Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation (original) (raw)
Abstract
Accurate chromosome segregation requires the execution and coordination of many processes during mitosis, including DNA replication, sister chromatid cohesion, and attachment of chromosomes to spindle microtubules via the kinetochore complex. Additional pathways are likely involved because faithful chromosome segregation also requires proteins that are not physically associated with the chromosome. Using kinetochore mutants as a starting point, we have identified genes with roles in chromosome stability by performing genome-wide screens employing synthetic genetic array methodology. Two genetic approaches (a series of synthetic lethal and synthetic dosage lethal screens) isolated 211 nonessential deletion mutants that were unable to tolerate defects in kinetochore function. Although synthetic lethality and synthetic dosage lethality are thought to be based upon similar genetic principles, we found that the majority of interactions associated with these two screens were nonoverlapping. To functionally characterize genes isolated in our screens, a secondary screen was performed to assess defects in chromosome segregation. Genes identified in the secondary screen were enriched for genes with known roles in chromosome segregation. We also uncovered genes with diverse functions, such as RCS1, which encodes an iron transcription factor. RCS1 was one of a small group of genes identified in all three screens, and we used genetic and cell biological assays to confirm that it is required for chromosome stability. Our study shows that systematic genetic screens are a powerful means to discover roles for uncharacterized genes and genes with alternative functions in chromosome maintenance that may not be discovered by using proteomics approaches.
Keywords: chromosome stability, synthetic genetic array, kinetochore
Cells have developed highly coordinated processes to ensure that chromosomes are faithfully duplicated and segregated during mitosis. During S-phase, chromosomes are duplicated, and the resultant sister chromatids are held together by the cohesin complex (1). The centromere (CEN) is the chromosomal assembly site of a multiprotein kinetochore complex that serves to link the chromosomes with spindle microtubules (MTs) (2). Once all chromosomes have formed a bipolar attachment with the spindle, the metaphase to anaphase transition proceeds by dissolving the cohesin complex between sister chromatids and chromosome segregation occurs. If kinetochores do not attach properly to spindle MTs, highly conserved spindle checkpoint proteins halt cell cycle progression at the metaphase to anaphase transition (3). Defects in any of these processes can result in chromosome imbalance, or aneuploidy, which is central to the accumulation of multiple alterations required for tumorigenesis (4).
In recent years, great strides have been made in identifying structural components of the kinetochore complex. The kinetochore consists of three protein layers that assemble in a hierarchical fashion onto CEN DNA: inner kinetochore proteins bind directly to CEN DNA, outer kinetochore proteins associate with MTs, and central kinetochore proteins link the inner and outer kinetochore (2, 5). In addition to kinetochore proteins, numerous proteins are integral to chromosome stability, including spindle checkpoint proteins, motor proteins, MT-associated proteins, regulatory proteins, and proteins implicated in CEN chromatin dynamics, structure, and sister chromatid cohesion (1, 2, 6, 7). Many of these proteins localize to CEN regions; however, CEN localization or physical interaction with the kinetochore is not a requirement for proteins that affect chromosome stability. Indeed, proteomic approaches have mainly identified structural components of the kinetochore but not other proteins that have a role in chromosome segregation. In contrast, genetic screening has successfully identified a myriad of proteins that are important for chromosome segregation in yeast. For example, a chromosome transmission fidelity (ctf) screen, which isolated mutants that are unable to stably maintain a nonessential chromosome fragment (CF), identified mutations in genes encoding DNA replication, cohesin, and kinetochore proteins (8). Similarly, synthetic dosage lethal (SDL) screens, in which mutants are isolated that cannot tolerate overexpression of kinetochore proteins, have also successfully identified chromosome stability genes, many of which are not components of the kinetochore, such as chromatin-modifying or tubulin-folding proteins (9–12).
The development of synthetic genetic array (SGA) analysis has enabled genetic screens to be performed systematically on a genome-wide scale in yeast (13). The first application of SGA analysis permitted high-throughput screening for synthetic lethality (SL) whereby two mutants, each individually viable, cause a significant fitness defect when combined (13). Mutants that are defective in the same essential pathway or parallel nonessential pathways often display SL interactions. SL screens using cohesin, spindle checkpoint, tubulin folding, MT binding, and motor protein mutants as query strains have all identified genetic interactions with genes encoding central kinetochore components (13–16). Because components of the kinetochore share numerous SL or SDL genetic interactions with multiple pathways involved in chromosome segregation, we hypothesized that systematic genome-wide kinetochore SL and SDL screening would likely identify genes that have important roles in chromosome stability.
Here, we adapt the SGA method to include other types of genetic screens (such as SDL) or functional screens (such as CF loss). We combine genome-wide SL and SDL screens as well as a secondary ctf screen to isolate mutants that have a role in chromosome segregation. The resulting data sets reveal that SL and SDL screens tend to uncover unique, rather than overlapping, genetic interactions, which likely reflects the distinct properties associated with loss-of-function mutation (SL) and potential gain-of-function (gene overexpression; SDL). Nonetheless, the intersecting data set from our genome-wide SL and SDL was still enriched for genes involved in chromosome segregation, suggesting that the overlap between multiple screens provides unanticipated clues about gene function. For example, we identified the iron transcription factor Rcs1p/Aft1p in all three screens. Rcs1p induces gene expression upon iron depletion but was not suspected to play a role in chromosome transmission (17). We show that rcs1 mutants are defective in chromosome maintenance and that Rcs1p interacts genetically and physically with the Cbf1p inner kinetochore protein, demonstrating the success of genome-wide screens in isolating unique determinants of chromosome stability.
