The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens (original) (raw)
Abstract
We have determined the complete 1,694,969-nt sequence of the GC-rich genome of Methanopyrus kandleri by using a whole direct genome sequencing approach. This approach is based on unlinking of genomic DNA with the ThermoFidelase version of M. kandleri topoisomerase V and cycle sequencing directed by 2′-modified oligonucleotides (Fimers). Sequencing redundancy (3.3×) was sufficient to assemble the genome with less than one error per 40 kb. Using a combination of sequence database searches and coding potential prediction, 1,692 protein-coding genes and 39 genes for structural RNAs were identified. M. kandleri proteins show an unusually high content of negatively charged amino acids, which might be an adaptation to the high intracellular salinity. Previous phylogenetic analysis of 16S RNA suggested that M. kandleri belonged to a very deep branch, close to the root of the archaeal tree. However, genome comparisons indicate that, in both trees constructed using concatenated alignments of ribosomal proteins and trees based on gene content, M. kandleri consistently groups with other archaeal methanogens. M. kandleri shares the set of genes implicated in methanogenesis and, in part, its operon organization with Methanococcus jannaschii and Methanothermobacter thermoautotrophicum. These findings indicate that archaeal methanogens are monophyletic. A distinctive feature of M. kandleri is the paucity of proteins involved in signaling and regulation of gene expression. Also, M. kandleri appears to have fewer genes acquired via lateral transfer than other archaea. These features might reflect the extreme habitat of this organism.
M_ethanopyrus kandleri_ was isolated from the sea floor at the base of a 2,000-m-deep “black smoker” chimney in the Gulf of California (1). The organism is a rod-shaped Gram-positive methanogen that grows chemolithoautotrophically at 80–110°C in the H2–CO2 atmosphere (2). The discovery of Methanopyrus showed that biogenic methanogenesis is possible above 100°C and could explain isotope discrimination at temperatures that had been thought to be unfavorable for biological methanogenesis (1).
Certain aspects of M. kandleri biochemistry place this organism aside from other archaea. First, the membrane of M. kandleri consists of a terpenoid lipid (3), which is considered the most primitive lipid in the evolution of membranes (4) and is the direct precursor of phytanyl diethers found in the membranes of all other archaea. Another unusual feature of M. kandleri is the high intracellular concentration (1.1 M) of a trivalent anion, cyclic 2,3-diphosphoglycerate, which has been reported to confer activity and stability at high temperatures on enzymes from this organism (5). Indeed, enzymes isolated from M. kandleri require high salt concentrations (>1 M) for stability and activity (6, 7). Finally, M. kandleri has several unique enzymes, the most notable being type 1B DNA topoisomerase V and the two-subunit reverse gyrase (8–12).
Perhaps the most distinctive feature of M. kandleri is its apparent position in the archaeal phylogeny. Several analyses, based on phylogenetic trees for 16S rRNA and the presence/absence of an 11-aa insertion in EF-1α, placed M. kandleri close to the root of the Euryarchaeota and did not suggest any specific affinity with other archaeal methanogens (13–15). Furthermore, some signatures shared with Crenarchaeota were noticed in the 16S RNA sequence of M. kandleri (13). In contrast, analysis of the methyl coenzyme M reductase operon of M. kandleri identified a group of genes unique to archaeal methanogens (15). The genome comparison reported here reveals clustering of M. kandleri with other methanogens in phylogenetic trees based on concatenated alignments of ribosomal proteins, which is mimicked by congruence of the sets of predicted genes, suggesting monophyly of this group. However, we found that M. kandleri is a “minimalist” archaeon whose regulatory and signaling systems are generally scaled down compared with those of other archaea. Comparative genome analysis of M. kandleri, Methanococcus jannaschii, and Methanothermobacter thermoautotrophicum resulted in the delineation of a distinct set of genes characteristic of archaeal methanogens.
Materials and Methods
Directed Genomic Sequencing.
A genome sequencing strategy was adopted to sequence M. kandleri strain AV19 (DSM 6324).
Skimming Shotgun Phase.
A small insert (2–4 kb) shotgun library in pUC18 cloning vector (SeqWright, Houston) was prepared from 150 μg of genomic DNA isolated as described (16). Approximately 1,000 purified plasma clones and 3,000 unpurified clones (aliquots of overnight cultures) were sequenced from both ends by using dye-terminator chemistry (Applied Biosystems), ThermoFidelase I (10), and standard end Fimers (17, 18) (Fidelity Systems) on ABI 377. A total of 3,986 sequences corresponding to ≈0.5× coverage were assembled into 901 contigs by using phred/phrap/consed software (www.phrap.org) (refs. 19–21).
Directed Sequencing Phase.
