CDK Phosphorylation of a Novel NLS-NES Module Distributed between Two Subunits of the Mcm2-7 Complex Prevents Chromosomal Rereplication (original) (raw)
Abstract
Cyclin-dependent kinases (CDKs) use multiple mechanisms to block reassembly of prereplicative complexes (pre-RCs) at replication origins to prevent inappropriate rereplication. In Saccharomyces cerevisiae, one of these mechanisms promotes the net nuclear export of a pre-RC component, the Mcm2-7 complex, during S, G2, and M phases. Here we identify two partial nuclear localization signals (NLSs) on Mcm2 and Mcm3 that are each necessary, but not sufficient, for nuclear localization of the Mcm2-7 complex. When brought together in cis, however, the two partial signals constitute a potent NLS, sufficient for robust nuclear localization when fused to an otherwise cytoplasmic protein. We also identify a Crm1-dependent nuclear export signal (NES) adjacent to the Mcm3 NLS. Remarkably, the Mcm2-Mcm3 NLS and the Mcm3 NES are sufficient to form a transport module that recapitulates the cell cycle-regulated localization of the entire Mcm2-7 complex. Moreover, we show that CDK regulation promotes net export by phosphorylation of the Mcm3 portion of this module and that nuclear export of the Mcm2-7 complex is sufficient to disrupt replication initiation. We speculate that the distribution of partial transport signals among distinct subunits of a complex may enhance the specificity of protein localization and raises the possibility that previously undetected distributed transport signals are used by other multiprotein complexes.
INTRODUCTION
The faithful transmission of genetic information during cell division requires that complete duplication of the genome during S phase strictly alternate with accurate segregation of the duplicated genome during M phase. Eukaryotic cells ensure that their genome is duplicated precisely once per cell cycle by enforcing a single round of replication initiation at each of the hundreds to thousands of replication origins scattered throughout their genome. We and others have shown that reinitiation in the budding yeast Saccharomyces cerevisiae causes a rapid and serious insult to the genome, triggering a DNA damage response and cell cycle arrest (Archambault et al., 2005; Green and Li, 2005). In other metazoans, rereplication also induces checkpoint responses and, in some cases, leads to apoptosis (Mihaylov et al., 2002; Melixetian et al., 2004; Zhu et al., 2004). Thus, restricting DNA replication initiation to a single round per cell cycle is critical for genome integrity and cell survival.
Cyclin-dependent kinases play a critical role in the cell cycle regulation of replication initiation by controlling both the activation and formation of the prereplicative complex (pre-RC), a critical intermediate in the initiation reaction (reviewed in Bell and Dutta, 2002; Diffley, 2004). Assembly of the pre-RC in G1 phase, when CDK activity is low, makes origins competent for replication initiation later in the cell cycle when CDK activity is induced. The pre-RC is assembled when the origin recognition complex (ORC) binds origins and recruits Cdc6 and Cdt1 to help load the putative replicative helicase, the heterohexameric Mcm2-7 complex. Activation of the pre-RC by CDKs and Cdc7-Dbf4 kinase then leads to recruitment of additional replication proteins and the triggering of initiation. This activation is accompanied by disassembly of the pre-RC: Cdc6 and Cdt1 dissociate from the origin and the Mcm2-7 complex is thought to move with the replication fork as part of the replisome.
In addition to triggering initiation, CDKs can inhibit reinitiation by blocking reassembly of the pre-RC (Diffley, 2004). This inhibitory function has been investigated most extensively in S. cerevisiae, where at least three inhibitory targets of the CDK Cdc28 have been identified: ORC, Cdc6, and the Mcm2-7 complex (Nguyen et al., 2001). The mechanism by which this inhibition occurs is best understood for Cdc6, which is regulated by Cdc28 both transcriptionally and post-translationally (Moll et al., 1991; Piatti et al., 1995; Drury et al., 1997; Mimura et al., 2004). Cdc28 kinase inhibits ORC function by phosphorylation of Orc2 and Orc6 (Nguyen et al., 2001) and binding of the S-phase cyclin Clb5 to Orc6 (Wilmes et al., 2004). In other eukaryotes, CDK phosphorylation has also been shown to promote the ubiquitin-mediated degradation of Cdt1 (Liu et al., 2004; Sugimoto et al., 2004; Thomer et al., 2004).
We and others have shown that Cdc28 promotes the nuclear exclusion of the Mcm2-7 complex (Labib et al., 1999; Nguyen et al., 2000). In G1 phase, when Cdc28 kinase levels are low, the Mcm2-7 complex accumulates in the nucleus independent of its loading onto origins. After Cdc28 kinase becomes active in late G1 phase, the Mcm2-7 complex experiences net nuclear export until it is excluded from the nucleus. Little is understood about how Cdc28 controls localization of the Mcm2-7 complex. For example, it is not known whether Cdc28 promotes Mcm2-7 nuclear export by directly phosphorylating the Mcm complex, by targeting some of the many replication proteins known to interact with the complex, or by modulating the activity of the transport machinery.
Most transport of proteins across the nuclear envelope is mediated by a family of nucleocytoplasmic transport receptors, which shuttle proteins through the nuclear pore in a unidirectional manner (reviewed in Weis, 2003). Nuclear import is mediated by the binding of nuclear import receptors to nuclear localization signals (NLSs) and nuclear export is mediated by the binding of nuclear export receptors to nuclear export signals (NESs). Proteins that shuttle between the nucleus and cytoplasm, like the Mcm2-7 complex, often contain both NLSs and NESs, with the relative rates of nuclear import and export specified by these signals determining the steady state localization of a protein. Hence, mechanisms that directly or indirectly affect the activity of NLSs and NESs can control the nucleocytoplasmic localization a protein (reviewed in Jans et al., 2000). Proteins that do not contain nuclear transport signals can also be nuclear localized by associating with a protein that does contain these signals. In fact, studies that have dissected the transport signals responsible for localization of multiprotein complexes have uncovered numerous examples of a single subunit providing the transport signal for the entire complex (Maridor et al., 1993; Pereira et al., 1998; Leslie et al., 2004; Subramaniam and Johnson, 2004; Wendler et al., 2004).
In this study, we identify two partial nuclear localization signals on two distinct subunits of the Mcm2-7 complex, Mcm2 and Mcm3. These elements are each necessary for nuclear localization of the entire Mcm2-7 complex, but unlike canonical NLSs, neither is sufficient for this localization. Together, however, they exhibit strong import activity when fused to a heterologous protein. We have also identified an NES adjacent to the Mcm3 NLS. All three signals effectively constitute a transport module that, when fused to a heterologous protein, can recapitulate the cell cycle-regulated localization of the Mcm2-7 complex. We show that CDK phosphorylation of the Mcm3 portion of this transport module promotes the net nuclear export of both a heterologous fusion protein and the Mcm2-7 complex. We also demonstrate that this CDK directed export is sufficient to disrupt the initiation of DNA replication, establishing that this regulation contributes significantly to the control of replication initiation. Finally, we suggest that distribution of transport modules among distinct subunits of a complex may couple protein localization to complex assembly and could be used by other multiprotein complexes.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth
S. cerevisiae strains used in this study were derivatives of YJL310 (Detweiler and Li, 1998; Nguyen et al., 2000) or W303. YEP medium and synthetic complete medium (Guthrie and Fink, 1991) were supplemented with 2% dextrose (YEPD; SDC), or 2% raffinose (YEPR; SRafC). The GAL10 promoter (pGAL) was induced by addition of 2% galactose unless otherwise indicated.
For in vivo labeling of Mcm3, low-phosphate YEPD was prepared as follows: 10 g yeast extract and 20 g bacto peptone were dissolved in 1 l of water and 10 ml 1 M MgSO4, and 10 ml concentrated NH4OH was added. A cloudy precipitate, formed during 30 min of stirring, was removed by filtration through Whatman No. 1 filter. The pH was adjusted to 5.8 with concentrated HCl, and the medium was autoclaved and then supplemented with 10 ml ADE/TRP (each 5 mg/ml).
Plasmids and Strains
Plasmids and strains used in this study are described in detail in the supplementary information.
Cell Growth, Arrest, and Release
To arrest cells, α-factor was used at a final concentration of 50 ng/ml (all strains were bar1; see Figures 2, 3, 4, 6, and 10), and nocodazole (NOC) was used at a final concentration of 15 μg/ml (see Figures 2, 3, 4, 6, and 8). For Figure 2, cell cycle blocks were relieved by filtering the cells, washing them three times with an equal volume of resuspension medium prewarmed to the appropriate temperature, and then resuspending them in the appropriate medium. For Figures 4B and 6, cells were released from the α-factor arrest by addition of Pronase at 100 μg/ml. To inactivate Crm1-T539C, Leptomycin B (a gift of Minoru Yoshida; Nishi et al., 1994; Kudo et al., 1999) was added to a final concentration of 100 ng/ml.
Figure 2.