Materials and Methods
Yeast Strains. The two starting strains, Y2454 and Y3084 and media used in the SL analysis have been described (13, 15). We used a switcher/replica plating method (13) to create full ORF deletions using the natR cassette in strain Y3084. The temperature sensitive (ts) query strains were constructed by PCR-based integration of the ts alleles into Y2454 as described (13). Deletion strains and 13-Myc C-terminal tagged strains for this study were designed as described (18). See Table 4, which is published as supporting information on the PNAS web site, for strains used in study.
SL Screens. The robotic manipulation of the deletion mutant array and SL screens was performed as described (13). The resultant double mutants were scored for SL interactions by visual inspection. Genome-wide SL screens were conducted a minimum of two times at 25°C for the following query strains: _ctf3_Δ, _ctf19_Δ, _mcm16_Δ, _mcm21_Δ, _mcm22_Δ, _cep3_-_1, cep3_-_2, cse4_-_1, ndc10_-_1, okp1_-5, and _skp1_-3. For each query gene, all deletion mutants isolated in one or both SL screens were condensed onto a miniarray, and an additional SL screen was conducted. Putative SL interactions were confirmed first by random spore analysis (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site) and then by tetrad analysis (Table 5, which is published as supporting information on the PNAS web site). The SL screening for the _chl4_Δ, _iml3_Δ, and _mif2_-3 query strains was performed as described (15). After random spore analysis, 31 deletion mutants were shown to cluster by 2D hierarchical cluster analysis. These mutants were directly tested for genetic interactions with all remaining query mutants by tetrad dissection (Table 5). 2D hierarchical clustering was performed as described (13, 15), and statistical analysis was performed as described (19).
SDL Screens. The SGA starting strain Y2454 was transformed with either _PGAL1_-SKP1 (BPH562) (20), _PGAL1_-CTF13 (pKF88) (10), _PGAL1_-NDC10 (pKH2) (11), or p415GEU2 (vector control, BPH546) (20). The resulting query strains were mated to the MATa deletion mutant array, and SGA methodology (13, 15) was used with modifications described in Supporting Materials and Methods. The genome-wide SDL screen was performed two times, and all deletion mutant array strains that scored positive in both screens were confirmed by reintroducing plasmids into each strain by means of traditional yeast transformation methods (Table 6, which is published as supporting information on the PNAS web site). To eliminate the possibility that a deletion strain was nonspecifically sensitive to over expression of any protein, we also introduced a plasmid containing _GAL1_-inducible VPS54C (pCC5).
ctf Screen. _ade2_-101::natR (YPH1724) was constructed by PCR-based integration into Y2454 as described (13). YPH1724 was mated with either YPH255 or YPH1124, which contain CFVII(RAD2.d) or CFIII(CEN3.L), respectively (8). The resulting diploids were sporulated, and _MAT_α progeny were recovered (YPH1725 and YPH1726). The ctf assay was performed in duplicate by mating query strains YPH1725 or YPH1726 to the 211 deletion mutants identified in the SL and SDL screens. The ctf SGA assay was performed as described (13), with modifications described in Supporting Materials and Methods. Quantitative half-sector analysis was performed as described (21, 22).
Chromatin Spreads. Chromatin spreads were performed as described (23). The antibodies used were mouse anti-Myc (9E10; 1:1,000, Roche Diagnostics) rabbit Ndc10p (1:1,000), Cy3-goat anti-mouse antibodies (1:3,000), and fluorescein goat anti-rabbit antibodies (1:1,000), Jackson ImmunoResearch). Chromatin spreads were visualized with a Zeiss Axioplan 2 Fluorescence Microscope and imaged with a COOLSNAPHQ-M camera (PerkinElmer).
Yeast Two-Hybrid. To construct pBD-RCS1, an NcoI-XhoI fragment encoding amino acids 1–413 of Rcs1p was inserted into the NcoI-SalI site of pGBKT7 (Clontech_). pAD-CBF1_ was created by inserting an NcoI-BamHI fragment containing the ORF of CBF1 into pACT2 (Clontech). The strain PJ69-4A (24) was first transformed with pBD-RCS1 and then transformed with a yeast genomic library fused to the transactivation domain of Gal4. Transformants were tested for growth on SC-Ade-His plates. The prey plasmids from positive clones were rescued and retransformed into PJ69-4A cells carrying pGBKT7 (to eliminate self-activation clones) and into PJ69-4A cells carrying pBD-RCS1 (to verify the bait–prey interaction). Of 30,000 total clones screened, 6 positives were identified (R.U. and Y.Y.-I., unpublished data). Quantification of β-galactosidase activity was performed as described (25).
Polyclonal Ndc10p Antibodies. Anti-Ndc10p polyclonal antibodies were generated in rabbits (Covance Research Products, Denver, PA) as described (26). The construct used to generate an Ndc10p fragment for expression in Escherichia coli strain BL21 was made by cloning a 1,014-bp EcoRI fragment from NDC10 (343 bp from the ATG to 1,357 bp) into the pRSET-C vector (Invitrogen) EcoRI multiple cloning site, thereby creating an in-frame fusion protein containing a 6xHis tag and amino acids 115–452 of Ndc10p.