Using the assembled contigs from the previous phase as islands to select Fimers for directed sequencing off genomic DNA, a total of 11 rounds of Fimer selection-sequencing assembly were performed, which allowed assembly of the genome into 29 contigs, with a 2.5× sequencing redundancy. At this stage, we synthesized 5,499 Fimers, from which 6,470 chromatograms were obtained. To select priming sites at the ends of contigs, we used a publicly available program primou (http://www.genome.ou.edu/informatics/primou.html.)
Gap Closure and Assembly Verification.
At this step, we isolated DNA from 293 λ clones of the M. kandleri European Molecular Biology Laboratory 3 λ library (12, 16). Remaining gaps in the genome, as well as low-quality and single-stranded regions, were closed by directed reads from genomic and λ DNA. For this step, we used the autofinish program (21, 22), with options allowing the choice of Fimers sequences for whole-genome reads and λ clone custom reads. After generating 1,585 chromatograms, the genome was assembled in a unique contig with an estimated error rate of 0.4/10 kb, which was done with 12,046 reads (≈3.0× coverage). With an additional 2,147 genomic and λ walking reads, the accuracy of less than one error per 40,000 bases was achieved (total 14,139 reads, 3.3× coverage). λ clones covered 85% of genome, with an average insert size of 14,500 bp (min 12,230, max 19,324). There were no discrepancies between the expected insert lengths in λ clones and the corresponding regions in the final genome sequence.
Detailed sequencing protocols are published as supporting information on the PNAS web site (www.pnas.org).
Computational Genome Analysis.
The tRNA genes were identified by using the trna-scan program (23), and rRNA genes were identified by using the blastn program (24) with archaeal rRNA as search queries. For identification of protein-coding genes, the genome sequence was conceptually translated in six frames to generate potential protein products of ORFs longer than 100 codons (from stop to stop). The potential protein sequences were compared with the database of Clusters of Orthologous Groups (COGs) of proteins by using cognitor (25). After manual verification of the COG assignments, the validated COG members from M. kandleri were called protein-coding genes. The COG assignment procedure was repeated with ORF products greater than 60 codons from intergenic regions. Additionally, the potential protein sequences were compared with the nonredundant protein sequence database by using the blastp program and to a six-frame translation of unfinished microbial genomes using the tblastn program; those that produced hits with E (expectation) values <0.01 were added to the protein set after examination of the alignments. Finally, protein-coding regions were predicted by using the genemarks (26) and syncod (27) programs. The genes predicted with these methods in the regions between evolutionarily conserved genes were added to produce the final protein set.
Protein function prediction was based primarily on the COG assignments. In addition, searches for conserved domains were performed by using the conserved domain database option of blast (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), the smart system (http://smart.embl-heidelberg.de/) (28), and customized position-specific score matrices for different classes of DNA-binding proteins. In-depth iterative database searches were performed by using the psi-blast program (24). The kegg database (http://www.genome.ad.jp/kegg/metabolism.html) (29) was used, in addition to the COGs, for the reconstruction of metabolic pathways. Paralogous protein families were identified by single-linkage clustering of M. kandleri proteins after comparing the predicted protein set to itself by using the blastp program (30). Signal peptides in proteins were predicted by using the singalp (31) program, and transmembrane helices were predicted by using the memsat program (32).
Gene orders in archaeal and bacterial genomes were compared by using the lamarck program (33). For phylogenetic analysis, multiple alignments of ribosomal protein sequences were constructed by using the t-coffee program (34) and concatenated head-to-tail. Maximum likelihood (ML) trees were generated by exhaustive search of all possible topologies by using the protml program of the molphy package, with the Jones–Thornton–Taylor-Frequencies model of amino acid substitutions (35). Bootstrap analysis was performed for each ML tree by using the Resampling of Estimated Log-Likelihoods method (10,000 replications) (36, 37). The likelihoods of alternative placements of M. kandleri in ML trees were compared by using the Kishino–Hasegawa test (37).
Results and Discussion
Directed Genomic Sequencing of M. kandleri.
The M. kandleri genome was sequenced directly from genomic DNA. The approach consisted in selecting primer sequences at the contig ends and around low-quality regions, followed by sequencing off genomic DNA to extend into gap regions and to cover low-quality areas. The skimming shotgun phase was required only to provide starting points for genome-wide Fimer selection. λ clones were used mainly to verify the genome assembly. With this method, the complete 1,694,969-bp high-quality sequence (less than one error per 40 kb) was obtained from just 14,139 sequencing reads, significantly exceeding the sequencing quality established by “Bermuda statement” (http://www.ornl.gov/hgmis/research/bermuda.html). The method has certain advantages over the whole shotgun approach: the overall workflow is simple; there is no need for storing plasmids and “cherry-picking” individual clones for finishing because, once isolated, genomic DNA is used throughout the entire project; and finally, Fimers synthesized for one project can be used for comparative sequencing of many related strains, which can greatly reduce costs of sequencing projects (see supporting information on the PNAS web site for examples).