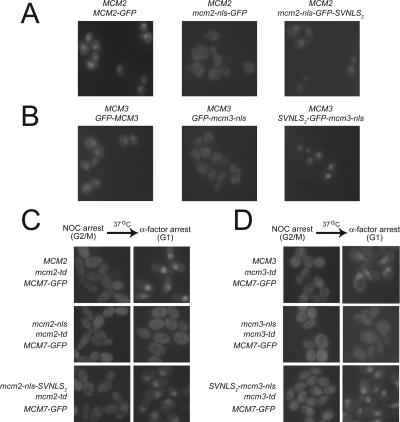
The Mcm2 and Mcm3 NLSs are each required for nuclear localization of the Mcm2-7 complex. (A) The Mcm2 NLS is required for nuclear localization of Mcm2-GFP. YJL3265 (MCM2::{MCM2-GFP}), YJL1231 (MCM2::{mcm2-nls-GFP}), and YJL1228 (MCM2::{mcm2-nls-GFP-SVNLS2}) were arrested in G1 phase with α-factor then examined by fluorescence microscopy. (B) The Mcm3 NLS is required for nuclear localization of GFP-Mcm3. YJL2669 (MCM3::{GFP-MCM3}), YJL2675 (MCM3::{GFP-mcm3-nls}), and YJL2665 (MCM3::{SVNLS2-GFP-mcm3-nls}) were arrested in G1 phase with α-factor then examined by fluorescence microscopy. (C) The Mcm2 NLS is required for nuclear localization of Mcm7-GFP. Cultures of YJL3765 (MCM7-GFP mcm2-td MCM2), YJL3840 (MCM7-GFP mcm2-td mcm2-nls), and YJL3799 (MCM7-GFP mcm2-td mcm2-nls-SVNLS2) growing exponentially at 23°C were arrested in G2/M phase by addition of nocodazole for 3 h. Galactose was added and cultures were shifted to 37°C for 30 min to degrade Mcm2-td. Cells were then released from G2/M phase into a G1 phase arrest by shifting them to fresh medium containing α-factor for 2 h (still in the presence of galactose and at 37°C). Cells examined by fluorescence microscopy are shown just before the G2/M phase release (NOC arrest) and at the G1-phase block (α-factor arrest). (D) The Mcm3 NLS is required for nuclear localization of Mcm7-GFP. Cultures of YJL3464 (MCM7-GFP mcm3-td MCM3), YJL3469 (MCM7-GFP mcm3-td mcm3-nls), and YJL3474 (MCM7-GFP mcm3-td SVNLS2-mcm3-nls) were subjected to the same experimental protocol described for Figure 2C.
Figure 3.
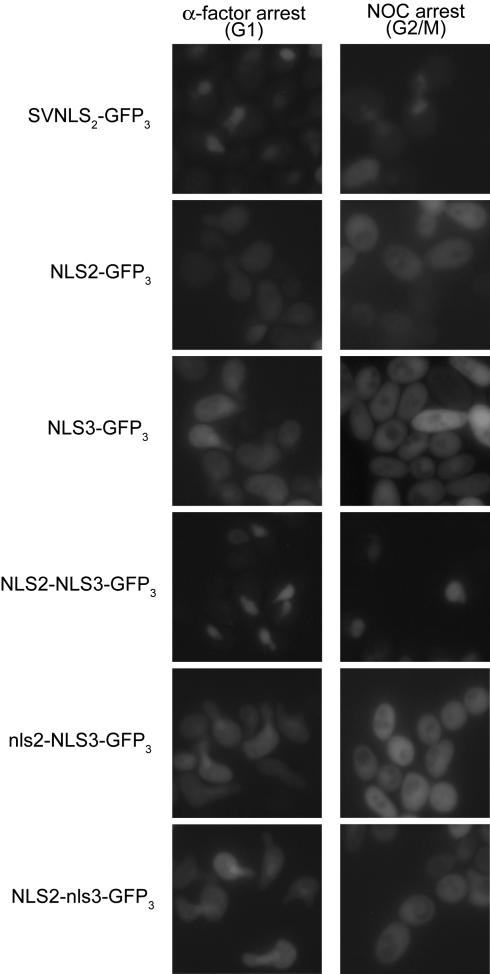
Together the Mcm2 and Mcm3 NLSs are sufficient to direct the nuclear localization of a heterologous protein. Overnight cultures of YJL310 containing _URA3_-marked centromeric plasmids pAR109 (pGAL-SV40NLS-GFP3), pAR110 (pGAL-NLS2-GFP3), pAR101 (pGAL-NLS3-GFP3), pAR113 (pGAL-NLS2-NLS3-GFP3), pAR126 (pGAL-nls2-NLS3-GFP3), or pAR127 (pGAL-NLS2-nls3-GFP3) and growing in SRaf-Ura medium were shifted to YEPRaf medium for 90 min before splitting each culture in two and adding α-factor to one-half and nocodazole to the other. One hour later, as the cultures were approaching a complete arrest, galactose was added to induce synthesis of the GFP3 fusion proteins. After 2 h of induction, cells were examined by fluorescence microscopy.
Figure 4.
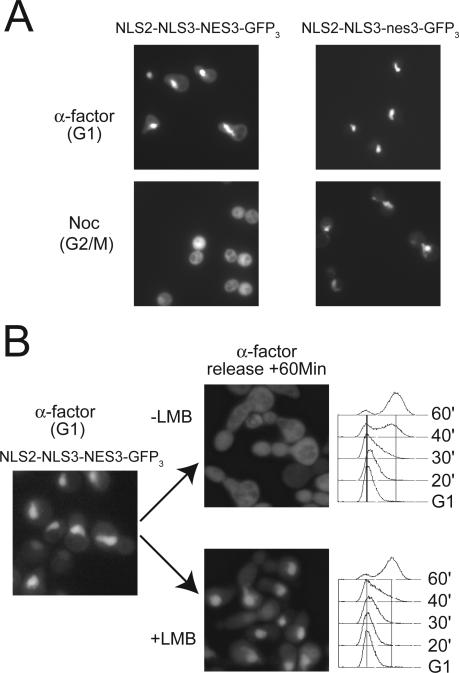
Mcm3 contains an NES, which in combination with the Mcm2 NLS and Mcm3 NLS, directs the cell cycle-regulated localization of GFP3. (A) The cytoplasmic localization of NLS2-NLS3NES3-GFP3 in G2/M phase is dependent on the leucine-rich motif of the Mcm3 NES. Cultures of YJL4662 (_crm1_Δ trp1::{pGAL-NLS2-NLS3NES3-GFP3),TRP1} [_crm1-T539C_]) and YJL4860 (_crm1_Δ trp1::{pGAL-NLS2-NLS3nes3-GFP3),TRP1} [_crm1-T539C_]) constitutively expressing their GFP3 fusion proteins in YEP-Gal medium were arrested for 90 min in either G1 phase with α-factor or G2/M phase with nocodazole before being examined by fluorescence microscopy. (B) NLS2-NLS3NES3-GFP3 is exported from the nucleus by a Crm1-dependent mechanism. YJL4662 constitutively expressing NLS2-NLS3NES3-GFP3 in YEPGal medium was arrested in G1 phase with α-factor. At time 0, cells were released from the arrest in the presence or absence of 100 ng/ml leptomycin B (LMB). Samples were collected at the indicated times for flow cytometry and fluorescence microscopy.
Figure 6.
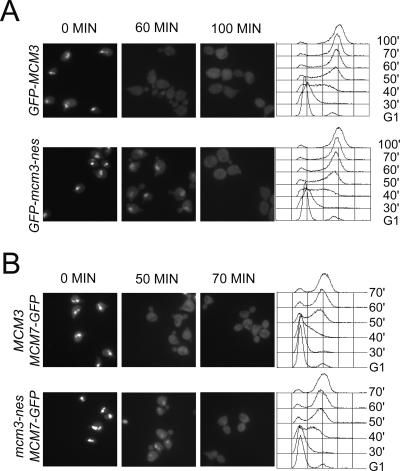
The Mcm3 NES is required for the timely nuclear export of Mcm3 and Mcm7. (A) Mutation of the Mcm3 NES delays the nuclear export of GFP-Mcm3. YJL2162 (GFP-MCM3) and YJL2741 (GFP-mcm3-nes) cells were arrested in G1 phase with alpha factor and at time 0 synchronously released into a G2/M phase arrest with nocodazole. At the indicated times, samples were taken for flow cytometry and fluorescence microscopy. (Left) Representative images from 0, 60, and 100 min. (Right) Flow cytometry profiles. (B) Mutation of the Mcm3 NES delays the nuclear export of Mcm7-GFP. YJL1979 (MCM3 MCM7-GFP) and YJL5439 (mcm3-nes MCM7-GFP) were treated and analyzed as described in C, except microscopic images from the 0-, 50-, and 70-min time points are shown.
