Search tools were then used to identify genes with similar phylogenetic profiles among 2,189 patterns present in COG. One protein, MsrA (COG0225), had the closest (with respect to SelR) expression pattern. The MsrA cluster was represented by 37 sequences in 31 genomes. Subsequently, BLAST analyses were used to scan sequences of interest (SelR and MsrA) in all completely and incompletely sequenced genomes, using the NCBI Microbial Genomes BLAST. At the time of analysis, 127 bacterial genomes including 51 completely sequenced, and 14 archaeal genomes including 11 completely sequenced, were available (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi). SelR and MsrA genes were also analyzed in completely sequenced eukaryotic genomes.
Domain fusion events were identified by searching the Protein Families (Pfam) database (http://www.sanger.ac.uk/Software/Pfam), using the Domain Organization tool. SelR was designated in Pfam as Domain of Unknown Function 25 and formed a PF01641 cluster, whereas MsrA was designated as PMSR and was a member of the PF01625 cluster. This analysis was complemented by analysis of COG fusion proteins containing either SelR or MsrA domains and by case-by-case analyses of SelR and MsrA protein sequences in 141 completely and incompletely sequenced microbial genomes. The latter analyses were also used to identify genes that flanked SelR and MsrA genes (gene clustering). Expression patterns of yeast and Escherichia coli SelR and MsrA homologs were determined by analyzing publicly available expression data generated with cDNA microarrays and provided by ExpressDB (http://arep.med.harvard.edu/ExpressDB) and Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces).
Construct of SelR Cysteine Mutant for Overexpression in E. coli.
PCR fragment containing the full-size coding region of mouse SelR (Cys mutant) was amplified with primers U_SELRM68 (5′-GGTGCG_CATATG_TCGTTCTGCAGCTTCTTCGGAGGC-3′) and L_SELRM397 (5′-CGTA_CTCGAG_GTGCCCCTGGGAGGCAGCAGC-3′), and cloned into the _Nde_I and _Xho_I sites of pET 21d(+) expression vector (Novagen).
Isolation of Recombinant SelR.
Mouse Cys-for-Sec SelR mutant was expressed in the BL21(DE3)E. coli strain. For protein isolation, cells were resuspended at 4°C in 25 mM Tris⋅HCl (pH 7.5), containing 1 mM EDTA, 1 mM PMSF, 1 mM DTT, 5 μg/ml leupeptin, and 5 μg/ml aprotinin, and further treated by sonication. Insoluble material was removed by centrifugation, and the supernatant was applied to a 100-ml DEAE-Sepharose column equilibrated with 25 mM Tris⋅HCl (pH 7.5), containing 1 mM EDTA and 1 mM DTT (buffer A). The flow-through fraction was collected and applied directly to a Heparin HPLC column equilibrated in 25 mM Tris⋅HCl (pH 6.0; buffer B). After washing the column with 2 volumes of buffer B, the bound proteins were eluted by application of a linear gradient from buffer B to 0.5 M NaCl in buffer B. SelR was eluted from the column as a major peak at ≈150 mM NaCl. The protein was analyzed for purity by SDS/PAGE and stored at −80°C until ready for use.
Metal Content of SelR.
Various mouse and Drosophila SelR samples were analyzed for the presence of 20 biologically relevant metals, using inductively coupled argon plasma (ICAP) at the Chemical Analysis Laboratory, University of Georgia. In parallel, control (buffer) samples were prepared and analyzed by ICAP. Zn was not detected in the control samples.
Determination of Methionine Sulfoxide Activity.
The reduction of protein-bound methionine sulfoxides was assayed using dabsyl methionine sulfoxide as described by Moskovitz et al.(14). Dabsyl derivatives of L-methionine and L-methionine-R,_S_-sulfoxides were prepared as described by Minetti et al. (18). L-Methionine-_S_-sulfoxide and L-methionine-_R_-sulfoxide were prepared from L-methionine-R,_S_-sulfoxide according to the method of Lavine (19).
Growth of Yeast Strains.