A key component of the method is a robust sequencing reaction customized for genomic DNA that gives high-quality reads. This reaction is based on big dye terminator sequencing chemistry (38) and incorporates ThermoFidelase 2 (39) and 2′-modified oligonucleotides (Fimers) with greatly improved specificity and template-annealing characteristics (18).
During this project, we experimented with the basic sequencing reaction to achieve the best results in terms of cost effectiveness, ability to pass high GC-rich regions (average 62.1% GC), and success rate. The 10-μl sequencing reaction with Fimers, ThermoFidelase 2, 0.1 mM deaza dGTP, and 200 cycles gave the best results. We achieved the overall success rate of ≈90% and increased average phred quality 20 read length from 370 bases in the beginning of the project to 500 bases at the end. All artifacts typically associated with increased number of cycles, such as primer–dimer extensions and nonspecific PCRs (18, 40), were eliminated by using Fimers.
The Genome, the Genes, and Their Phyletic Patterns.
The genome of M. kandleri is a single circular chromosome that consists of 1,694,969 bp. Using a combination of sequence database searches and coding potential prediction, 1,691 protein-coding genes and 39 genes for structural RNAs were identified. Using a combination of GC-skew and gene context analysis, the origin of replication site of M. kandleri was predicted to reside in the intergenic spacer between nucleotides 1694501–747 (see supporting information on the PNAS web site for details).
When compared with proteins from other archaea, the predicted proteins of M. kandleri had certain notable properties that appeared to correlate with its extremely high intracellular salinity (>3 M K+). With the sole exception of Halobacterium sp., M. kandleri has the highest ratio among all archaea of negatively to positively charged residues and the lowest isoelectric point of ≈5 (see supporting information on the PNAS web site.).
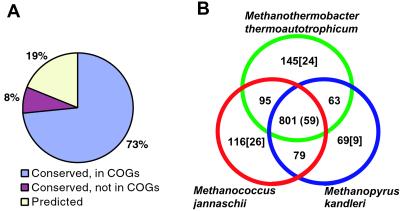
The analysis of evolutionary relationships and predicted functions of M. kandleri proteins was based on the COGs of proteins from sequenced genomes (25). Similar to other archaea and bacteria, 73% of the gene products of M. kandleri are conserved proteins that belong to COGs. A substantial fraction of the remaining proteins are shared with one of the archaeal methanogens. A much smaller number of proteins were conserved in M. kandleri and other archaea or bacteria, and ≈19% of the predicted proteins had no detectable homologs in current databases and were identified on the basis of predicted structural features (such as transmembrane helices or signal peptides) or by coding potential prediction alone (Fig. 1A).
Figure 1.
The gene repertoires of M. kandleri and other archaeal methanogens. (A) Conserved and unique proteins in M. kandleri. (B) A Venn diagram of the shared and unique portions of the proteomes of archaeal methanogens. The number of COGs that are unique for the three methanogens is shown in parentheses in the central section. The number of COGs that include a given archaeal species and two or more bacteria, but no other archaea, is shown in brackets.
Examination of the phyletic patterns of the COGs that include M. kandleri and other archaea showed remarkable coherence between the three available genomes of archaeal methanogens. The great majority of the COGs, in which one of these species is represented, also include the other two species; many of the remaining COGs include two methanogens (Fig. 1B). Notably, the number of COGs, in which M. kandleri is the only methanogenic archaeon, is much smaller than the corresponding numbers for the other two methanogens (Fig. 1B). Only nine COGs include M. kandleri as the sole archaeal species, suggesting that there are fewer traces of horizontal gene exchange with bacteria in this genome than in other archaeal genomes (Fig. 1B) (41). The three methanogens share 59 COGs that are not represented in any other archaea or bacteria and therefore seem to comprise a genomic signature of this group. This notion is supported by the fact that a much smaller number of COGs or no COGs at all are uniquely shared by M. kandleri and any two nonmethanogenic archaeal species (see supporting information on the PNAS web site). In addition, the methanogens seem to have a specific relationship with Archaeoglobus fulgidus, an archaeon with a similar metabolism; 34 COGs are uniquely shared by the three methanogens and A. fulgidus.
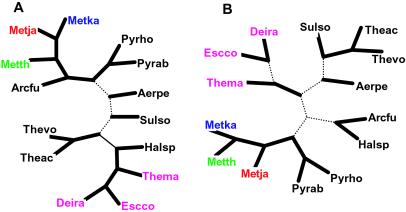
Overlap between gene sets can be used to compute distances between genomes; this measure seems to reflect a combination of phylogenetic affinity and similarities in the life styles of the corresponding organisms (25). The gene content tree for the sequenced archaeal genomes shows tight clustering of the three methanogens, which is strongly supported by bootstrap replication, emphasizing the coherence of their gene repertoires, to the exclusion of other species (Fig. 2A).