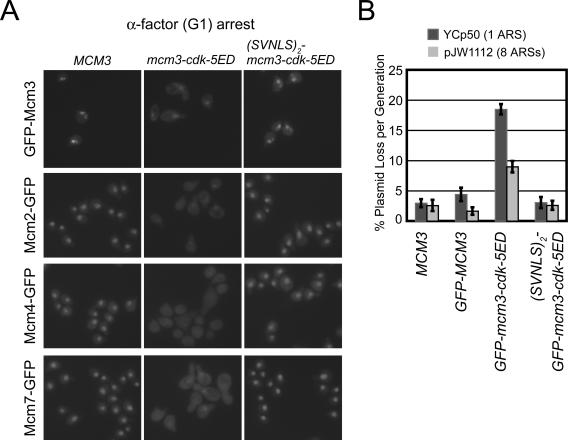
Figure 10.
Phosphomimic mutations of the Mcm3 CDK consensus sites promote net nuclear export of Mcm proteins in G1 phase and this mislocalization impairs replication initiation. (A) (First Row) YJL2160 (GFP-MCM3), YJL1265 (GFP-mcm3-cdk5ED), YJL1260 (SVNLS2-GFP-mcm3-cdk5ED). (Second Row) YJL2033 (mcm2::{MCM2-GFP}), YJL4162 (mcm2::{MCM2-GFP} mcm3-cdk5ED), YJL4094 (mcm2::{MCM2-GFP} SVNLS2-mcm3-cdk5ED). (Third Row) YJL2037 (mcm4::{MCM4-GFP}), YJL4103 (mcm4::{MCM4-GFP} mcm3-cdk5ED), YJL4098 (mcm4::{MCM4-GFP} SVNLS2-mcm3-cdk5ED). (Fourth Row) YJL2217 (mcm7::{MCM7-GFP}), YJL4108 (mcm7::{MCM7-GFP} mcm3-cdk5ED), YJL4096 (mcm7::{MCM7-GFP} SVNLS2-mcm3-cdk5ED). All strains were arrested in G1 phase with α-factor for 90 min (>95% unbudded) before being examined by fluorescence microscopy. (B) Mcm mislocalization due to the mcm3-cdk5ED mutation impairs replication initiation. Plasmid loss rates per were measured over 12-20 generations for plasmids YCp50 (1 ARS) and pJW1112 (8 ARSs) in YJL310 (MCM3), YJL2160 (GFP-MCM3), YJL1265 (GFP-mcm3-cdk5ED), and YJL1259 (_SVNLS2_-GFP-mcm3-cdk5ED). Histogram shows average loss rate per generation and SE for four independent isolates of each plasmid-yeast pair. Complete failure to replicate a plasmid would result in a theoretical loss rate of 50% per generation.
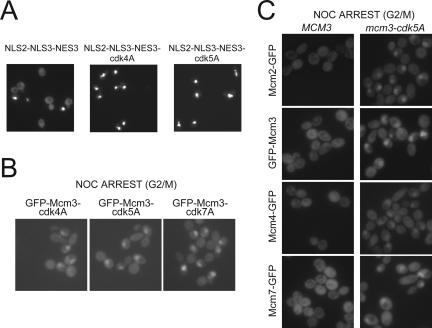
Figure 8.
The Mcm3 consensus CDK phosphorylation sites are required for nuclear exclusion of the Mcm2-7 complex. (A) Net nuclear export of NLS2-NLS3NES3-GFP3 is dependent on the consensus CDK phosphorylation sites in NLS3NES3. YJL4662 (_crm1_Δ trp1::{pGAL-NLS2-NLS3NES3-GFP3),TRP1} [_crm1-T539C_]), YJL5750 (trp1::{pGAL-NLS2-NLS3NES3-cdk5A-GFP3),TRP1}) and YJL5753 (trp1::{pGAL-NLS2-NLS3NES3-cdk4A-GFP3), TRP1}) growing exponentially and constitutively expressing their GFP3 fusion proteins in YEPGal medium were examined by fluorescence microscopy. (B) Mcm3 CDK consensus sites are required for nuclear exclusion of Mcm3. Fluorescent microscopy of nocodazole-arrested YJL2714 (GFP-mcm3-cdk4A), YJL2720 (GFP-mcm3-cdk5A), and YJL2314(GFP-mcm3-cdk7A). (C) Mcm3 Cdk consensus sites are required for nuclear exclusion of other Mcm subunits. Fluorescence microscopy of nocodazole-arrested YJL2033 (mcm2::{MCM2-GFP}), YJL4165 (mcm2::{MCM2-GFP} mcm3-cdk5A), YJL2160 (GFP-MCM3), YJL2720 (GFP-mcm3-cdk5A), YJL2037 (mcm4::{MCM4-GFP}), YJL4169 (mcm4::{MCM4-GFP}, mcm3-cdk5A), YJL2217 (mcm7::{MCM7-GFP}), and YJL4167 (mcm7:: {MCM7-GFP}, mcm3-cdk5A).
Immunoblot Analysis
Immunoblot analysis was performed as described (Nguyen et al., 2001). Blots were probed with c-Myc polyclonal antibody at a 1:200 dilution (sc-789, Santa Cruz Biotechnology, Santa Cruz, CA).
Flow Cytometry Analysis
Samples were fixed in ethanol and processed as described in (Green and Li, 2005).
Fluorescence Microscopy
For fluorescence microscopy of live cells for Figures 2, 3, and 10, cells were rapidly washed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 43 mM Na2HPO4, 14 mM KH2PO4 at pH 7.4) and visualized using a Leica DMLB fluorescence microscope (Deerfield, IL) with a 100× PL Fluotar oil immersion objective. Images were acquired with an Optronics DEI-750 CCD camera (Goleta, CA) using the Scion Image Software program. Live cell GFP fluorescence microscopy for Figures 4, 5, 6B, 8, and 9 were performed as follows: cells (OD600 ≈ 0.2) were washed with 1 ml SDC, pelleted, and resuspended in 40 μl of SDC and visualized within 8 min of sampling using 60× PlanApo oil objective on an Olympus BX60 microscope (Melville, NY) with EndowBandpass GFP filter cube (Chroma, Rockingham, VT; Cat. no. 41017). For Figure 6A time point samples were fixed in 100% ethanol. Samples were pelleted and washed in PBS with 50 ng/ml DAPI and visualized using the Olympus microscope described above. Images were acquired with Openlab 3.1.7 software (Improvision, Lexington, MA) driving a Hamamatsu ORCA ER CCD (Bridgewater, NJ; exposure time was on the order of 300 ms). Figure panels were assembled using Openlab 3.1.7 and Illustrator 10.0 (Adobe Systems, San Jose, CA).
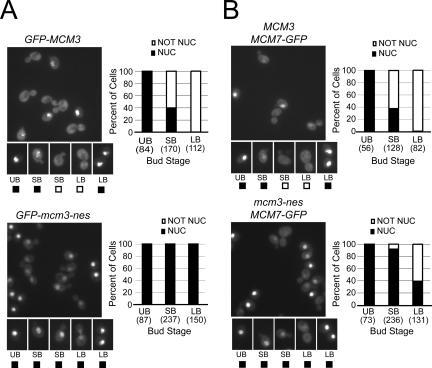
Figure 5.
The Mcm3 NES is required for efficient nuclear export of Mcm3 and Mcm7 in cycling cells. (A) Mutation of the Mcm3 NES increases the population of cells containing nuclear GFP-Mcm3 during exponential growth. Exponentially growing cultures of YJL2162 (GFP-MCM3) (top) and YJL2741 (GFP-mcm3-nes) (bottom) were examined by fluorescence and DIC microscopy. Representative fluorescent fields are shown. Cells were categorized as unbudded (UB), small budded (SB), or uninucleate large budded (LB) based on their DIC image and DAPI fluorescence. Binucleate large budded cells, which comprise approximately half of all large budded cells, are unseparated postmitotic cells (many in G1 phase) and were thus not included in the analysis. Each of these categories was further subclassified into cells with or without detectable nuclear GFP fluorescence above cytoplasmic levels as exemplified by the pictures of individual cells. Bar graphs show the percent of cells with nuclear or nonnuclear GFP fluorescence and the total number of cells counted (in parentheses) for each bud stage. (B) Mutation of the Mcm3 NES increases the population of cells containing nuclear Mcm7-GFP during exponential growth. The same experiment and analysis described in Figure 5A was performed for YJL1979 (MCM3 MCM7-GFP) and YJL5439 (mcm3-nes MCM7-GFP).
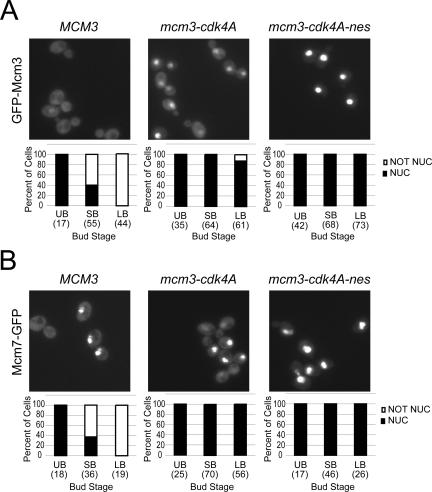
Figure 9.