Results
Kinetochore SL Screen. In an effort to identify unique determinants of chromosome segregation, we performed 14 SL screens with query genes of both the inner and central kinetochore. The query strains, which carried either deletions of nonessential kinetochore genes or temperature-sensitive mutations of essential kinetochore genes, comprised mutants of genes encoding central kinetochore components (COMA complex mutants, _ctf19_Δ, _mcm21_Δ, _okp1_-5 and _chl4_Δ, _ctf3_Δ, _iml3_Δ, _mcm16_Δ, and _mcm22_Δ) or inner kinetochore components [_cep3_-_1, cep3_-_2, cse4_-_1, mif2_-_3, ndc10_-_1_, and _skp1_-_3_ (2)]. Inviable double mutants were scored as SL interactions and slow growing double mutants were scored as synthetic sick (SS) interactions. The resulting confirmed data set contains 230 genetic interactions among 84 genes, with the number of interactions per query gene varying from 8 to 22 (Table 5). Of the 230 genetic interactions, 18% (42) were SL interactions and the remaining were SS interactions.
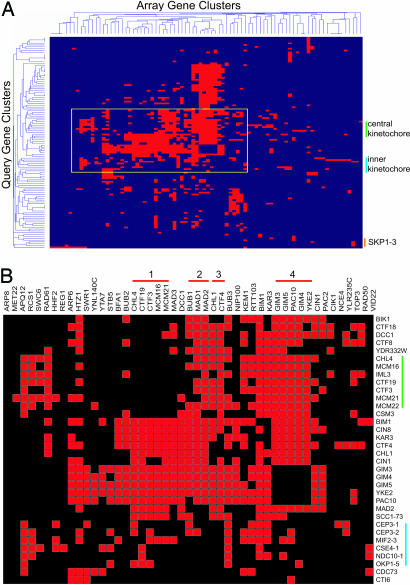
2D hierarchical clustering of SL interaction profiles has been used successfully to group sets of genes that function within the same pathway or complex (15). We clustered our kinetochore SL data with current SL data sets that contain the same 84 genes identified here (Fig. 1_A_). Thus, in addition to our 14 kinetochore query strains, we included 102 query strains in the clustergram. Our analysis revealed that query genes encoding the central kinetochore components cluster together, based on a shared genetic interaction profile (with the exception of _okp1_-5). Query genes encoding the inner kinetochore components cluster separately from the central kinetochore (Fig. 1). The _skp1_-3 query strain did not cluster with the other inner kinetochore query strains. In addition to its role in the CBF3 inner kinetochore complex, Skp1p is a component of the SCF (Skp1p-Cdc53p-F-box protein) E3 ubiquitin ligase complex that degrades proteins to mediate cell cycle transitions (27–29). The _skp1_-3 mutant arrests in G1 phase due to defects in SCF function but has a wild-type rate of chromosome segregation (20, 30). Thus, _skp1_-3 is expected to have a different SL interaction profile than the kinetochore mutants. Many of the array genes that are components of known pathways cluster together, indicating that the SL interaction profiles between members of the same complex are similar. Examples of clustered array genes include the central kinetochore (CHL4, CTF19, CTF3, MCM16, and MCM21), the spindle check point pathway (BUB1, MAD1, and MAD2), the sister chromatid cohesion pathway (CHL1 and CTF4), and the GimC/Prefoldin complex (GIM3, GIM5, PAC10, GIM4, and YKE2) (Fig. 1_B_).
Fig. 1.
2D hierarchical clustering of the synthetic genetic interactions determined by SL analysis. (A) Rows display 116 query genes; columns indicate 84 deletion mutant array genes. The central kinetochore query mutant cluster is indicated by a green line, the inner kinetochore query mutant cluster is indicated by a blue line, and the _skp1_-3 query mutant is indicated by an orange line. The cluster trees organize query and deletion mutant array genes that show similar patterns of genetic interactions. The yellow box outlines a cluster of array genes identified in the central and inner kinetochore query mutant SL screens. (B) The yellow outline of cluster in A is expanded to allow visualization of specific array genes. Clustered array genes are indicated by red lines (1, central kinetochore; 2, spindle checkpoint; 3, sister chromatid cohesion; 4, GimC/Prefoldin complex).
Kinetochore SDL Screen. An SDL interaction occurs when overexpression of a gene is compatible with viability in a wild-type strain but is lethal or causes a slow growth defect in a target mutant strain (11). SDL screens in which proteins of the CBF3 inner kinetochore complex have been overexpressed in a subset of deletion mutants have been particularly successful in identifying mutants defective in chromosome segregation (9). Therefore, in a parallel effort to our SL screens, we developed a genome-wide SDL screen to identify deletion mutants that have a role in chromosome segregation. We used SGA methodology to introduce _PGAL1_-inducible plasmids carrying one of three components of the CBF3 complex (CTF13, NDC10, and SKP1) and a vector control into the set of ≈4,700 viable deletion mutants. CTF13, NDC10, or SKP1 overexpression was induced on galactose medium, and strains were incubated at three different temperatures (16°C, 25°C, and 37°C). An SDL or synthetic dosage sickness (SDS) phenotype was scored by comparing growth of the deletion mutant overexpressing CTF13, NDC10, or SKP1, to the vector control plasmid. One hundred forty-one deletion mutants displayed one or more SDL or SDS genetic interactions upon increased dosage of CTF13, NDC10, and/or SKP1, resulting in a total of 382 genetic interactions (Table 6). The majority of these interactions were SDS (358) whereas the rest were SDL (24) interactions, and 68% of the identified mutants displayed more than one interaction.