S. cerevisiae wild-type (WT) strain BY4741 (MAT_α his3D1leu2D0met15D0ura3D0) and its isogenic SelR:KANR mutant (YCL033c) were purchased from ResGen, Huntsville, AL. To isolate_MsrA single mutant (GY4) and MsrA SelR double mutant (GY5), two primers (P1, P2) containing 45-bp and 52-bp_MsrA_-specific sequences and 22-bp and 20-bp plasmid-specific sequences were designed to amplify URA3 marker from pRS306 by PCR. The PCR product was used to transform the WT and_SelR_:KANR mutant strains to isolate MsrA single mutants and MsrA SelR double mutants, respectively. URA_+ transformants for single and_URA+/KANR transformants for double mutants were selected and screened by PCR using primer P1 and an out of ORF primer (P3) to confirm the deletion of MsrA. The sequences of the primers are as follows: P1, AGTTATATTCTAACGTCAAAAACTTAACAAGTAAACGAAGTTGCCTGTGCGGTATTTCACACCG; P2, CATTCATGCACTTGACTTTTTTTCATAAATAAGGGCACGTACACTAAAAAAGAGATTGTACTGAGAGTGCAC; and P3, ACAGCTAACAATCCTATCACAG. Cells from stationary phase cultures were grown aerobically in yeast extract/peptone/dextrose or yeast nitrogen base minimal medium (YNB) containing indicated supplements at 30°C. The growth was monitored by absorbance at 600 nm.
Drosophila SelR.
Drosophila SelR cDNA sequence was determined by sequencing Drosophila EST clones (ResGen) that contained SelR ORF. The expression construct pET28_SelR was obtained by cloning the SelR gene into the pET28 a(+) vector (Novagen) designed for expression of N-terminal 6-His tagged proteins. The SelR coding region was amplified by PCR using oligonucleotides AGTCTAACATATGGATAACAAGAGCGAGAAGG and AGACAACTCGAGTCACTGCTGGGCAATGGGCG. The PCR product was inserted into pET28a(+) by using the_Xho_I and _Nde_I sites and verified by sequencing. The N-terminal His-tagged Drosophila SelR was purified from an E. coli strain BL21 (DE3). Cells were resuspended in 50 mM sodium phosphate (pH 7.0), containing 300 mM NaCl and 1 mM PMSF, and lysed by sonication. The resulting crude extract was centrifuged at 30,000 × g for 30 min, and the supernatant loaded on a 2-ml Talon metal affinity column (CLONTECH) equilibrated with the extraction buffer. Proteins were eluted by an imidazole elution buffer and the protein fractions were analyzed for purity by SDS/PAGE.
Results
Occurrence of SelR in Completely Sequenced Genomes.
SelR, being a member of the original minimal gene set (20), was recognized as one of the proteins required for cellular life. However, as the sequences of additional genomes became available, it was clear that SelR homologs are absent in several organisms. We analyzed in detail the pattern of occurrence of SelR genes in all available completely sequenced genomes and identified 15 bacterial and 9 archaeal genomes that lacked SelR genes (Table 1). These organisms were not organized in specific groups based on a kingdom-level phylogeny, suggesting that SelR homologs were lost independently during evolution. Further analysis revealed that these organisms could be classified as either hyperthermophiles or obligatory parasites. In addition, within thermophiles, SelR was absent in all acidophilic microorganisms. No clear correlation was observed between the presence of SelR and the aerobic/anaerobic nature of the organisms.
Table 1.