Together the Mcm3 NES and the consensus CDK sites in Mcm3 NLS are required for nuclear export of GFP-Mcm3 and Mcm7-GFP. (A) GFP-Mcm3 is strongly nuclear throughout the cell cycle when both the leucine-rich motif of the Mcm3 NES and the four CDK consensus sites flanking the Mcm3 NLS are mutated. Exponentially growing cultures of YJL2162 (GFP-MCM3), YJL2714 (GFP-mcm3-cdk4A), and YJL5216 (GFP-mcm3-cdk4A-nes) were examined by fluorescence and DIC microscopy. Cells were categorized as unbudded (UB), small budded (SB), or uninucleate large budded (LB) based on their DIC image and DAPI fluorescence. Binucleate large budded cells, which comprise approximately half of all large budded cells, are unseparated postmitotic cells (many in G1 phase) and were thus not included in the analysis. Each bud category was further subclassified into cells with or without detectable nuclear GFP fluorescence above cytoplasmic levels. Bar graphs show the percent of cells with nuclear or nonnuclear GFP fluorescence and the total number of cells counted (in parentheses) for each bud category. (B) Mcm7-GFP is strongly nuclear throughout the cell cycle when both the leucine-rich motif of the Mcm3 NES and the four CDK consensus sites flanking the Mcm3 NLS are mutated. Exponentially growing cultures of YJL1979 (MCM3 MCM7-GFP), YJL5691 (mcm3-cdk4A mcm7::{MCM7-GFP}), and YJL5221 (mcm3-cdk4A-nes MCM7-GFP) were examined by fluorescence and DIC microscopy as described in Figure 9A.
In Vitro Phosphorylation of Mcm3
One nanogram of purified Cdc28-His6 and 10 ng purified Clb2-MBP (a gift of Jeff Ubersax, Morgan lab, UC San Francisco, CA), mixed in 2 μl of storage buffer (300 mM NaCl, 25 mM HEPES-KOH, pH 7.4, 10% glycerol) for 5 min on ice, were added to 23 μl kinase buffer (50 mM HEPES-KOH, pH 7.4, 2 mM MgCl2, 1 mM dithiothreitol) containing 0.1 mM ATP, 2 μCi [γ-32P]ATP (Amersham, Piscataway, NJ), and 1 μg purified GST-Mcm3, 1 μg purified GST-Mcm3-cdk-5A, or 1 μg purified GST-Mcm3-cdk-7A and incubated at 25°C for 15 min. The reaction was stopped by adding 10 μl 4× SDS sample buffer (250 mM Tris-HCl, pH 6.8, 2.8 M β-mercaptoethanol, 10% SDS, 0.06% bromophenol blue, and 40% glycerol) and boiling for 5 min. The reaction products were resolved on an 8% SDS-PAGE, stained with Coomassie, then dried, and subjected to autoradiography.
In Vivo Phosphate Labeling of Mcm3
YJL4313, YJL4315, and YJL4324 expressing Myc-Mcm3 were grown in low-phosphate YEPD to OD600 of 0.5-0.8 at 30°C. 1 OD unit was spun down, resuspended in 1 ml low-phosphate YEPD containing 1 mCi 32P-orthophosphate (Amersham; PBS.13), and incubated for 60 min at 30°C. The labeled cells were pelleted in a 1.5-ml screw cap tube and placed on ice. In parallel, 15 OD units of exponentially growing YJL2160 expressing untagged Mcm3 were pelleted, resuspended in 1 ml dH20, and added to the labeled cell pellet. After thorough mixing, the labeled and unlabeled cells were repelleted and frozen in liquid nitrogen. Frozen cell pellets were resuspended in 350 μl lysis buffer (25 mM HEPES, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM Na3VO4, 1 mM EDTA, pH 8.0, 80 mM β-glycerol phosphate, and 50 mM NaF) containing 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 10 μg/ml aprotinin, 2 mM benzamidine, and 1 μg/ml pepstatin. Cells were lysed by bead beating in a Mini Bead Beater (Biospec Products, Bartlesville, OK) for two 1-min pulses. After pelleting the cell debris, the supernatant was clarified by centrifugation for 10 min at 20,800 × g at 4°C and then incubated with 1 μl of c-Myc monoclonal antibody (mAb; 9E10, MMS-150R, Covance, Madison, WI) prebound to 10 μl of magnetic beads (Dynabead Protein G, 100.03, Dynal Biotech, Lake Success, NY). After 2-h incubation the magnetic beads were washed and resuspended in 25 μl of 2× SDS sample buffer. The sample was loaded on a 4-15% SDS-PAGE gradient gel (Bio-Rad, Richmond, CA), and the dried gel was developed on a phosphorimager.
RESULTS
Mcm2 and Mcm3 Each Contain a Sequence Required for Nuclear Localization of Their Respective Proteins
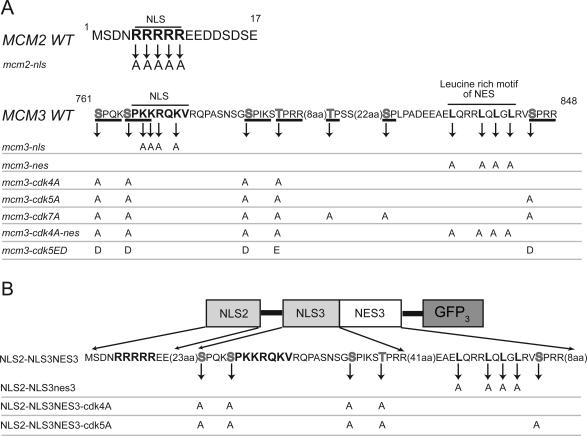
As a first step to understanding the CDK regulation of Mcm2-7 nucleocytoplasmic localization we first sought to identify the NLSs and NESs responsible for nucleocytoplasmic transport of the Mcm proteins. Our previous work, suggesting that Mcm proteins colocalize as a complex (Nguyen et al., 2000), raised the possibility that NLSs or NESs on one or more of the Mcm proteins could be responsible for nuclear or cytoplasmic localization of the entire complex. We scanned the amino acid sequences of the six Mcm proteins (Mcm2-Mcm7) for the two NLS sequence motifs that are recognized and bound by the import receptor adapter, importin alpha (reviewed in Jans et al., 2000). One of these motifs, represented by the SV40 NLS (PKKKRKV), contains a single cluster of highly basic residues, and the other motif, represented by the nucleoplasmin NLS (KRPAATKKAGQAKKKKL) contains two smaller clusters of basic residues separated by 7-22 amino acids. Four good matches to these motifs were identified on Mcm2 (residues 5-9, RRRRR; residues 150-155, RRRRRR), Mcm3 (residues 766-772, PKKRQRV), and Mcm7 (residues 199-219, RR-13aa-RRYRKK).
To determine whether any of these sequences were required for nuclear transport of the Mcm proteins, we initially examined whether these sequences were essential for cell viability. We reasoned that a sequence required for nuclear localization of any of the Mcm proteins would also be essential for viability, because the Mcm proteins must perform their essential replication function in the nucleus. We generated mutant mcm genes with alanines substituting for multiple basic residues in the identified sequences and attempted to replace the endogenous genes with these mutant genes by two-step gene replacement. Haploid strains expressing mutant Mcm proteins with alanine substitutions on Mcm2 (at amino acid residues 150-155) or on Mcm7 (at residues R214, R216, and K217) were viable and exhibited growth rates indistinguishable from wild-type strains. Thus, these sequences are not required for nuclear localization of Mcm proteins. In contrast, we could not isolate haploid strains expressing mutant Mcm proteins containing alanine substitutions in Mcm2 (at residues 5-9) or Mcm3 (at residues 766-772) (Figure 1A; mcm2-nls and mcm3-nls; Supplementary Figure 1). Fusing sequences encoding two tandem copies of the SV40 NLS onto the mutant mcm2-nls or mcm3-nls genes did allow isolation of gene replacement strains expressing these NLS-tagged mutant genes. Tetrad analysis confirmed that mutations in residues 5-9 of Mcm2 or 766-772 of Mcm3 resulted in inviability and that fusion of the SV40 NLS to these mutated Mcm proteins restored viability. Together these results demonstrate that Mcm2 residues 5-9 and Mcm3 residues 766-772 sequences are essential for viability and that their essential role may be to direct the nuclear localization of their respective proteins.
Figure 1.
Wild-type and mutant Mcm2 and Mcm3 nucleocytoplasmic transport signals. (A) Key amino acid sequences of wild-type and mutant Mcm2 NLS and Mcm3 NLS-NES transport module. Putative NLSs and leucines in leucine-rich motif are in bold. Consensus CDK phosphorylation sites are underlined with putative phosphoacceptor residues in gray. Amino acids not spelled out are indicated in parentheses. Amino acid substitutions in the mutant Mcm proteins used in this study are indicated below the wild-type sequences. (B) A schematic of the MCM transport module containing the Mcm2 NLS and the Mcm3 NLS-NES. Below the schematic are the WT and corresponding mutant alleles of this construct that are discussed in the text.