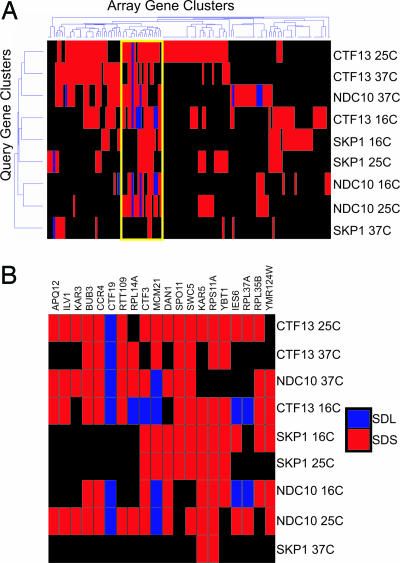
We performed 2D hierarchical clustering analysis to group the query genes and array genes according to their SDL interaction profiles (Fig. 2_A_). We identified a cluster of array genes that display a high density of SDL and SDS interactions (Fig. 2_B_). The KAR3, BUB3, CTF19, CTF3, and MCM21 genes are present in this cluster, suggesting that the cluster may be enriched with mutants defective in chromosome attachment and movement. The cluster also contains SWC5 and IES6, genes encoding components of the SWR1 and Ino80 chromatin remodeling complexes, respectively. The SWR1 complex has been linked to chromosome stability, raising the possibility that the Ino80 complex may also have a role at the kinetochore (31).
Fig. 2.
2D hierarchical clustering of the synthetic genetic interactions determined by SDL analysis. (A) Rows, nine SDL screens conducted at the indicated temperature; columns, 141 deletion mutant array genes. Blue indicates SDL interactions, and red indicates SDS interactions. (B) Yellow outline of cluster in A is expanded to allow visualization of specific array genes.
High-Throughput ctf Screen. The genome-wide SL and SDL kinetochore screens identified a total of 612 genetic interactions involving 211 nonessential genes, of which only 14 genes were isolated in both screens (Table 1). Despite this apparent low degree of overlap, statistical analysis suggests that the overlap is greater than expected from comparing random screens (see Discussion). We were interested in determining how many of the 211 nonessential deletion mutants exhibit a reduced fidelity of chromosome transmission using a color-based CF loss assay (21). To circumvent the task of deleting 211 genes in a ctf tester strain, we developed a high-throughput CF loss procedure based on the SGA method by introducing a CF into the 211 mutants. To eliminate the possibility of deletion mutant suppression by genes located on a CF, two different screens were performed, one with a CF created from chromosome III and the other with a CF created from chromosome VII (8). Deletion mutants were struck for single colonies to qualitatively assess the rate of red sector formation, or CF loss. Twenty-eight of the 211 deletion strains tested displayed a detectable sectoring phenotype: 15 genes isolated from only the SL screen, 5 genes isolated from only the SDL screen, and 8 genes isolated from both the SL and SDL screens (Table 2). CF loss mutants identified in both the SL and SDL screens were significantly enriched for genes with roles in chromosome segregation (Table 2).
Table 1. Genes identified in both SL and SDL screens.
| ORF | Gene name | Functional role* |
|---|---|---|
| YIL040W | APQ12 | mRNA-nucleus export |
| YHR013C | ARD1 | Protein amino acid acetylation |
| YOR026W | BUB3 | Mitotic spindle checkpoint |
| YPL018W | CTF19 | Chromosome segregation |
| YLR381W | CTF3 | Chromosome segregation |
| YPR135W | CTF4 | Mitotic sister chromatid cohesion |
| YOL012C | HTZ1 | Chromatin structure |
| YPR141C | KAR3 | Microtubule motor |
| YDR318W | MCM21 | Chromosome segregation |
| YOR350C | MNE1 | Unknown |
| YGR078C | PAC10 | Tubulin Folding |
| YGL071W | RCS1 | High-affinity iron transport |
| YGR063C | SPT4 | Chromosome architecture/segregation |
| YGR270W | YTA7 | Protein catabolism |
Table 2. Deletion mutants isolated in high-throughput ctf screen.