Patterns of gene occurrence, clustering, and domain fusion involving MsrA and SelR genes in completely sequenced genomes
| Organisms | MsrA/SelR occurrence/ clustering/fusion | Optimal growth temperature | Anaerobic | Optimal growth pH | Obligatory parasite |
|---|---|---|---|---|---|
| Eukaryotes | |||||
| Homo sapiens | M R R R | ||||
| Drosophila melanogaster | M R | ||||
| Caenorhabditis elegans | M R | ||||
| Arabidopsis thaliana | M (5) R (9) | ||||
| Saccharomyces cerevisiae | M R | ||||
| Archaea | |||||
| Aeropyrum pernix | − − | 90 | − | 7 | − |
| Sulfolobus solfataricus | M − | 70–85 | − | 4 | − |
| Sulfolobus tokodaii | − − | 80 | − | 2–3 | − |
| Archaeoglobus fulgidus | − − | 80–85 | + | 7 | − |
| Halobacterium sp. NRC-1 | M R | 37 | − | 7 | − |
| Methanothermobacter thermautotrophicus | M R | 60–65 | + | 7 | − |
| Methanococcus jannaschii | − − | 80–85 | + | 6 | − |
| Pyrococcus (abyssi,horikoshii) | − − | 95 | + | 7 | − |
| Thermoplasma (acidophilum,volcanium) | − − | 55–60 | − | 2 | − |
| Bacteria | |||||
| Aquifex aeolicus | − − | 85 | − | − | |
| Chlamydia (muridarum,trachomatis) | − − | + | |||
| Chlamydophila pneumoniae | − − | + | |||
| Synechocystis sp. PCC 6803 | M M R | − | |||
| Corynebacterium glutamicum | M R | 30 | − | 7 | − |
| Mycobacterium leprae | M − | + | |||
| Mycobacterium tuberculosis | M M | 37 | − | 7 | + |
| Bacillus halodurans | M M…R | 30 | − | 9.7 | − |
| Bacillus subtilis | M…R | 30 | − | 7 | − |
| Clostridium acetobutylicum | M R | 37 | + | − | |
| Mycoplasma(genitalium, pneumoniae,pulmonis) | M R | 37 | − | 7 | − |
| Ureaplasma urealyticum | M − | 37 | − | 7 | |
| Lactococcus lactis subsp.lactis | M M R | 30 | − | 7 | − |
| Staphylococcus aureus | M M M…R | 37 | − | 7 | − |
| Streptococcus pneumoniae | MR MR | 37 | − | 7 | − |
| Streptococcus pyogenes | M MR R | 37 | − | 7 | − |
| Caulobacter crescentus | M M R | 30 | − | 7 | − |
| Agrobacterium tumefaciens | M R | 26–30 | − | 7 | − |
| Mesorhizobium loti | M M R R | 26 | − | 7.2 | − |
| Sinorhizobium meliloti | M M R R | 26 | − | 7.2 | − |
| Rickettsia(conorii, prowazekii) | − − | + | |||
| Neisseria meningitidis | TMR | 37 | − | 7 | − |
| Campylobacter jejuni | M R | 37 | m | 7 | − |
| Helicobacter pylori | MR | 37 | m | 7 | − |
| Escherichia coli | M R | 37 | − | 7 | − |
| Yersinia pestis | M R | 37 | − | 7 | + |
| Buchnera sp. APS | − − | + | |||
| Vibrio cholerae | M MR R R | 28 | − | 7.6 | − |
| Xylella fastidiosa 9a5c | M R | 26 | − | 7 | − |
| _Haemophilus influenzae_Rd | MR | 37 | m | 7 | − |
| Pasteurella multocida | M R | 37 | − | 7 | − |
| Pseudomonas aeruginosa | M R | 30 | − | 7 | − |
| Borrelia burgdorferi | − − | + | |||
| Treponema pallidum | RM | 37 | m | 7 | − |
| Thermotoga maritima | − − | 80 | + | 6.5 | − |
| Deinococcus radiodurans | M R | 30 | − | 7 | − |
SelR− organisms with high optimal growth temperature were largely archaea, but two unrelated bacterial hyperthermophiles (Aquifex aeolicus and Thermotoga maritima) also lacked SelR genes. The absence of essential genes is not unusual for intracellular parasites, which have access to essential metabolic processes of the host. Thus, it is feasible that SelR or a set of genes involving SelR and its metabolic partners are essential for life of free-living organisms at moderate temperatures.
Correlated Evolution Functionally Links SelR to Methionine Sulfoxide Reduction.
SelR homology analyses revealed that all nonvertebrate SelR homologs contained cysteine in place of Sec (Fig.1). However, neither homology to functionally characterized proteins nor previously defined functional motifs/patterns were found in SelR. To obtain insight into SelR function, we attempted to identify proteins exhibiting patterns of gene occurrence similar to that of SelR. Surprisingly, one protein, MsrA, had a nearly identical phylogenetic profile, and no other proteins matched this pattern. This finding of correlated evolution of SelR and MsrA strongly suggested that these two proteins are functionally linked. SelR and MsrA gene occurrence patterns differed in that MsrA was present and SelR was absent in three genomes: archaeon hyperthermophile Sulfolobus solfataricus, and bacterial parasites Mycobacterium leprae and_Ureaplasma urealyticum_. This difference in phylogenic profiles was consistent with the loss of SelR in parasites and hyperthermophiles.