Figure 2.
Genome trees of archaea. (A) Gene content tree. (B) Tree based on conserved gene pairs. Solid lines show terminal branches and internal branches with >90% bootstrap support, and dotted lines show internal branches with <90% support. The three methanogen species are highlighted by red, green, and blue. The bacterial species included as an outgroup are shown in magenta. Species abbreviations: Euryarchaeota: A. fulgidus (Arcfu), M. thermoautotrophicum (Metth), M. jannaschii (Metja), M. kandleri (Metka), Pyrococcus horikoshii (Pyrho), Pyrococcus abyssi (Pyrab), Thermoplasma volcanium (Thevo), Thermoplasma acidophilum (Theac), Halobacterium sp_._ (Halsp); Crenarchaeota: Aeropyrum pernix (Aper), Sulfolobus solfataricus (Sulso); bacteria: Thermotoga maritima (Thema), Deinococcus radiodurans (Deira), Escherichia coli (Escco).
Conservation of Gene Order and Operons in the Three Methanogens.
Gene order is generally poorly conserved in prokaryotic evolution (33, 42): synteny breaks down even between closely related species, and only a handful of operons are conserved, for example, between archaea and bacteria (33). Nevertheless, local gene order tends to roughly reflect the phylogenetic distance between moderately diverged genomes as well as preferential horizontal gene transfer (43, 44). When the number of shared gene pairs was used to generate a tree of archaeal genomes (44), clustering of the three methanogens was observed with a strong support (Fig. 2B). Within the methanogen cluster, M. kandleri confidently grouped with M. thermoautotrophicum (Fig. 2B), emphasizing the specific conservation of gene order in these two genomes.
The three methanogens contain the gene clusters that, to a varying degree, are conserved in all archaea, including the ribosomal superoperon, the predicted exosomal superoperon, and the gene cluster coding for a predicted thermophile-specific DNA repair system. In addition, the methanogens share several large conserved gene clusters (including one or more predicted operons) that are missing in other genomes. These clusters consist of genes coding for enzymes and structural membrane proteins known or inferred to be involved in methanogenesis or hydrogenogenesis. In particular, both M. kandleri and M. jannaschii possess readily identifiable orthologs of all genes in a large operon recently described in M. thermoautotrophicus, which includes two hydrogenase subunits (45) (see supporting information on the PNAS web site). However, M. jannaschii and M. thermoautotrophicum also have a partial duplication of this operon, which is missing in M. kandleri. M. kandleri has two disjointed genes for the α and C subunits of methylcoenzyme M reductase; the genome surroundings of the gene for the truncated α subunit suggest that the second copy of this operon was disrupted in the M. kandleri lineage (see supporting information on the PNAS web site). Another methanogen-specific gene cluster contains genes for a set of predicted enzymes implicated in the biosynthesis of cofactors for specific hydrogenation reactions involved in methanogenesis (see supporting information on the PNAS web site). Thus, gene order comparison supports a specific relationship between the three archaeal methanogens. Greater conservation of predicted operons was consistently observed between M. kandleri and M. thermoautotrophicum than between any of them and M. jannaschii; an example is the considerable collinearity of the exosomal superoperons in the former two, but not in the latter (data not shown).
Phylogeny: Strong Support for a Methanogen Clade Among Archaea.
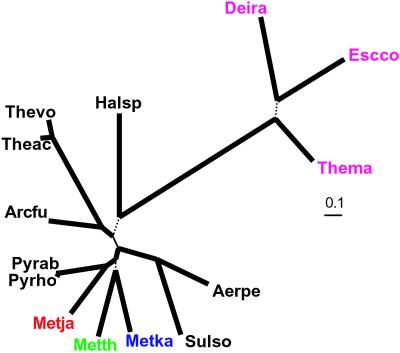
Until recently, phylogenetic analysis of rRNA, in some cases supplemented by analysis of selected protein families, remained the principal means of deciphering the evolutionary relationship between organisms (44, 46). As of late, several approaches to “genome-wide” phylogeny were explored (44, 47). A comparison of several such measures suggested that the most reliable phylogenies were produced by using concatenated alignments of ribosomal proteins, whose genes are less prone to horizontal gene transfer than other genes and therefore are suitable for phylogenetic analysis (44). In ML trees constructed by using this approach, the three methanogens formed a strongly supported clade with the Pyrococci. The topology within this cluster was less well defined, but the preferred one placed M. kandleri in a clade with M. thermoautotrophicum, whereas M. jannaschii clustered with the Pyrococci (Fig. 3). The only alternative placement of M. kandleri in the tree with comparable likelihood was in the root of the methanogen–pyrococci clade (data not shown). A strongly supported methanogen–pyrococci clade was also observed in a tree constructed using the median distances between orthologs (data not shown; ref. 44).