| ORF | Gene name | Screen | Functional role* |
|---|---|---|---|
| YER016W | BIM1 | SL | Microtubule nucleation |
| YGL003c | CDH1 | SL | Mitotic metaphase/anaphase transition |
| YPL008W | CHL1 | SL | Mitotic sister chromatid cohesion |
| YDR254W | CHL4 | SL | Chromosome segregation |
| YMR198W | CIK1 | SL | Mitotic spindle orientation |
| YFR046C | CNN1 | SL | Chromosome segregation |
| YGL086W | MAD1 | SL | Mitotic spindle checkpoint |
| YPR046W | MCM16 | SL | Chromosome segregation |
| YOL064C | MET22 | SL | Methionine biosynthesis |
| YPL024W | NCE4 | SL | Unknown |
| YDR014W | RAD61 | SL | Response to radiation |
| YDR289C | RTT103 | SL | mRNA processing |
| YGR184C | UBR1 | SL | Protein ubiquitination |
| YLR235C | YLR235C | SL | Unknown |
| YNL140C | YNL140C | SL | Unknown |
| YPL055C | LGE1 | SDL | Histone ubiquitination |
| YDR378C | LSM6 | SDL | Nuclear mRNA splicing |
| YGL066W | SGF73 | SDL | Histone acetylation |
| YLR079W | SIC1 | SDL | Regulation of G1/S cell cycle transition |
| YGR064W | YGR064W | SDL | Unknown |
| YOR026W | BUB3 | SL/SDL | Mitotic spindle checkpoint |
| YPL018W | CTF19 | SL/SDL | Chromosome segregation |
| YLR381C | CTF3 | SL/SDL | Chromosome segregation |
| YPR135W | CTF4 | SL/SDL | Mitotic sister chromatid cohesion |
| YPR141C | KAR3 | SL/SDL | Microtubule motor |
| YDR318W | MCM21 | SL/SDL | Chromosome segregation |
| YGL071W | RCS1 | SL/SDL | High-affinity iron transport |
| YGR063C | SPT4 | SL/SDL | Chromosome architecture/segregation |
Rcs1p Is Required for Chromosome Stability. Our genome-wide SL and SDL screens identified 211 candidate genes involved in the fidelity of chromosome segregation, and deletion of 28 of these genes resulted in chromosome segregation defects. RCS1 was the only gene identified in all three screens that did not already have a known role in chromosome segregation (Table 2). Rcs1p is a well characterized transcription factor that regulates expression of genes involved in iron uptake but has not been previously linked to chromosome segregation (17). To begin our characterization of the potential role for Rcs1p in chromosome stability, we performed a quantitative CF loss assay (21) in an _rcs1_Δ homozygous diploid deletion strain. A central kinetochore mutant that was isolated in both our SL and ctf screen, called cnn1, was used as a positive control for chromosome loss (32). Consistent with the qualitative high-throughput ctf screen, both _cnn1_Δ and _rcs1_Δ homozygous diploid strains displayed a 46-fold and 22-fold increase in CF loss events, respectively, compared with wild-type diploid strains (Table 3). In addition, _rcs1_Δ mutants exhibited an 8.9-fold increase in chromosome nondisjunction events, which is even higher than the 4.4-fold increase of the _cnn1_Δ mutant. Thus, Rcs1p has an unexpected requirement for a high fidelity of chromosome transmission.
Table 3. Rates of chromosome missegregation events.
| Strain no. | Genotype | Rate of chromosome loss, 1:0 events | Rate of nondisjunction, 2:0 events | Total colonies |
|---|---|---|---|---|
| YPH982 | ± | 8.7 × 10-5 (1.0) | 8.7 × 10-5 (1.0) | 22,800 |
| + | ||||
| YPH1727 | _rcsI_Δ | 1.9 × 10-3 (22.2) | 7.7 × 10-4 (8.9) | 25,913 |
| _rcsI_Δ | ||||
| YPH1737 | _cnnI_Δ | 4.0 × 10-3 (46) | 3.9 × 10-4 (4.4) | 31,133 |
| _cnnI_Δ |
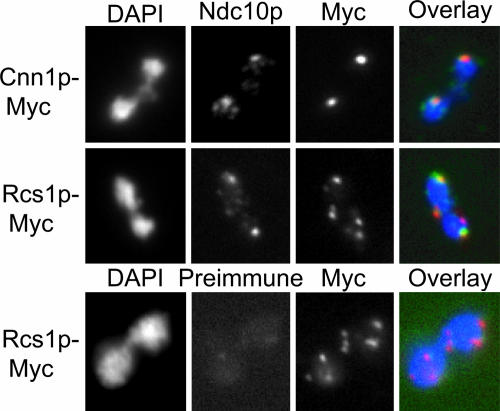
Rcs1p Colocalizes with the Ncd10p Kinetochore Protein. Because rcs1 deletion mutants exhibit increased rates of chromosome loss, we were interested in determining the localization of Rcs1p on chromatin in relation to a kinetochore protein. In iron-replete cells, Rcs1p localizes throughout the cell, including diffuse nuclear staining (33, 34). To specifically analyze Rcs1p localization on chromatin, we performed indirect immunofluorescence analysis on chromosome spreads of Rcs1p-Myc-expressing cells. We used antibodies specific to the CBF3 component Ndc10p, which stain brightly at the centromere, and costained the slides with Myc antibodies. Resolution of the fluorescence signal is optimal in bilobed chromatin masses where chromosomes are partially separated. To test our Ndc10p antibodies, we demonstrated that both Ndc10p and Cnn1p-Myc localize to two discrete signals that overlap in bilobed masses, consistent with colocalization of both proteins at the centromere (Fig. 3). We found that Rcs1p-Myc localizes to between 5 and 10 discrete foci, and we repeatedly visualized two of these foci colocalizing with Ndc10p foci (Fig. 3). Additional Rcs1p-Myc foci are expected due to Rcs1p binding to multiple genomic loci when grown in rich media (35). We also found that Rcs1p-Myc colocalizes with Ndc10p in single chromatin masses, and thus their colocalization is not specific to bilobed cells (data not shown).
Fig. 3.