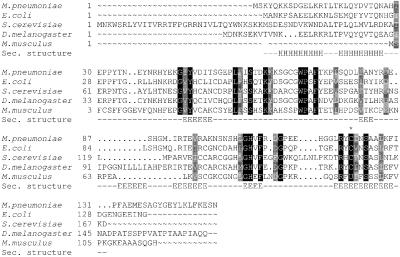
Figure 1.
Alignment of SelR homologs. SelR proteins relevant to the studies reported herein are shown in the figure. Sec. structure, secondary structure consensus for the SelR family (generated withjpred2 and sspro). In the secondary structure, H show α-helixes, and E β-strands. A star above the sequence shows the location of conserved selenocysteine (shown as U) and Cys.
Gene Clustering and Domain Fusion Link SelR and MsrA Functions.
All available completely sequenced genomes were analyzed to determine identities of genes that flanked SelR and MsrA genes. Interestingly, we found multiple examples of clustering of SelR and MsrA genes, suggesting a correlated expression of these proteins. Additional multiple examples of MsrA/SelR gene clustering were revealed by analyses of over 100 incompletely sequenced genomes, and in the majority of these clusters, MsrA genes were located immediately upstream of SelR genes.
MsrA and SelR were also often linked to each other forming two-domain fusion proteins (Table 1). Apparently, the two proteins could form fusions such that SelR could be located either downstream or upstream of MsrA. The strong tendency for independent domain fusion was also illustrated by the fact that the two related bacteria had different order of domains in the fusion: SelR-MsrA in Treponema pallidum and MsrA-SelR in Treponema denticola. In addition, we observed several examples of gene clustering versus domain fusion in closely related microorganisms (Table 1).
Neisseria fusion protein containing SelR and MsrA domains also included a thioredoxin-fold domain (Table 1). Thioredoxin is a previously characterized electron donor for MsrA. A fusion protein was also identified that contained two glutaredoxin domains upstream of MsrA, suggesting that the glutathione system may also serve as an electron source for methionine sulfoxide reduction in certain organisms. Finally, an Arabidopsis thaliana protein was identified that contained two SelR domains.
SelR Is Implicated in Protection Against Oxidative Stress.
SelR function was also studied by computational analyses of gene expression of bacterial and yeast SelR homologs in response to various treatments and growth conditions. The analyzed data sets were publicly available, previously generated expression profiles of E. coli and S. cerevisiae genes on a genome-wide scale by using cDNA microarrays (9, 21–28). In parallel with SelR, expression of MsrA gene was analyzed. We found that expression of SelR and MsrA was not affected by various treatments and conditions, such as different alpha-factor concentrations, progression through the cell cycle, and varying zinc levels. However, expression of both genes was elevated in response to several agents that cause oxidative stress, such as hydrogen peroxide and diamide, suggesting a role of SelR and MsrA in protection against oxidative stress and/or in redox regulation of cellular processes. Expression of SelR and MsrA was also altered in response to other agents that cause environmental stress, although these responses were not always identical for the two proteins.
Growth of SelR and MsrA Mutant Strains Is Delayed in the Presence of Hydrogen Peroxide or Methionine Sulfoxides.