To directly examine the role of residues 5-9 of Mcm2 and 766-772 of Mcm3 in the localization of their respective proteins, we used fluorescence microscopy to examine the subcellular distribution of mutant Mcm2 and Mcm3 fused to GFP during the G1 phase of the cell cycle (Figure 2, A and B). We previously reported that wild-type Mcm2, 3, 4, 6, and 7 tagged with GFP are fully functional when expressed as the sole copy of their respective Mcm proteins, indicating that these fusion proteins can complex with other Mcms (Nguyen et al., 2000). In Figure 2, A and B, the fusion proteins were expressed in addition to the endogenous wild-type MCM genes. The latter supported the viability of these cells but did not interfere with the fluorescence analysis of the GFP fusion proteins. As expected, GFP fusions to wild-type Mcm2 or Mcm3 accumulated in the nucleus during G1 phase. In contrast, Mcm2-nls-GFP and GFP-Mcm3-nls were distributed throughout the cytoplasm. This mislocalization could be rescued by fusing two tandem copies of the SV40 NLS to the mutant fusion proteins; both Mcm2-nls-GFP-SVNLS2 and SVNLS2-GFP-Mcm3-nls were constitutively nuclear. These results directly demonstrate that residues 5-9 of Mcm2 and residues 766-772 of Mcm3 are required for the nuclear localization of their respective proteins. Hence, we refer to these residues as the Mcm2 NLS and Mcm3 NLS, respectively. Our results corroborate previously published results in S. cerevisiae, implicating the Mcm3 residues in nuclear localization of Mcm3 (Young et al., 1997), and in S. pombe, implicating N-terminal Mcm2 residues in nuclear localization of Mcm2 (Pasion and Forsburg, 1999).
The Mcm2 and Mcm3 NLSs Are Each Required for Localization of the Mcm2-7 Complex
We have previously shown that fusion of two tandem SV40 NLSs to any Mcm subunit promotes the constitutive nuclear localization of both that subunit and each of the other Mcm subunits (Nguyen et al., 2000). Hence, we suspected the lethality arising from mutation of the Mcm2 or Mcm3 NLSs could be rescued by fusing the SV40 NLS to other Mcm subunits. To examine this possibility, we reattempted two-step gene replacement of MCM2 and MCM3 with mcm2-nls and mcm3-nls, respectively, in haploid strains containing two tandem copies of the SV40 NLS fused to other Mcm proteins (Supplementary Figure 1). We could successfully replace MCM2 with mcm2-nls when the SV40 NLSs were fused to Mcm3, Mcm4, Mcm5, or Mcm6 and could successfully replace MCM3 with mcm3-nls when the SV40 NLSs were fused to Mcm2, Mcm4, Mcm5, or Mcm6. Furthermore, the mutant Mcm2-GFP or GFP-Mcm3 in these strains displayed constitutive nuclear localization (unpublished data). These results indicate that mislocalization of the mutant Mcm2-GFP or GFP-Mcm3 can be rescued by ensuring the nuclear localization of one other Mcm subunit. These results further suggest that mutating the Mcm2 or Mcm3 NLS disrupts the nuclear localization of all other Mcm subunits; otherwise an Mcm subunit that could retain its nuclear localization would have rescued the NLS mutations without requiring fusion to the SV40 NLS.
To directly examine whether the Mcm2 or Mcm3 NLS is required for the nuclear localization of other Mcm subunits, we performed a set of experiments exemplified by the one shown in Figure 2C. In this experiment, we examined the effect of mutating the NLS in Mcm2 on the nuclear import of Mcm7-GFP during the transition from G2/M to G1 phase. Because the NLS mutation is lethal, we complemented the mutant mcm2-nls gene with an MCM2 gene encoding a conditionally degraded version of the Mcm2 protein (mcm2-td). Mcm2-td is targeted for ubiquitin-mediated degradation by both raising the temperature to 37°C and shifting cells into galactose containing media to induce the E3 ubiquitin ligase Ubr1; under these restrictive conditions there is no detectable Mcm2-td after 60 min (Labib et al., 2000). In this conditionally complemented strain we could examine how the NLS-defective Mcm2 protein directs the localization of Mcm7-GFP after Mcm2-td is degraded. In parallel, we generated control strains where either the wild-type MCM2 gene or the suppressed mutant gene mcm2-nls-GFP-SVNLS 2 was introduced into the mcm2-td parent strain.
Experimental cells expressing Mcm7-GFP, Mcm2-nls, and Mcm2-td under permissive conditions for Mcm2-td (rich medium containing raffinose at 25°C) were arrested in metaphase with nocodazole. Once arrested, we induced degradation of Mcm2-td by shifting the cells to restrictive conditions (adding galactose at 37°C). After 30 min, the cells were released from the nocodazole arrest into an α-factor G1 arrest, still under restrictive conditions. Mcm proteins normally enter the nucleus during this G2/M to G1 phase transition, but Mcm7-GFP failed to accumulate in the nucleus of these cells (Figure 2C). In contrast, Mcm7-GFP strongly accumulated in the nucleus of the control strains expressing either Mcm2 or Mcm2-nls-SVNLS2 (Figure 2C). Moreover, when cells were maintained at permissive conditions for the Mcm2-td proteins, Mcm7-GFP accumulated in the nucleus in all three strains (unpublished data). These results indicate that the Mcm2 NLS is required for the nuclear localization of Mcm7 in G1 phase. Similar experiments with Mcm3 (Figure 2D) demonstrate that the Mcm3 NLS is also required for nuclear localization of Mcm7. Finally, by repeating these experiments with GFP fused to other Mcm subunits, we have been able to show that the Mcm2 NLS is required for nuclear localization of Mcm3 and Mcm4, and the Mcm3 NLS is required for nuclear localization of Mcm2 (unpublished data). Taken together, these results suggest that the Mcm2 and Mcm3 NLSs are required, not just for nuclear localization of their respective proteins, but of the entire Mcm2-7 complex.
Together the Mcm2 and Mcm3 NLS Are Sufficient for Strong Nuclear Localization Activity
The classical definition of an NLS is a sequence that is both necessary and sufficient for directing the nuclear localization of proteins. To determine whether the Mcm2 and Mcm3 NLSs are sufficient to direct the nuclear localization of proteins, we fused them to three tandem copies of GFP (GFP3). With a combined molecular mass of 82 kDa, these tandem GFPs are larger than the 60-kDa upper limit for proteins to diffuse through nuclear pores and therefore must be actively transported to enter and exit the nucleus (reviewed in Weis, 2003). These fusion proteins were placed under the control of the regulatable GAL10 promoter. To ensure that the observed localization was not inherited from a previous stage of the cell cycle, yeast cells containing these fusion constructs were first arrested in G1 phase or G2/M phase before the fusion proteins were induced by galactose.
When the SV40 NLS was fused to GFP3, strong nuclear localization was observed. In contrast, sequences containing the Mcm2 NLS (amino acids 1-17) or the Mcm3 NLS (amino acids 760-789), and hereafter referred to as NLS2 and NLS3, respectively, showed only a very weak ability to localize GFP3 to the nucleus at either stage of the cell cycle (Figure 3). Hence, individually, these two sequences are insufficient to confer robust nuclear localization on a heterologous protein. This conclusion is consistent with the observation that neither NLS is sufficient to direct the Mcm2-7 complex into the nucleus in the absence of the other. Together, however, the NLS2 and NLS3 strongly directed GFP3 into the nucleus in both G1 and G2/M phases (Figure 3). Thus, the weak Mcm2 and Mcm3 NLSs can functionally act together as a single strong NLS. Using a larger segment spanning the Mcm3 NLS (amino acids 746-789), which increases the spacing between the Mcm2 and Mcm3 NLSs (to 30 amino acids), also resulted in strong composite NLS activity.
Our findings differ from the previous study in S. cerevisiae that identified the Mcm3 NLS (Young et al., 1997). In that study, the sequence was reported to be not only necessary for nuclear localization of Mcm3, but also sufficient for nuclear localization of a heterologous protein. That conclusion, however, was based on a slightly different segment spanning the Mcm3 NLS (amino acids 755-781 vs. our segment of amino acids 760-789) examined in combination with 50 amino acids of the Leu2 protein. When we fused that Mcm3 segment without the Leu2 segment to our tandem GFP reporter, we still observed poor NLS activity relative to the SV40 NLS or the combined NLS2-NLS3 (unpublished data). Thus, our examination of an isolated Mcm3 NLS indicates that this NLS only has weak activity.