Rcs1p colocalizes with the Ndc10p kinetochore protein. Shown is a chromosome spread analysis of a bilobed chromatin mass carrying either Myc-tagged Cnn1p (YPH1733) or Rcs1p (YPH1731). Spreads were imaged for DNA (DAPI), Ndc10p (Ndc10p), and Myc-tagged protein (Myc). The preimmune panel is a control for the Ndc10p polyclonal antibodies. In the merged images (Overlay), DNA is colored blue, Ndc10p is colored green, and Myc-tagged proteins are colored red. Overlap of Ndc10p and Myc signal appears yellow. In 100 chromatin masses examined, 85% of single chromatin masses showed overlap between Rcs1p signal and Ndc10p signal, and 80% of bilobed chromatin masses showed overlap between Rcs1p and at least one Ndc10p signal.
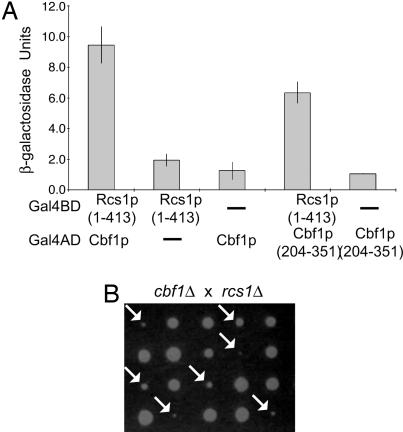
Interaction of Rcs1p with the Inner Kinetochore Protein Cbf1p. The colocalization of Rcs1p with a centromere-binding protein encouraged us to identify Rcs1p-interacting partners that may have a role in chromosome segregation. We performed a yeast two-hybrid library screen using the N-terminal 413 aa of Rcs1p as bait [full-length Rcs1p self-activates in the two-hybrid system (34)] and identified the inner kinetochore protein Cbf1p as an Rcs1p-binding partner (Fig. 4_A_). This result is intriguing because, like Rcs1p, Cbf1p is a transcription factor that responds to environmental cues to mediate gene expression. Cbf1p localizes to CDEI elements that are present both at CEN regions and upstream of genes required for methionine biosynthesis (36). Because the _cbf1_Δ mutant was not a query strain in our kinetochore SGA screens, we mated a _cbf1_Δ mutant to an _rcs1_Δ mutant to test whether the _cbf1_Δ _rcs1_Δ double mutant showed reduced fitness. We found that the _cbf1_Δ _rcs1_Δ mutant exhibited severe growth defects compared with either single mutant and that the size of the double mutant was variable, possibly due to chromosome missegregation events or suppressor activity (Fig. 4_B_). Thus, Rcs1p displays both a genetic and physical interaction with the Cbf1p inner kinetochore protein.
Fig. 4.
Interaction of Rcs1p with the inner kinetochore protein Cbf1p. (A) Rcs1p interacts with Cbf1p in a yeast two-hybrid assay. Amino acids 1–413 of Rcs1p were fused to the Gal4 DNA-binding domain (Gal4BD), and full-length Cbf1p or amino acids 204–351 of Cbf1p were fused to the Gal4 activation domain (Gal4AD). Cbf1p (204–351) is the original clone isolated in the Rcs1p two-hybrid screen. Gal4BD and Gal4AD fusions were expressed either alone or together in PJ69-4A cells, and quantitative β-galactosidase assays were performed (see Materials and Methods). β-Galactosidase values represent the averages of at least two independent experiments with three independent colonies, and error bars indicate the standard deviations. (B) _cbf1_Δ _rcs1_Δ double mutants exhibit reduced fitness compared with either single mutant. An agar plate containing five dissected tetrads from a mating of an _rcs1_Δ mutant (YPH1735) to a _cbf1_Δ mutant (YPH792) is shown. The plate was grown at 25°C for 3 days. Arrows indicate _cbf1_Δ _rcs1_Δ double mutants.
Discussion
In an effort to identify determinants of chromosome segregation, we used SGA methodology to perform complementary genome-wide screens, including a series of SL and SDL screens and a functional secondary ctf screen. We identified a total of 612 genetic interactions that encompass 211 genes involved in a diverse range of biological processes; the SL screens identified 230 genetic interactions among 84 genes, whereas the SDL screens identified 382 interactions amongst 141 genes. We find that performing multiple primary screens and a secondary functional screen to prioritize genes of interest is a valuable method to identify genes with no prior connections to the process being studied. We isolated the Rcs1p iron transcription factor in all three of our screens and discovered that Rcs1p has an unexpected role in chromosome transmission.
Comparison of SL and SDL Genome-Wide Screening. The overlapping SL/SDL data set identified 14 genes that were highly enriched for genes encoding kinetochore proteins (Table 1). The chance of finding 14 overlapping genes in two random screens with the haploid yeast deletion set is P = 3.1 × 10–9, suggesting that the overlap between the SL and SDL screens, although relatively small, is significant. Proof of principle experiments suggested that SDL may occur due to perturbation of protein complexes already defective in one component by titrating out second component, thus mimicking a SL effect (10). However, because the majority of genes in the SL and SDL data sets do not overlap, these two genetic screens must assess different genetic relationships in general. The results from our SL and SDL screens are consistent with the hypothesis that different subsets of mutants are sensitive to loss of kinetochore function versus increased dosage of kinetochore proteins. The distinctly different data sets obtained through SL and SDL screens and the ability of both screens to identify genes with roles in chromosome segregation demonstrate that SL and SDL screening are largely complementary genomics approaches to identify genes of biological relevance.