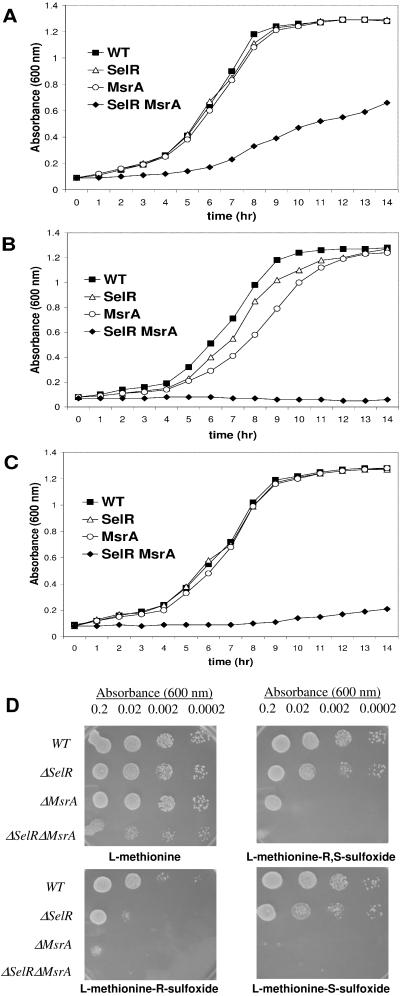
To test the prediction that SelR is involved in protection against oxidative stress, we characterized the _S. cerevisiae_strain that lacked the SelR gene and a yeast strain lacking the MsrA gene. We also isolated and characterized a strain that lacked both SelR and MsrA genes. The three knockout strains were viable, but the double knockout strain grew slower than the wild-type and single-mutant strains (Fig. 2A). The growth rate of single mutant strains was inhibited by the treatment with 2 mM hydrogen peroxide (Fig. 2B), and the double mutant could not grow under these conditions. Even at 0.5 mM H2O2, the double mutant was extremely sensitive to the presence of hydrogen peroxide (Fig.2C). The growth of yeast mutant strains was further inhibited when methionine sulfoxide was included in the growth medium (Fig. 2D). Because L-methionine sulfoxide occurs as a mixture of two stereoisomers, L-methionine-_R_-sulfoxide and L-methionine-_S_-sulfoxide, we also tested the ability of the mutants to grow on these compounds. Interestingly, both SelR and MsrA appeared to contribute to the reduction of both isomers, although the role of MsrA appeared to be more pronounced.
Figure 2.
Growth of yeast strains in the presence of hydrogen peroxide and methionine sulfoxides. Overnight cultures of wild-type (WT), SelR knockout (SelR), MsrA knockout (MsrA), or SelR/MsrA double knockout (SelR MsrA) yeast strains were diluted to 0.05 OD600 in supplemented yeast nitrogen base minimal medium (YNB) and incubated at 30°C with 250 rpm shaking. OD600 of the cultures was monitored during the incubation until cells reached stationary phase. Cells were growing in the absence of hydrogen peroxide (A), in 2 mM hydrogen peroxide (B), and in 0.5 mM hydrogen peroxide (C). For utilization of methionine sulfoxides (D), cultures were diluted to 0.2, 0.02, 0.002, and 0.0002 OD600 and 10 μl of each cell suspension was applied to spots. Each plate contained 0.7 mM hydrogen peroxide and 0.14 mM of methionine or indicated methionine sulfoxides. Cells were allowed to grow for three days at 30°C, and plates were pictured.
SelR Is a Zinc-Containing Protein.
To characterize further SelR function, we expressed the Cys-for-Sec mutant of mouse SelR in E. coli. The purified protein was colorless and spectrophotometric analysis revealed no absorption besides a peak at 280 nm. Metal analysis has been performed on a protein fraction taken from the last step of the isolation procedure, which revealed the presence of 1.08 equivalent of Zn (Table2). To test whether Zn was stably associated with the protein, a separate protein preparation was subjected to extensive dialysis either in the presence or absence of EDTA. This procedure resulted in lower but still significant content of Zn (Table 2). The presence of Se and Zn in one protein has not been described previously.
Table 2.
Metal content of Cys-for-Sec mutant mouse SelR and_Drosophila_ SelR
| Sample number | Solvent | Protein concentration | Zn, equivalents |
|---|---|---|---|
| Mouse SelR as isolated | 25 mM Tris⋅HCl, 1 mM EDTA, 0.15 M NaCl, pH 6.0 | 6 mg/ml | 1.08 |
| Mouse SelR dialyzed | 10 mM Tris⋅HCl, pH 7.5 | 1.8 mg/ml | 0.33 |
| Mouse SelR dialyzed (EDTA) | 10 mM Tris⋅HCl, 4 mM EDTA, pH 7.5 | 1.7 mg/ml | 0.42 |
| _Drosophila_SelR | 50 mM sodium phosphate, 300 mM NaCl, 150 mM imidazole, pH 7.0 | 1.08 mg/ml | 0.82 |
SelR Is Methionine-_R_-Sulfoxide Reductase.