Mcm3 Contains a Crm1-dependent NES, Which Cooperates with the Mcm2 and Mcm3 NLSs to Form a Cell Cycle-regulated Transport Module
We have previously shown that the Mcm2-7 complex undergoes net nuclear export when cells activate Cdc28 kinase activity (Nguyen et al., 2000). Because each subunit is too large to diffuse through the nuclear pore, we suspected the complex contains nuclear export signals on one or more subunits. The most recognizable NES motif identified to date is a leucine-rich motif that recruits the nucleocytoplasmic export receptor Crm1 (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Stade et al., 1997) and is exemplified by the NESs of the HIV REV and PKIα (LQLP-PLERLTL and LALKLAGLDI respectively; Fischer et al., 1995; Wen, et al., 1995). One such motif (LQRRLQLGL; aa 834-842) is present in a 67-amino acid segment (aa 790-856) immediately C-terminal of the Mcm3 NLS segment. To test the export activity of this potential NES, we added it to a GFP3 reporter construct containing a composite Mcm2-Mcm3 NLS. In this new construct (Figure 1B) the adjacent Mcm3 NLS and NES are derived from one contiguous 111 amino acid segment of Mcm3 (aa 746-856).
When the resulting fusion protein was induced in exponentially growing cells, it displayed cell cycle-regulated localization that was reminiscent of the CDK regulation of Mcm2-7 localization (Labib et al., 1999; Nguyen et al., 2000). The protein was distributed throughout the cytoplasm of budded cells, which contain active Cdc28 kinase, and was predominantly nuclear in unbudded G1 phase cells, which contain little or no active Cdc28 kinase. Moreover, the fusion protein was strongly nuclear in G1 cells arrested with α-factor and was distributed throughout the cytoplasm in G2/M cells arrested with nocodazole (Figure 4A, two left panels). The cytoplasmic localization in nocodazole-arrested cells was dependent on the leucine-rich motif (Figures 1B and 4A).
To demonstrate that this cytoplasmic localization was due to net nuclear export during passage through the cell cycle, we introduced the GFP3 fusion construct into cells containing a leptomycin B-sensitive allele of CRM1, crm1-T539C (Neville and Rosbash, 1999). Cells expressing the fusion protein were released from a G1 arrest into a G2/M arrest, either in the presence or absence of 100 ng/ml leptomycin B. By the time both cultures had completed S phase (60 min), the GFP3 fusion protein was distributed throughout the cytoplasm in the absence of leptomycin B, but remained strongly nuclear in its presence (Figure 4B). Similarly, addition of leptomycin B to exponentially growing cells expressing the fusion protein resulted in constitutive nuclear localization of the protein (unpublished data). Thus, the redistribution of this fusion protein from nucleus to cytoplasm is indeed due to nuclear export and is dependent on the Crm1 export receptor as well as the leucine-rich motif. We henceforth refer to the 67-amino acid segment downstream of the Mcm3 NLS as the Mcm3 NES or NES3. Importantly, the Mcm2 NLS and the contiguous Mcm3 NLS and NES behave as a minimal transport module (NLS2-NLS3NES3) that recapitulates the cell cycle-regulated localization of the entire Mcm2-7 complex.
The Mcm3 NES Promotes the Nuclear Export of Mcm Subunits
We next examined whether the Mcm3 NES promotes nuclear export in the context of the full Mcm2-7 complex. To do this, we generated a haploid strain in which the wild-type endogenous MCM3 gene was replaced by a mutant GFP-Mcm3-nes gene, which contains alanine substitutions at the leucine residues of the Mcm3 leucine-rich motif (AQR-RAQAGA). The ability to generate such a strain indicates the Mcm3 leucine-rich motif does not perform an essential function. As a control, we generated a GFP-MCM3 strain, which expresses a wild-type fusion protein. Both experimental and control strains divided at identical rates and contained a similar distribution of cells throughout the cell cycle by both budding indices and flow cytometry (unpublished data). However, the mutant GFP-Mcm3-nes could be detected in the nuclei of virtually all uninucleate budded cells, whereas GFP-Mcm3 could only be detected in the nuclei of 40% of small budded cells and 0% of uninucleate large budded cells (Figure 5A). Similar results were observed if GFP was fused to Mcm7 instead of Mcm3 (Figure 5B), suggesting that the Mcm3 NES also promotes the nuclear export of other Mcm subunits.
To confirm this role for the Mcm3 NES, we compared the distribution of GFP-Mcm3 with mutant GFP-Mcm3-nes protein in cells synchronously released from an α-factor arrest into a nocodazole arrest (Figure 6A). Both flow cytometry and budding indices confirmed that cell cycle progression was not affected by mutation of the Mcm3 NES. As expected, at the beginning of the time course (Figure 6A, 0 min), when all cells were unbudded G1 cells, the GFP fusion proteins of both experimental and control strains were strongly nuclear. However, 60 min after release from α-factor, when most cells were small-budded and in late S phase, there was a dramatic difference. In the GFP-MCM3 strain, few of the small budded cells (5/100 cells) retained detectable nuclear levels of the fusion protein, whereas in the GFP-mcm3-nes strain, residual nuclear accumulation of the fusion protein could be detected in almost all small budded cells (122/123). Similar, but less striking differences were observed at 50 and 70 min after release from α-factor arrest (unpublished data). These data indicate that Mcm3 NES promotes the nuclear export of Mcm3. Eventually, as cells remained at the nocodazole arrest and became large-budded (Figure 6A, 100 min), nuclear accumulation of the GFP fusion proteins gradually became undetectable (0/71 for GFP-Mcm3 and 1/54 for GFP-Mcm3-nes), indicating that the NES is not absolutely required for the net nuclear export of Mcm3. Nonetheless, the NES is essential for the timely export of Mcm3, and without this timely export, Mcm3 is not effectively cleared from the nuclei of cycling cells (Figure 5A). A very similar delay in nuclear export was observed in a GFP-Mcm3 crm1-T539C leptomycin B-sensitive strain, if leptomycin B was added upon release from alpha factor arrest (unpublished data). These results suggest that the Mcm3 NES functions through Crm1 in the full Mcm2-7 complex as it does in the GFP3 fusion protein.
Nuclear export of Mcm7-GFP is also delayed in a mcm3-nes strain relative to an MCM3 strain (Figure 6B). Fifty minutes after release from α-factor arrest, 42% (56/134) of the small budded MCM3 cells retained barely detectable nuclear accumulation of Mcm7-GFP, whereas almost 90% (94/110) of the small budded mcm3-nes cells retained residual nuclear accumulation of Mcm7-GFP. Similar but smaller differences could be seen at 40 and 60 min after release from α-factor (unpublished data). By 70 min, however, most cells were large-budded, and almost no wild-type (0/77) or mutant (3/172) large budded cells exhibited detectable nuclear accumulation. Again, the MCM7-GFP crm1-T539C strain showed a similar delay in Mcm7 nuclear export (unpublished data). Thus, although there appears to be another partially redundant export signal(s) for the Mcm2-7 complex, we conclude that the Mcm3 NES functions as a Crm1-dependent export signal for at least two subunits of the complex. Also, because the export defect in the crm1-T539C mutant pheno-copies the export defect of the mcm3-nes mutant, it appears that the partially redundant export signal(s) may function through a different export receptor besides Crm1.
Mcm3 Is a Substrate of Cdc28 Kinase
We and others have previously shown that Cdc28 kinase activity promotes the net nuclear export of Mcm proteins (Labib et al., 1999; Nguyen et al., 2000), raising the possibility that this regulation is through direct phosphorylation of the Mcm2-7 complex. A scan of the amino acid sequences of all six Mcm proteins for the full consensus CDK phosphorylation site (S/T)-P-X-(K/R; Nigg, 1993) only identified two sites on Mcm4 and five sites on Mcm3. The sites on Mcm3 were of particular interest because they are all located within the Mcm3 portion of the NLS-NES transport module (Figure 1A). Four sites flank the basic region of the Mcm3 NLS, and the fifth site is adjacent to the leucine-rich motif of the Mcm3 NES. Two additional sites that satisfy a more degenerate CDK phosphorylation site consensus ((S/T)-P) are positioned between the basic region and leucine-rich motif. For the experiments discussed below, we generated mutations in these sites that substitute alanine for the phosphoacceptor serine or threonine of these consensus sites.
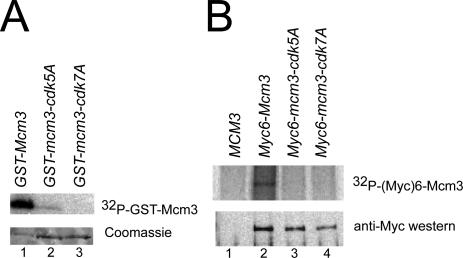
Figure 7A shows that recombinant Clb2-Cdc28 kinase can phosphorylate purified GST-Mcm3 in vitro, confirming previous reports of Mcm3 phosphorylation by purified Clb2-Cdc28 and Clb5-Cdc28 kinases (Ubersax et al., 2003; Loog and Morgan, 2005). Importantly, GST-Mcm3-cdk5A and GST-Mcm3-cdk7A, which contain mutations in the five full CDK consensus sites and all seven potential CDK sites, respectively (Figure 1A), were both poorly phosphorylated. We also examined Mcm3 phosphorylation in vivo by metabolically labeling cells with 32P-orthophosphate and observed that Mcm3 displayed significantly more phosphorylation than Mcm3-cdk5A and Mcm3-cdk7A (Figure 7B). Together these results suggest that the Mcm3 portion of the NLS-NES transport module is a target of Cdc28 kinase in vitro and in vivo.