Figure 3.
The ML phylogenetic tree of archaea constructed by using concatenated alignments of ribosomal proteins. Thick solid lines indicate internal branches with Resampling of Estimated Log-Likelihoods bootstrap support >95%, thin solid lines indicate branches with 90–95% bootstrap support, and dotted lines indicate branches with <70% support. Distances are indicated in substitutions per site. Methanogens are highlighted. Species name abbreviations are as in Fig. 2.
Predicted Proteins and Functional Systems: M. kandleri Is a Typical Archaeal Methanogen.
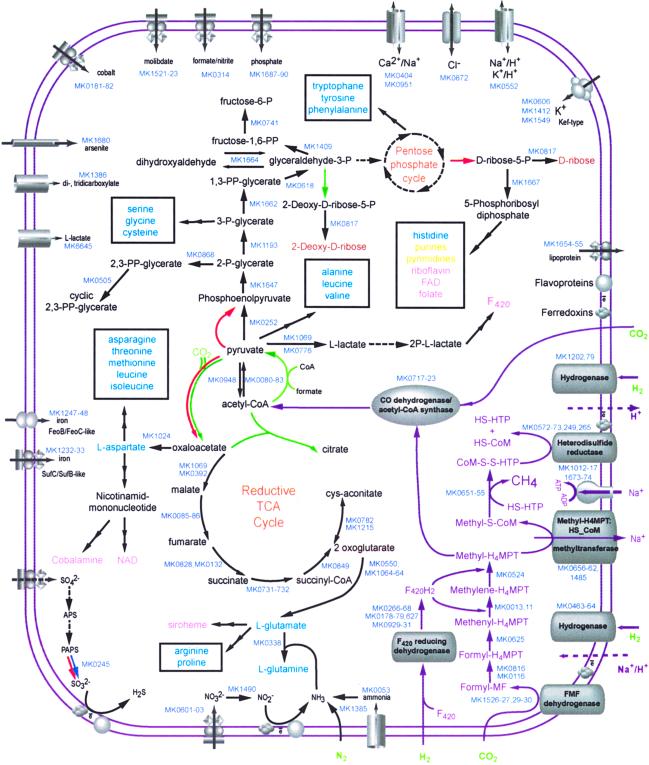
Comparison of the predicted proteome of M. kandleri to those of other archaea and bacteria allowed a reconstruction of the principal functional systems and metabolic pathways of this organism (Fig. 4). M. kandleri encodes all functional systems typical of archaea, including certain characteristic complexes, such as the proteasome (MK0385, MK0878, MK1228), the predicted exosome (MK0375-MK0389), and, notably, the potential thermophile-specific repair system (MK1296-MK1299, MK1314-MK1319) (48–50).
Figure 4.
Predicted functional systems and general metabolic pathways of M. kandleri compared with the counterparts in other archaeal methanogens. Specific enzymatic reactions predicted in a given species but not in the other two are color-coded blue (M. kandleri), green (M. thermoautotrophicus), and red (M. jannaschii). Gene identifiers are shown only for M. kandleri. The methanogenesis pathway is shown by magenta arrows. Reactions for which no candidate enzyme was confidently predicted are shown by dashed arrows. Final biosynthetic products are shown as follows: light blue for amino acids, dark yellow for nucleotides, brown for sugars, pink for cofactors. MF, methanofuran; FMF, formyl-methanofuran; H4MPT, tetrahydromethanopterin; CoM, coenzyme M.
The general metabolic schemes of M. kandleri, M. jannaschii, and M. thermoautotrophicum are highly similar (Fig. 4), which is not surprising because they are all H2-dependent autotrophic methanogens and have anaerobic respiration with CO2 as the electron acceptor (51). In addition to central metabolic pathways and biosynthetic pathways, which, with some variations, are conserved in all autotrophic archaea, all three have the same set of genes for proteins involved in methanogenesis and the coupled electron transfer (Fig. 4).
As with any other sequenced genome, several “gaps” remain in the predicted proteome of M. kandleri whereby an enzyme for an essential step in a pathway could not be identified. The most notable case in M. kandleri is the apparent absence of an enzyme for coupling pyruvate metabolism with the tricarboxylic acid cycle (Fig. 4). Several different enzymes can make malate or oxaloacetate from pyruvate or phosphoenolpyruvate, but we failed to identify orthologs for any of them in M. kandleri; both M. thermoautotrophicum and M. jannaschii produce oxaloacetate from pyruvate in a reaction catalyzed by pyruvate carboxylase. An uncharacterized predicted ATP-dependent carboligase (COG2232), which is present in all methanogenes and A. fulgidus, could be an alternative enzyme for this reaction and the only one in M. kandleri. Additional details about shared and unique features of methanogens are published as supporting information on the PNAS web site. The most likely explanation for found anomalies is nonorthologous gene displacement (52).