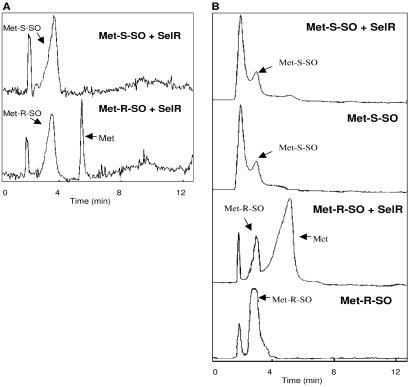
Having determined a close functional relationship between SelR and MsrA and a role of SelR homologs in methionine sulfoxide reduction and in protection against oxidative stress, we tested the ability of this protein to directly catalyze reactions relevant to the methionine sulfoxide reduction pathway. The mouse recombinant protein reduced neither dabsyl methionine-_S_-sulfoxide, a substrate of MsrA, nor methionine sulfone, an oxidation product of methionine sulfoxide in the presence of DTT. Remarkably, we found that SelR reduced a dabsyl derivative of methionine-R_-sulfoxide to dabsyl methionine in_in vitro HPLC assays (Fig.3A).
Figure 3.
SelR is methionine-_R_-sulfoxide reductase. (A) Methionine sulfoxide reductase activity of mouse SelR. HPLC assays of reduction of dabsyl L-methionine-_S_-sulfoxide (upper chromatogram) and L-methionine-R_-sulfoxide (lower chromatogram) are shown as detected by absorbance at 436 nm. (B) Methionine sulfoxide reductase activity of_Drosophila SelR. HPLC assays of reduction of dabsyl L-methionine-_S_-sulfoxide (two top chromatograms) and L-methionine-_R_-sulfoxide (two lower chromatograms) are shown as detected by absorbance at 436 nm. Locations of dabsyl methionine sulfoxide substrates (Met-_S_-SO and Met-_R_-SO) and dabsyl methionine product (Met) are indicated. The peak appearing at ≈1.5 min corresponds to dabsyl chloride impurities.
Cysteine mutants of Sec-containing proteins typically preserve substrate specificity but exhibit decreased activity compared with wild-type enzymes (1), suggesting that the wild-type SelR could also be more active than the cysteine mutant in methionine sulfoxide reduction. To test the unlikely possibility that the mutation affected stereo-specific activity of the mouse protein, we wanted to examine one of the SelR homologs in its natural form. For this purpose, we cloned a gene for Drosophila SelR (Fig. 1). Like all nonvertebrate SelR homologs, Drosophila SelR had Cys in place of Sec. Interestingly, this protein of 155 aa differed from the ORF that was predicted from genome sequence analyses in that the protein lacked a stretch of 11 aa in the middle of protein sequence. The fruit fly protein was expressed in E. coli in form of a His-tagged protein, and the affinity-purified protein was tested for its metal content and ability to reduce methionine sulfoxides. Similarly to the mouse protein, it contained 0.82 equivalents of zinc and reduced dabsyl methionine-_R_-sulfoxide, but not dabsyl methionine-_S_-sulfoxide (Fig. 3B), suggesting that this stereo-specific reductase activity may be common for all SelR homologs.
Discussion
Methionine sulfoxide reduction is an important process, by which cells regulate biological processes and cope with oxidative stress (29). MsrA, a protein involved in the reduction of methionine sulfoxides in proteins, has been known for four decades (30, 31) and has been extensively characterized with respect to structure and function (32–35). However, recent studies revealed that MsrA is only specific for methionine-S_-sulfoxides (16, 17). Because oxidized methionines occur in a mixture of R and_S isomers in vivo, it was unclear how stereo-specific MsrA could be responsible for the reduction of all protein methionine sulfoxides.
Our study provides an explanation for this puzzle. It appears that a second methionine sulfoxide reductase, SelR, evolved that is specific for methionine-_R_-sulfoxides, the activity that is different but complementary to that of MsrA. Thus, these proteins, working together, could reduce both stereoisomers of methionine sulfoxide. A complementary function of SelR and MsrA was initially predicted from our computational analyses of patterns of gene occurrence and expression, gene clustering, and domain fusion in organisms, for which completely sequenced genomes are currently available (36). Typically, functional linkages between proteins have strong predictive power if two or more of these methods link the proteins. Remarkably, in the case of SelR and MsrA, all four methods pointed to a functional linkage.