Figure 7.
In vitro and in vivo phosphorylation of Mcm3 is dependent on the consensus CDK phosphorylation sites in the Mcm3 NLS3NES3 module. (A) GST-Mcm3 is phosphorylated by Cdc28-Clb2 kinase in vitro. In vitro kinase reactions were performed with purified Cdc28-Clb2 kinase mixed with purified GST-Mcm3, GST-Mcm3-cdk5A, or GST-Mcm3-cdk7A. Reaction products electrophoresed on SDS-PAGE were subjected to autoradiography (top) and Coomassie staining (bottom). (B) The CDK consensus sites in the Mcm3 NLS3NES3 are required for in vivo phosphorylation of Mcm3. YJL4110 (MCM3), YJL4313 (Myc6-MCM3), YJL4324 (Myc6-mcm3-cdk5A), and YJL4315 (Myc6-mcm3-cdk7A) were metabolically labeled with 32P-orthophosphate for 1 h before lysis and immunoprecipitation with 9E10 anti-Myc mAb. Immunoprecipitates were electrophoresed on SDS-PAGE and either subjected to autoradiography (top) or immunoblotted with rabbit anti-Myc polyclonal antibodies (bottom).
The Mcm3 CDK Consensus Phosphorylation Sites Regulate the Transport Activity of the NLS-NES Module
We next asked whether Cdc28 phosphorylation of the Mcm3 NLS3NES3 segment regulates the activity of the NLS-NES transport module. We examined the effect of mutating the Mcm3 consensus CDK phosphorylation sites on the localization of the GFP3 fusion protein containing the transport module. We introduced alanine substitutions in all five full consensus CDK sites or just the four sites flanking the Mcm3 NLS (Figure 1B). Representative cells from exponentially growing cultures expressing the wild-type or mutant fusion proteins are shown in Figure 8A. As described earlier, the wild-type fusion protein displayed a range of subcellular distributions from nuclear to cytoplasmic depending on the cell cycle position of individual cells. In contrast, both mutant proteins were constitutively nuclear. These results show that Cdc28 promotes the net nuclear export of the GFP3 fusion protein through phosphorylation of the NLS-NES transport module. They suggest that phosphorylation acts as a switch that flips the activity of the NLS-NES transport module from directing net nuclear import to directing net nuclear export.
Phosphorylation of the NLS-NES Transport Module Promotes the Net Nuclear Export of the Mcm2-7 Complex
To determine whether Cdc28 regulation of the NLS-NES transport module contributes to the cell cycle-regulated export of the entire Mcm2-7 complex, we investigated the effect of mutating the CDK consensus sites of the transport module in the endogenous MCM3 gene. We first examined three strains where the wild-type MCM3 gene was replaced by GFP-mcm3-cdk4A, GFP-mcm3-cdk5A, or GFP-mcm3-cdk7A (Figure 1A). At a metaphase arrest imposed by nocodazole, all three strains displayed partial nuclear retention of their mutant GFP-Mcm3 (Figure 8B), in contrast to wild-type GFP-Mcm3, which showed no such retention (Figure 8C, GFP-Mcm3 and Nguyen et al., 2000). This inability to fully export the mutant GFP-Mcm3 proteins was also observed in exponentially growing cells. Figure 9A (first two panels and accompanying bar graphs) shows a quantitative analysis of the nuclear localization of GFP-Mcm3-cdk4A and GFP-Mcm3 in unbudded, small budded, and uninucleate large budded cells. GFP-Mcm3-cdk4A persists in the nuclei of small budded and uninucleate large budded cells while GFP-Mcm3 is disappearing. These observations show that the CDK consensus sites in the Mcm3 NLS-NES module are required for the efficient cytoplasmic localization of Mcm3.
To examine the effect of the CDK consensus site mutations on the net nuclear export of other Mcm proteins, MCM3 was replaced by mcm3-cdk5A in MCM2-GFP, MCM4-GFP, and MCM7-GFP strains. At a nocodazole arrest, partial nuclear retention of the GFP fusion protein was observed in all three mcm3-cdk5A strains, in contrast to the full cytoplasmic distribution observed in the congenic MCM3 strains (Figure 8C). Similarly, in exponentially growing cells, the Mcm-GFP fusion proteins in these strains were never fully cleared from the nucleus. Partial nuclear retention of these three Mcm-GFP proteins was also observed in both nocodazole-arrested and exponentially growing cells when mcm3-cdk5A was replaced by mcm3-cdk4A strains (Figure 9B and unpublished data). These results suggest that the CDK consensus sites in the Mcm3 portion of the NLS-NES transport module are required for efficient nuclear export of each Mcm protein. We conclude that Cdc28 phosphorylation of NLS-NES module promotes the net nuclear export of the Mcm2-7 complex.
Because some nuclear export of Mcm proteins was still observed in mcm3-cdk4A, mcm3-cdk5A, and mcm3-cdk7A strains, it appears that phosphorylation of the Mcm3 NLS-NES module is not the sole mechanism by which Cdc28 promotes the net nuclear export of Mcm2-7. Whatever the additional mechanism, it presumably requires one or more NES(s) in the Mcm2-7 complex. To determine whether the Mcm3 NES contributes to this residual export, we replaced the MCM3 ORF with mcm3-cdk4A-nes in GFP-MCM3 and MCM7-GFP strains. The mutant Mcm3 expressed in these strains contains alanine substitutions in both the four CDK consensus sites flanking the NLS and the leucine-rich repeat of the NES (Figure 1A). The GFP-Mcm fusion proteins in these strains were strongly nuclear throughout the cell cycle, indicating that the combination of CDK consensus site and NES mutations in Mcm3 could completely abrogate the net nuclear export of Mcm3 and Mcm7 (Figure 9, A and B, last panels and graph). These results provide further evidence of the importance of the NLS-NES transport module in the regulation of Mcm protein localization.
Phosphomimic Mutation of Mcm3 Promotes the Net Nuclear Export of Mcm Proteins and Impairs Replication Initiation
We have shown that phosphorylation of the CDK consensus sites in the NLS-NES transport module is necessary for the efficient net nuclear export of the Mcm2-7 complex after G1 phase. To examine whether phosphorylation of these CDK consensus sites is sufficient to promote this export during G1 phase, we mutated the phosphoacceptor residues of all five full consensus sites to aspartic acid or glutamic acid to mimic constitutive phosphorylation of these sites (Figure 1A). This mutant mcm3 allele, mcm3-cdk5ED, was substituted for the wild-type endogenous MCM3 gene by two-step gene replacement in MCM2-GFP, GFP-MCM3, MCM4-GFP, or MCM7-GFP strains. As described earlier (Figure 6A, and Labib et al., 1999; Nguyen et al., 2000), these GFP fusion proteins normally concentrate in the nucleus during G1 phase. In the mcm3-cdk5ED mutant background, in contrast, much of these proteins were redistributed to the cytoplasm at a G1 phase arrest (Figure 10A). Some residual nuclear localization was still observed, although this is not surprising given that 1) phosphomimic mutations only partially resemble phosphorylated residues, 2) CDK phosphorylation of sites beyond the CDK consensus sites on Mcm3 may be necessary for full exclusion of the Mcm2-7 complex, and 3) the loading of Mcm2-7 onto chromatin makes these proteins refractory to cytoplasmic redistribution. Despite these limitations, the phosphomimic mutations on Mcm3 were sufficient to promote significant, albeit incomplete, export of the Mcm2-7 complex in G1 phase.
Such incomplete export may account for the ability to isolate mcm3-cdk5ED strains, as complete exclusion of Mcm2-7 from the nucleus during G1 phase would presumably be lethal. Nonetheless, the phosphomimic mutations clearly compromised the cell cycles of these cells. Although exponentially growing liquid cultures of wild-type GFP-MCM3 control strains doubled every 90 min, GFP-mcm3-cdk5ED mutant strains doubled every 140-150 min. Analysis of microcolonies derived from individually plated GFP-mcm3-cdk5ED cells not only confirmed that a significant percent of these cells divide much slower than wild-type GFP-MCM3 cells, but also established that 10-12% of the mutant cells generated fully arrested microcolonies after <3-4 divisions (Supplementary Figure 2A). Hence, although a mutant cell lineage could be propagated, considerable inviability was experienced every generation. Flow cytometry and budding indices of exponentially growing GFP-mcm3-cdk5ED cells showed that they spend a greater proportion of their cell cycle in G2/M phase relative to GFP-MCM3 cells (Supplementary Figure 2B). The simplest interpretation of these observations is that poor nuclear accumulation of Mcm2-7 during G1 phase compromises the initiation of DNA replication, making it difficult to complete a full S phase in a timely manner and triggering a checkpoint delay or arrest at G2/M phase. Many known replication initiation mutants, including the originally isolated mcm mutants, display a similar accumulation of G2/M cells (Gibson et al., 1990; Hennessy et al., 1991; Foiani et al., 1994; Merchant et al., 1997; Jacobson et al., 2001).