Analysis of the metabolic capabilities of the three methanogens suggested that M. kandleri has the lowest diversity of characterized metabolic pathways and enzymes among the three species (see supporting information on the PNAS web site). The predicted M. kandleri proteome also shows a substantially smaller diversity of predicted membrane transporters compared with other archaeal methanogens (see Table 1, which is published as supporting information on the PNAS web site). However, we identified several specific membrane proteins and even protein families that potentially could be novel transporters (MK1144, MK0913, MK0914, MK0278, MK0279, and some others).
In addition to the conserved enzymatic systems implicated in methanogenesis, M. kandleri shares several unusual features of essential proteins and systems previously noticed in M. jannaschii and M. thermoautotrophicum (see supporting information on the PNAS web site). These organisms are the only ones so far that lack a predicted cysteinyl–tRNA synthetase. In M. jannaschii, the role of this enzyme is taken over by the bifunctional prolyl-tRNA synthetase (53), and the same can be predicted for M. kandleri, especially given that the prolyl-tRNA synthetases of the three methanogens form a tight cluster in the corresponding phylogenetic tree (data not shown). The three methanogens also have unusual seryl-tRNA synthetases that are highly conserved among themselves but are only distantly related to the orthologs from other species.
M. kandleri has only a few recognizable unique domain architectures and predicted operons. Previously, two protein domain architectures of M. kandleri were described that are different from the architectures of orthologs in other archaea: the split of the reverse gyrase into distinct genes for the helicase and the topoisomerase domains and fusion of the genes for archael orthologs of histones H3 and H4 (11, 12, 16). Genome analysis revealed a few additional unusual domain arrangements, e.g., the unique fusion of archaeal fructose-1,6-bisphosphatase and an enzymatic domain related to phosphoribosyl-ATP pyrophosphohydrolase, a histidine biosynthesis enzyme (MK0741). Several other unique domain architectures involve uncharacterized domains. Examples include an archaea-specific paralog of the histidine biosynthesis enzyme HisA (MK0492 protein), 2-phosphoglycerate kinase (MK0033), and hydrolase of the metallo-β-lactamase superfamily (MK1258), each of which is fused to a domain of unknown function.
Unusual features of operon organization in M. kandleri include the fission of certain conserved operons, such as the rRNA and V-type ATPase operons. M. kandleri also has several unique predicted operons. The most notable one is a large gene cluster with 12 genes (MK0698-MK0708) that apparently encodes a type II/IV-like secretory system, which, in other prokaryotes, is involved in the secretion of protein and DNA–protein complexes. The readily recognizable genes in this cluster encode the traffic ATPase of the PilT superfamily (MK0707), a type IV signal peptidase (MK0703) and a ComM-like AAA+ ATPase. Also encoded by this operon are at least five uncharacterized proteins with predicted N-terminal signal peptides (MK0698-MK0702) that might be secreted by this system and could perform specialized extracellular functions.
Genome analysis showed that each species has lineage-specific expansions of paralogous protein families, which encompass from 3 to ≈30% of the genes (54). In M. kandleri, ≈12% of the genes belong to specific expansions. The largest family consists of uncharacterized proteins that have no detectable homologs in other genomes. Some of these proteins have predicted signal peptides and transmembrane helices and might be involved in unique transport mechanisms (families typified by MK1439, MK1354, and MK1158). Among the few specific expansions of known enzymes are ATP-dependent DNA ligases (in addition to the two conserved forms typical of all archaea, M. kandleri encodes two highly diverged predicted ligases), the SUN family of RNA cytosine-C5-methylases (9 paralogs versus one and none in M. jannaschii and M. thermoautotrophicum, respectively), and terpene cyclase/mutase family enzymes (five paralogs versus none in other methanogens). Also, M. kandleri has three lineage-specific paralogs of a predicted secreted polysaccharide hydrolase that is present in a single copy in M. jannaschii and several bacteria. This protein was recently claimed to be a cysteinyl tRNA synthetase (55), but the predicted catalytic activity and extracellular localization argue against this (56). These lineage-specific expansions are likely to be unique adaptations, but the available data are insufficient to offer specific biological interpretations.
Signaling and Regulatory Systems: M. kandleri Is a “Minimalist” Archaeon.