Subsequent analyses of expression patterns of SelR and MsrA homologs in E. coli and S. cerevisiae not only reinforced their functional linkage but also provided further evidence for the involvement of these proteins in redox regulation of cellular processes. For example, the expression of SelR and MsrA mRNAs was induced by the oxidative stress compounds, hydrogen peroxide, and diamide.
We experimentally tested the prediction of the involvement of SelR in protection against oxidative stress by using yeast strains lacking SelR, MsrA, or both these genes. The observation was that the knockout cells were more sensitive to oxidative stress caused by hydrogen peroxide than the corresponding wild-type strain, and their growth was also inhibited by culturing yeast cells in the presence of methionine-_S_-sulfoxide or methionine-_R_-sulfoxide. The double mutant grew poorly and was extremely sensitive to oxidative stress. Interestingly, single SelR and MsrA knockout strains could grow on either isomer of methionine sulfoxide and use these compounds as sulfur source. These data suggest that perhaps yet another cellular component(s) could contribute or participate in the pathway of methionine sulfoxide reduction.
With the previously established role of MsrA and thioredoxin in methionine sulfoxide reduction (11), and the computational evidence that SelR is an antioxidant protein functionally linked to methionine sulfoxide reduction, the function of SelR was further tested in_in vitro_ enzyme assays. We analyzed reactions in which this protein could be involved. The finding that mouse and_Drosophila_ SelRs had methionine-_R_-sulfoxide reductase activity was both consistent with the functional predictions and filled the missing link in the metabolic chain of reactions necessary for methionine sulfoxide reduction. Our current model of peptide and free methionine sulfoxide formation and reduction is shown in Fig. 4.
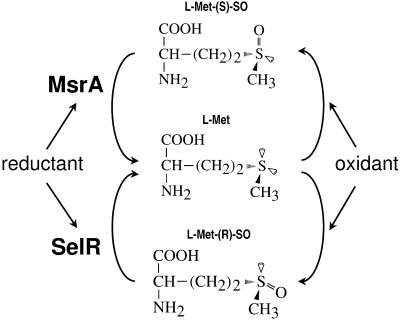
Figure 4.
A model for a pathway of methionine sulfoxide reduction.R and S isomers of methionine sulfoxide (chiral centers are located on sulfur as indicated in the figure) are formed directly or indirectly in free and protein methionines in response to oxidants, such as hydrogen peroxide. Oxidation of methionines may modulate activities of proteins, regulate cellular pathways, and disrupt redox homeostasis. Methionine-_S_-sulfoxides are reduced by MsrA and methionine-_R_-sulfoxides by SelR with reductants, such as thioredoxin.
Although both SelR and MsrA catalyze methionine sulfoxide reduction, these proteins have neither sequence homology nor common structure. SelR is a zinc-containing protein, and its β-rich secondary structure and therefore structural fold (Fig. 1) is clearly different from that of MsrA (αβ protein, in which an N-terminal active site Cys is located upstream of an α-helix dipole). Thus, SelR and MsrA have evolved independently of each other, apparently by convergent evolution of similar function in structurally distinct proteins.
Whereas MsrA does not employ metals or cofactors for methionine sulfoxide reduction, SelR is a selenium-containing protein. Because all functionally characterized selenoproteins are involved in redox reactions and Se is essential for catalytic activities of these enzymes, it is likely that Sec in SelR is also part of the enzyme active center. The presence of Se in SelR may be relevant to the previously proposed role of Se in aging. Aging cells accumulate higher levels of oxidatively damaged molecules (37), including elevated levels of protein methionine sulfoxides. Disruption of the MsrA gene in mice results in elevated levels of methionine sulfoxides and a shortened life span (15). Dietary selenium can act as an antioxidant by elevating expression of several redox selenium-containing proteins, including glutathione peroxidases and thioredoxin reductases. Further studies may test the prediction that dietary supplementation with selenium may protect mammals from oxidative stress and delay the aging process by decreasing the levels of methionine sulfoxides.
Acknowledgments
This work was supported by National Institutes of Health Grant GM61605 (to V.N.G.).