Adaptation of SGA to a ctf Screen. In addition to the SDL screen, we adapted the SGA methodology to introduce a point mutation and CF into the SGA starting strain, thereby enabling rapid screening of 211 deletion mutations for ctf defects. The 28 ctf mutants isolated from both the SL and SDL data sets were significantly enriched for chromosome segregation mutants (Table 2). The high-throughput ctf assay requires that a certain threshold of chromosome loss be caused by a single mutation; therefore, the screen may underestimate the number of deletion mutants with chromosome segregation defects. Further, mutations that are buffered by another gene may not display a ctf defect until combined with a secondary mutation. A portion of the 183 mutants from our SL and SDL screens that were not identified in the high-throughput ctf screen may have chromosome loss rates below detection or represent buffered mutations.
A Role for Rcs1p in Chromosome Stability. Genes identified in the overlapping data sets from our SL, SDL, and ctf screens were enriched for genes with roles in chromosome maintenance (Table 2). We were surprised to isolate RCS1, which encodes an iron transcription factor, in both of our genomic screens as well as our ctf screen. Rcs1p induces expression of genes involved in iron uptake upon iron depletion (37). Further, microarray studies have indicated that Rcs1p does not regulate expression of any known kinetochore genes (38, 39). Genome-wide chromatin immunoprecipitation (ChIP) experiments identified an enrichment of CEN DNA in Rcs1p IPs (35). We performed multiple Rcs1p-Myc ChIP experiments but were unable to detect specific binding of Rcs1p to three different CEN regions (data not shown). Rcs1p may still bind to CEN DNA, but the binding may be below the level of detection in our ChIP assay.
We detected both a two-hybrid and genetic interaction between Rcs1p and the inner kinetochore protein, Cbf1p. We performed co-IP experiments from yeast lysates but were unable to detect an interaction between Rcs1p and Cbf1p in log phase cells (data not shown). Thus, the interaction between Rcs1p and Cbf1p may be indirect or require different conditions to detect than those used in our IPs. The potential interaction of Rcs1p with Cbf1p is particularly intriguing in light of functional similarities between these proteins. In addition to its role at the kinetochore, Cbf1p forms a transcriptional activation complex with Met4p and Met28p to activate transcription of sulfur amino acid metabolism genes (36). Unlike cbf1 mutants, we found that rcs1 mutants are not methionine auxotrophs, suggesting that Rcs1p does not have a role in sulfur amino acid metabolism (data not shown).
Whether Rcs1p binds to CEN DNA or not, it clearly has a role in chromosome stability. rcs1 homozygous diploid deletion mutants have elevated rates of both chromosome loss and nondisjunction events (Table 3). Rcs1p is required for cell viability in cells defective for kinetochore function, interacts by two-hybrid with Cbf1p, and colocalizes on chromatin with Ndc10p (Figs. 1, 2, 3, 4). Future studies will determine whether Rcs1p has a structural or transcriptional role in maintaining chromosomes and whether there is a connection between iron regulation and chromosome segregation.
Supplementary Material
Supporting Information
Acknowledgments
We thank E. Conibear (University of British Columbia) for the gift of pCC5. This work was supported by a Collaborative Genomics Special Project Grant from the Canadian Institutes of Health Research (CIHR) and Genome Canada through the Ontario Genomics Institute (to B.A. and C.B.), by operating grants from the CIHR, by a U.S. National Institutes of Health grant (to P.H.), and by grants-in-aid from the Ministry of Education, Culture, Science and Technology of Japan (to Y.Y-I.). C.B. is a Canada Research Chair (CRC) in Proteomics, Bioinformatics, and Functional Genomics. V.M. was supported by a CIHR senior research fellowship and by a Michael Smith Foundation for Health Research (MSFHR) postdoctoral fellowship and is a CRC in Enology and Yeast Genomics and a MSFHR Scholar. K.B. was supported by a CIHR postdoctoral fellowship and a MSFHR postdoctoral fellowship and is a CRC in Functional and Chemical Genomics. K.Y. was supported by a Natural Sciences and Engineering Research Council of Canada postgraduate scholarship and a University of British Columbia graduate scholarship. R.U. was supported by a fellowship from the Japan Science Promotion Society. B.C. was supported by a CIHR Doctoral Research Award. I.P. was supported by a National Institute of Canada Research Studentship.
Author contributions: V.M., K.B., J.G., K.Y., I.P., A.T., Y.Y.-I., C.B., and B.A. designed research; V.M., K.B., J.G., K.Y., T.K., R.U., T.H., I.P., and A.T. performed research; V.M., K.B., J.G., K.Y., T.K., R.U., and B.C. contributed new reagents/analytic tools; V.M., K.B., J.G., K.Y., B.S., H.D., R.U., I.P., A.T., Y.Y.-I., C.B., P.H., and B.A. analyzed data; and V.M., K.B., C.B., and B.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CEN, centromere; CF, chromosome fragment; ctf, chromosome transmission fidelity; MT, microtubule; SDL, synthetic dosage lethal; SDS, synthetic dosage sickness; SGA, synthetic genetic array; SL, synthetic lethal; IP, immunoprecipitation.