Signal transduction systems are generally underrepresented in hyperthemophiles compared with mesophiles and moderate thermophiles, and M. kandleri is no exception. In particular, like M. jannaschii, it has no two-component histidine kinase regulatory systems, which are present in moderately thermophilic archaea, including M. thermoautotrophicum. However, an unexpected finding in the M. kandleri genome is a homolog of the bacterial P-loop-containing serine kinase that phosphorylates the Hpr protein (this protein has a unique insert of a Zn-ribbon), a component of the phosphotransferase system (PTS) of sugar transport, which is a key regulator of carbohydrate metabolism in Gram-positive bacteria. This is a key regulator of carbohydrate metabolism in Gram-positive bacteria, but so far has not been detected in archaea. Because archaea do not have a PTS system, this kinase might have a regulatory role distinct from its function in bacteria. This enzyme is one of the few apparent lateral acquisitions of bacterial genes in M. kandleri.
All archaeal genomes sequenced so far encode a substantial number of predicted DNA-binding proteins of the helix-turn-helix and Arc-MetJ classes. The abundance of these proteins in archaea is comparable to that in bacteria, and most of them are implicated in regulation of transcription of specific operons. As shown by sequence profile analysis, M. kandleri has markedly less DNA-binding proteins of each of these classes than most of the other archaea (see supporting information on the PNAS web site), which probably reflects an unusually low, for a free-living organism, diversity of regulatory responses.
Concluding Remarks.
Comparative analysis of the M. kandleri genome and other sequenced archaeal and bacterial genomes led to three principal conclusions. First, M. kandleri encodes the core of proteins that are conserved in other Euryarchaea and, accordingly, is predicted to have all major functional systems and pathways typical of this group of organisms. Second, M. kandleri closely resembles other archaeal methanogens in terms of gene content and local gene order, and this conservation parallels the reliable grouping of the methanogens in phylogenetic trees constructed from concatenated alignments of ribosomal proteins. Thus, comparative-genomic analysis appears to indicate that M. kandleri belongs to a monophyletic group of archaeal methanogens (or at least to a larger group that also includes Pyrococci) and is not a deep-branching species close to the root of the archaeal (or euryarchaeal) clade, as previously suspected. The congruence of different types of evidence seems to suggest that the result of rRNA analysis (13–15) (the main aspects of which we were able to reproduce; data not shown) might be an artifact, perhaps because of unusual base composition effects. Third, several features of the gene repertoire of M. kandleri characterize this organism as a “minimalist” archaeon, with a small number of genes transferred from bacteria, low diversity of transporters, and particularly underrepresentation of predicted regulatory proteins. This frugality might be related to the extreme habitat of M. kandleri, which could limit its ability to exchange genes with other organisms.
Supplementary Material
Supporting Information
Acknowledgments
This work was supported in part by Department of Energy and National Institutes of Health grants (DE-FG02-98ER82577, 00ER83009, R44GM55485, and R43HG02186) to S.A.K and A.I.S.
Abbreviations
COG
Cluster of Orthologous Groups
ML
maximum likelihood
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE009439).
References
- 1.Huber R, Kurr M, Jannasch H W, Stetter K O. Nature (London) 1989;342:833–836. [Google Scholar]
- 2.Kurr M, Huber R, Konig H, Jannasch H W, Fricke H, Trincone A, Kristjansson J K, Stetter K O. Arch Microbiol. 1991;156:239–247. [Google Scholar]
- 3.Hafenbradl D, Keller M, Thiericke R, Stetter K O. Syst Appl Microbiol. 1993;16:165–169. [Google Scholar]
- 4.Wachtershauser G. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shima S, Herault D A, Berkessel A, Thauer R K. Arch Microbiol. 1998;170:469–472. doi: 10.1007/s002030050669. [DOI] [PubMed] [Google Scholar]
- 6.Breitung J, Borner G, Scholz S, Linder D, Stetter K O, Thauer R K. Eur J Biochem. 1992;210:971–981. doi: 10.1111/j.1432-1033.1992.tb17502.x. [DOI] [PubMed] [Google Scholar]
- 7.Slesarev A I, Lake J A, Stetter K O, Gellert M, Kozyavkin S A. J Biol Chem. 1994;269:3295–3303. [PubMed] [Google Scholar]
- 8.Slesarev A I, Stetter K O, Lake J A, Gellert M, Krah R, Kozyavkin S A. Nature (London) 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 9.Belova G I, Prasad R, Kozyavkin S A, Lake J A, Wilson S H, Slesarev A I. Proc Natl Acad Sci USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slesarev A I, Belova G I, Lake J A, Kozyavkin S A. Methods Enzymol. 2001;334:179–192. doi: 10.1016/s0076-6879(01)34467-1. [DOI] [PubMed] [Google Scholar]