Abbreviations
SelR
selenoprotein R
MsrA
methionine sulfoxide reductase
COG
Clusters of Orthologous Groups
Note
While our paper was under review, two papers were published (38, 39) reporting that bacterial SelR homologs have methionine sulfoxide reductase activity. These data support our conclusions.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper for_Drosophila_ SelR cDNA has been deposited in the GenBank database (accession no. AF486578).
References
- 1.Stadtman T C. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 2.Low S C, Berry M J. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 3.Kryukov G V, Kryukov V M, Gladyshev V N. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 4.Lescure A, Gautheret D, Carbon P, Krol A. J Biol Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 5.Martin-Romero F J, Kryukov G V, Lobanov A V, Carlson B A, Lee B J, Gladyshev V N, Hatfield D L. J Biol Chem. 2001;276:29798–29804. doi: 10.1074/jbc.M100422200. [DOI] [PubMed] [Google Scholar]
- 6.Castellano S, Morozova N, Morey M, Berry M J, Serras F, Corominas M, Guigo R. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcotte E M, Pellegrini M, Ng H L, Rice D W, Yeates T O, Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 8.Enright A J, Iliopoulos I, Kyrpides N C, Ouzounis C A. Nature (London) 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 9.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 10.Marcotte E M, Pellegrini M, Thompson M J, Yeates T O, Eisenberg D. Nature (London) 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 11.Brot N, Weissbach H. Biopolymers. 2000;55:288–296. doi: 10.1002/1097-0282(2000)55:4<288::AID-BIP1002>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Levine R L, Moskovitz J, Stadtman E R. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 13.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moskovitz J, Bar-Noy S, Williams W M, Requena J, Berlett B S, Stadtman E R. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharov V S, Ferrington D A, Squier T C, Schoneich C. FEBS Lett. 1999;455:247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- 17.Moskovitz J, Poston J M, Berlett B S, Nosworthy N J, Szczepanowski R, Stadtman E R. J Biol Chem. 2000;275:14167–14172. doi: 10.1074/jbc.275.19.14167. [DOI] [PubMed] [Google Scholar]
- 18.Minetti G, Balduini C, Brovelli A. Ital J Biochem (Engl Ed) 1994;43:273–283. [PubMed] [Google Scholar]
- 19.Lavine T F. J Biol Chem. 1947;169:477–491. [PubMed] [Google Scholar]
- 20.Mushegian A R, Koonin E V. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 22.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 23.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferea T L, Botstein D, Brown P O, Rosenzweig R F. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons T J, Gasch A P, Gaither L A, Botstein D, Brown P O, Eide D J. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 28.Gasch A P, Huang M, Metzner S, Botstein D, Elledge S J, Brown P O. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshi T, Heinemann S. J Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brot N, Weissbach H. Biofactors. 1991;3:91–86. [PubMed] [Google Scholar]
- 31.Black S, Harte E M, Hudson B, Wartofsky L. J Biol Chem. 1960;235:2910–2916. [Google Scholar]
- 32.Tete-Favier F, Cobessi D, Boschi-Muller S, Azza S, Branlant G, Aubry A. Structure. 2000;8:1167–1178. doi: 10.1016/s0969-2126(00)00526-8. [DOI] [PubMed] [Google Scholar]
- 33.Lowther W T, Brot N, Weissbach H, Matthews B W. Biochemistry. 2000;39:13307–13312. doi: 10.1021/bi0020269. [DOI] [PubMed] [Google Scholar]
- 34.Lowther W T, Brot N, Weissbach H, Honek J F, Matthews B W. Proc Natl Acad Sci USA. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boschi-Muller S, Azza S, Sanglier-Cianferani S, Talfournier F, Van Dorsselear A, Branlant G. J Biol Chem. 2000;275:35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg D, Marcotte E M, Xenarios I, Yeates T O. Nature (London) 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 37.Stadtman E R. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 38.Grimaud R, Ezraty B, Mitchell J K, Lafitte D, Briand C, Derrick P J, Barras F. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 39.Singh V K, Moskovitz J, Wilkinson B J, Jayaswal R K. Microbiology. 2001;147:3037–3045. doi: 10.1099/00221287-147-11-3037. [DOI] [PubMed] [Google Scholar]