References
- 1.Uhlmann, F. (2003) Curr. Biol. 13**,** R104–R114. [DOI] [PubMed] [Google Scholar]
- 2.McAinsh, A. D., Tytell, J. D. & Sorger, P. K. (2003) Annu. Rev. Cell Dev. Biol. 19**,** 519–539. [DOI] [PubMed] [Google Scholar]
- 3.Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112**,** 407–421. [DOI] [PubMed] [Google Scholar]
- 4.Rajagopalan, H. & Lengauer, C. (2004) Nature 432**,** 338–341. [DOI] [PubMed] [Google Scholar]
- 5.Measday, V. & Hieter, P. (2004) Nat. Cell Biol. 6**,** 94–95. [DOI] [PubMed] [Google Scholar]
- 6.Biggins, S. & Walczak, C. E. (2003) Curr. Biol. 13**,** R449–R460. [DOI] [PubMed] [Google Scholar]
- 7.Sharp, J. A. & Kaufman, P. D. (2003) Curr. Top. Microbiol. Immunol. 274**,** 23–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer, F., Gerring, S. L., Connelly, C. & Hieter, P. (1990) Genetics 124**,** 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baetz, K. K., Krogan, N. J., Emili, A., Greenblatt, J. & Hieter, P. (2004) Mol. Cell. Biol. 24**,** 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroll, E. S., Hyland, K. M., Hieter, P. & Li, J. J. (1996) Genetics 143**,** 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Measday, V. & Hieter, P. (2002) Methods Enzymol. 350**,** 316–326. [DOI] [PubMed] [Google Scholar]
- 12.Measday, V., Hailey, D. W., Pot, I., Givan, S. A., Hyland, K. M., Cagney, G., Fields, S., Davis, T. N. & Hieter, P. (2002) Genes Dev. 16**,** 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294**,** 2364–2368. [DOI] [PubMed] [Google Scholar]
- 14.Mayer, M. L., Pot, I., Chang, M., Xu, H., Aneliunas, V., Kwok, T., Newitt, R., Aebersold, R., Boone, C., Brown, G. W. & Hieter, P. (2004) Mol. Biol. Cell 15**,** 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong, A. H., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., et al. (2004) Science 303**,** 808–813. [DOI] [PubMed] [Google Scholar]
- 16.Warren, C. D., Eckley, D. M., Lee, M. S., Hanna, J. S., Hughes, A., Peyser, B., Jie, C., Irizarry, R. & Spencer, F. A. (2004) Mol. Biol. Cell 15**,** 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutherford, J. C. & Bird, A. J. (2004) Eukaryot. Cell 3**,** 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14**,** 953–961. [DOI] [PubMed] [Google Scholar]
- 19.Parsons, A. B., Brost, R. L., Ding, H., Li, Z., Zhang, C., Sheikh, B., Brown, G. W., Kane, P. M., Hughes, T. R. & Boone, C. (2004) Nat. Biotechnol. 22**,** 62–69. [DOI] [PubMed] [Google Scholar]
- 20.Connelly, C. & Hieter, P. (1996) Cell 86**,** 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshland, D. & Hieter, P. (1987) Methods Enzymol. 155**,** 351–372. [DOI] [PubMed] [Google Scholar]
- 22.Hyland, K. M., Kingsbury, J., Koshland, D. & Hieter, P. (1999) J. Cell Biol. 145**,** 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelis, C., Ciosk, R. & Nasmyth, K. (1997) Cell 91**,** 35–45. [DOI] [PubMed] [Google Scholar]
- 24.James, P., Halladay, J. & Craig, E. A. (1996) Genetics 144**,** 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarente, L. (1983) Methods Enzymol. 101**,** 181–191. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, M. L., Gygi, S. P., Aebersold, R. & Hieter, P. (2001) Mol. Cell 7**,** 959–970. [DOI] [PubMed] [Google Scholar]
- 27.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86**,** 263–274. [DOI] [PubMed] [Google Scholar]
- 28.Feldman, R. M., Correll, C. C., Kaplan, K. B. & Deshaies, R. J. (1997) Cell 91**,** 221–230. [DOI] [PubMed] [Google Scholar]
- 29.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91**,** 209–219. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa, K., Skowyra, D., Elledge, S. J., Harper, J. W. & Hieter, P. (1999) Mol. Cell 4**,** 21–33. [DOI] [PubMed] [Google Scholar]
- 31.Krogan, N. J., Baetz, K., Keogh, M. C., Datta, N., Sawa, C., Kwok, T. C., Thompson, N. J., Davey, M. G., Pootoolal, J., Hughes, T. R., et al. (2004) Proc. Natl. Acad. Sci. USA 101**,** 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Wulf, P., McAinsh, A. D. & Sorger, P. K. (2003) Genes Dev. 17**,** 2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425**,** 686–691. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi-Iwai, Y., Ueta, R., Fukunaka, A. & Sasaki, R. (2002) J. Biol. Chem. 277**,** 18914–18918. [DOI] [PubMed] [Google Scholar]
- 35.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298**,** 799–804. [DOI] [PubMed] [Google Scholar]
- 36.Robinson, K. A. & Lopes, J. M. (2000) Nucleic Acids Res. 28**,** 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi-Iwai, Y., Dancis, A. & Klausner, R. D. (1995) EMBO J. 14**,** 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shakoury-Elizeh, M., Tiedeman, J., Rashford, J., Ferea, T., Demeter, J., Garcia, E., Rolfes, R., Brown, P. O., Botstein, D. & Philpott, C. C. (2004) Mol. Biol. Cell 15**,** 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford, J. C., Jaron, S. & Winge, D. R. (2003) J. Biol. Chem. 278**,** 27636–27643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information