- 11.Kozyavkin S A, Krah R, Gellert M, Stetter K O, Lake J A, Slesarev A I. J Biol Chem. 1994;269:11081–11089. [PubMed] [Google Scholar]
- 12.Krah R, Kozyavkin S A, Slesarev A I, Gellert M. Proc Natl Acad Sci USA. 1996;93:106–110. doi: 10.1073/pnas.93.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burggraf S, Stetter K O, Rouviere P, Woese C R. Syst Appl Microbiol. 1991;14:346–351. doi: 10.1016/s0723-2020(11)80308-5. [DOI] [PubMed] [Google Scholar]
- 14.Rivera M C, Lake J A. Int J Syst Bacteriol. 1996;46:348–351. doi: 10.1099/00207713-46-1-348. [DOI] [PubMed] [Google Scholar]
- 15.Nolling J, Elfner A, Palmer J R, Steigerwald V J, Pihl T D, Lake J A, Reeve J N. Int J Syst Bacteriol. 1996;46:1170–1173. doi: 10.1099/00207713-46-4-1170. [DOI] [PubMed] [Google Scholar]
- 16.Slesarev A I, Belova G I, Kozyavkin S A, Lake J A. Nucleic Acids Res. 1998;26:427–430. doi: 10.1093/nar/26.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polushin N N. Nucleic Acids Res. 2000;28:3125–3133. doi: 10.1093/nar/28.16.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polushin N, Malykh A, Malykh O, Zenkova M, Chumakova N, Vlassov V, Kozyavkin S. Nucleosides Nucleotides. 2001;20:507–514. doi: 10.1081/NCN-100002326. [DOI] [PubMed] [Google Scholar]
- 19.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 20.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 21.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 22.Gordon D, Desmarais C, Green P. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fichant G A, Burks C. J Mol Biol. 1991;220:659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 26.Besemer J, Lomsadze A, Borodovsky M. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogozin I B, D'Angelo D, Milanesi L. Gene. 1999;226:129–137. doi: 10.1016/s0378-1119(98)00509-5. [DOI] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 32.McGuffin L J, Bryson K, Jones D T. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 33.Wolf Y I, Rogozin I B, Kondrashov A S, Koonin E V. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 34.Notredame C, Higgins D G, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 35.Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phytogenetics 27. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 36.Hasegawa M, Kishino H, Saitou N. J Mol Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 37.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;31:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 38.Rosenblum B B, Lee L G, Spurgeon S L, Khan S H, Menchen S M, Heiner C R, Chen S M. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malykh A, Malykh O, Polouchine N, Kozyavkin S, Slesarev A. In: Methods in Molecular Biology. Stodolsky M, Zhao S, editors. Totowa, NJ: Humana; 2002. , in press. [Google Scholar]
- 40.Stump M D, Cherry J L, Weiss R B. Nucleic Acids Res. 1999;27:4642–4648. doi: 10.1093/nar/27.23.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin E V, Makarova K S, Aravind L. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mushegian A R, Koonin E V. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- 43.Suyama M, Bork P. Trends Genet. 2001;17:10–13. doi: 10.1016/s0168-9525(00)02159-4. [DOI] [PubMed] [Google Scholar]
- 44.Wolf Y I, Rogozin I B, Grishin N V, Tatusov R L, Koonin E V. BMC Evol Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tersteegen A, Hedderich R. Eur J Biochem. 1999;264:930–943. doi: 10.1046/j.1432-1327.1999.00692.x. [DOI] [PubMed] [Google Scholar]
- 46.Olsen G J, Woese C R, Overbeek R. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snel B, Bork P, Huynen M A. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 48.Macario A J, Lange M, Ahring B K, De Macario E C. Microbiol Mol Biol Rev. 1999;63:923–967. doi: 10.1128/mmbr.63.4.923-967.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koonin E V, Wolf Y I, Aravind L. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makarova K S, Aravind L, Grishin N V, Rogozin I B, Koonin E V. Nucleic Acids Res. 2001;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer G, Engelhard M, Muller V. Microbiol Mol Biol Rev. 1999;63:570–620. doi: 10.1128/mmbr.63.3.570-620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonin E V, Mushegian A R, Bork P. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 53.Stathopoulos C, Jacquin-Becker C, Becker H D, Li T, Ambrogelly A, Longman R, Soll D. Biochemistry. 2001;40:46–52. doi: 10.1021/bi002108x. [DOI] [PubMed] [Google Scholar]
- 54.Jordan I K, Makarova K S, Spouge J L, Wolf Y I, Koonin E V. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabrega C, Farrow M A, Mukhopadhyay B, de Crecy-Lagard V, Ortiz A R, Schimmel P. Nature (London) 2001;411:110–114. doi: 10.1038/35075121. [DOI] [PubMed] [Google Scholar]
- 56.Iyer L M, Aravind L, Bork P, Hofmann K, Mushegian A R, Zhulin I B, Koonin I V. Genome Biol. 2001;2:0051.1–0051.11. doi: 10.1186/gb-2001-2-12-research0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information