To determine whether replication initiation is indeed disrupted in the GFP-mcm3-cdk5ED mutant, we examined the rate of plasmid loss during multiple generations of nonselective growth (Figure 10B). Failure to initiate DNA replication on a plasmid will enhance its intrinsic loss rate. In control MCM3 and GFP-MCM3 strains, the plasmid YCp50 was lost at a rate of 3.0 and 4.4% per generation, respectively. In the GFP-mcm3-cdk5ED strain this rate was increased to 18.5%. Increased plasmid loss rates due to defective initiation can often be suppressed by increasing the number of origins on the plasmid. The plasmid pJW1112, which contains an additional seven tandem copies of the H4ARS origin inserted into Ycp50, specifically reduced the plasmid loss rate in the GFP-mcm3-cdk5ED strain (unpublished data), indicating that the elevated plasmid loss seen in the GFP-mcm3-cdk5ED strain is due to defective replication initiation.
This defect in initiation could be due to mislocalization of the Mcm2-7 complex or to localization-independent effects of these mutations on the initiation function of the complex. To distinguish between these possibilities, we fused the mcm3-cdk5ED to two tandem copies of the SV40 NLS (SVNLS2-mcm3-cdk5ED). This NLS fusion restored strong nuclear localization of Mcm2-GFP, GFP-Mcm3, Mcm4-GFP, and Mcm7-GFP in G1 phase (Figure 10A) and restored liquid culture doubling times and microcolony expansion to wild-type levels (unpublished data). In addition, this fusion restored plasmid loss rates in the SVNLS2-GFP-mcm3-cdk5ED strain to wild-type levels (Figure 10B). We conclude that the replication initiation defect observed in the GFP-mcm3-cdk5ED is primarily due to mislocalization of Mcm proteins in G1 phase. Considering the incomplete extent of this mislocalization arising from the phosphomimic mutations, these results suggest that bona fide ectopic phosphorylation of the Mcm3 portion of the NLS-NES module in G1 phase would severely impair replication initiation in S phase and that the normal CDK regulation of Mcm2-7 localization contributes significantly to restraining reinitiation of DNA replication during and after S phase.
DISCUSSION
The Mcm2-7 Complex Is Localized by a Transport Module Distributed over More than One Subunit
In our effort to understand CDK regulation of Mcm2-7 localization, we have mapped transport signals that promote the nuclear import and nuclear export of this complex. We show that three transport signals distributed across two subunits effectively cooperate to form a regulatory transport module that plays a key role in controlling the localization of the Mcm2-7 complex. Importantly, the isolated transport module fused to a heterologous protein is sufficient to recapitulate the cell cycle-regulated localization of the entire complex. Two of the transport signals in this module are weak NLSs on Mcm2 and Mcm3. Individually, each is required but not sufficient for robust nuclear accumulation of the Mcm2-7 complex and the heterologous protein. The third signal, positioned next to the Mcm3 NLS, is a Crm1-dependent NES containing a leucine-rich motif.
Previous analyses of nuclear localization of protein complexes have identified single subunits that are responsible for nuclear localization of an entire complex (Maridor et al., 1993; Pereira et al., 1998; Leslie et al., 2004; Subramaniam and Johnson, 2004; Wendler et al., 2004). To our knowledge, our analysis of Mcm2-7 nuclear localization in budding yeast provides the first documentation of a complex whose nuclear localization requires transport signals on more than one subunit. Distributing multiple required transport signals over distinct subunits provides one way to make complex formation a prerequisite for nuclear accumulation. Imposing such a prerequisite could be particularly important in preventing individual subunits or subcomplexes with unrestrained activity from accumulating in the nucleus, where they might threaten the integrity of the genome. The Mcm4-6-7 subcomplex, for example, exhibits helicase activity in vitro that is suppressed by other Mcm subunits in the full complex (Ishimi et al., 1998; Lee and Hurwitz, 2001). Preventing such an active subcomplex from accumulating in the nucleus unless it first associates with Mcm2 and Mcm3 may safeguard against inappropriate or uncontrolled unwinding of genomic DNA. Thus, using transport modules distributed over multiple subunits of a multiprotein complex could increase the specificity with which these subunits are localized by coupling their accumulation in either nucleus or cytoplasm to incorporation into a complex.
The use of weak transport signals distributed over more than one polypeptide potentially challenges the identification of transport signals directing the localization of multi-protein complexes. Although the weak NLSs identified in this work were somewhat evident based on their basic sequence composition, other weak or partial transport signals (for either import or export) may not be readily identified because of lack of sequence homology to known canonical signals. This raises the possibility that unrecognized distributed localization signals may program the localization of other multiprotein complexes. We also note that in addition to the transport module that we have identified on the Mcm2-7 complex, other NLSs or NESs may contribute to the localization of the complex; indeed our data suggest that other NESs that do not function through the Crm1 export receptor may work in parallel with the Mcm3 NES.
Future analysis of the mechanism by which the Mcm2 and Mcm3 NLS cooperate to promote nuclear localization of the Mcm2-7 complex should provide insight into how large multiprotein complexes are transported across the nuclear pore. Particularly interesting is the question of whether the NLSs act as a single bipartite NLS that strongly binds a single nuclear transport receptor, or whether they independently and weakly bind separate nuclear transport receptors. In the latter case, two transport receptors might still transport the entire complex as one unit across the nuclear pore, or they might transport separate subcomplexes that can only be retained and accumulate in the nucleus through mutual interaction. Whatever the precise mechanism, the end result, as suggested by this study, is to couple complex formation to nuclear accumulation.
Complex formation can also be coupled to nucleocytoplasmic localization if transport signals are masked or unmasked by protein interactions or conformational changes associated with complex formation (reviewed in Kaffman and O'Shea, 1999; Jans et al., 2000). Such a mechanism has been proposed for nuclear localization of Mcm2-7 in both Schizosaccharomyces pombe (Pasion and Forsburg, 1999) and S. cerevisiae (Labib et al., 2001), based on the interdependence among Mcm proteins for maintaining their nuclear localization. When the complex is nuclear (throughout the cell cycle in S. pombe and during G1 phase in S. cerevisiae), conditionally disrupting one Mcm subunit results in nuclear export of the remaining Mcm subunits. It has been suggested that complex formation may expose inaccessible NLSs or mask competing NESs on Mcm subunits such that nuclear import is favored over nuclear export (Pasion and Forsburg, 1999). Although direct evidence for this model is lacking, we cannot rule out the possibility that such a mechanism works in parallel with the distributed transport module reported here.
The NLS-NES Transport Module Is a Regulatory Target of CDKs
In addition to providing important transport signals for localization of the Mcm2-7 complex, we have shown that the distributed NLS-NES transport module provides a key target for CDK regulation of this localization. The unphosphorylated module promotes net nuclear import and the phosphorylated module promotes net nuclear export. Future experiments will be needed to address the precise mechanism by which CDK phosphorylation alters the activity of this module. In other examples of CDK-regulated protein localization, CDK phosphorylation has been implicated in modulating NLS and/or NES activity (Moll et al., 1991; Sidorova et al., 1995; Deng et al., 2004; Geymonat et al., 2004; Harreman et al., 2004). A classic and well defined example is provided by the related CDK, Pho80-Pho85, whose phosphorylation of the Pho4 NES and NLS promotes nuclear export and inhibits nuclear import, respectively (Komeili and O'Shea, 1999). Based on these precedents, phosphorylation of the Mcm3 transport module could down-regulate its NLS activity, up-regulate its NES activity, or both. Furthermore, in the context of the full Mcm2-7 complex, phosphorylation of the Mcm3 transport module could induce conformational changes that alter the activity or accessibility of other transport signals in the complex.
Some residual nuclear export of Mcm subunits persists after alanine substitutions in the Mcm3 CDK consensus sites, suggesting that CDKs may target more than the Mcm3 transport module to promote nuclear export. The only other full CDK consensus phosphorylation sites ((S/T)-X-P-(K/R)) in the Mcm2-7 complex are at the N-terminus of Mcm4. However, alanine substitution at these sites alone or in combination with mutation of the Mcm3 CDK sites did not have any striking effect on Mcm2-7 localization (unpublished data). Hence, further work will be necessary to refine our understanding of Cdc28 regulation of Mcm2-7 localization.
Nuclear Export of Mcm2-7 Provides a Significant Contribution to Replication Control
The cell cycle-regulated nuclear export of replication initiation proteins provides an appealing mechanism for preventing rereplication within a single cell cycle. Regulated localization of replication proteins has also been observed in metazoans, where Mcm proteins are constitutively nuclear. In both mammalian and Xenopus cells, ectopically expressed Cdc6 is exported from the nucleus in a CDK-dependent manner (Saha et al., 1998; Jiang et al., 1999; Petersen et al., 1999; Pelizon et al., 2000; Delmolino et al., 2001). Although interpretation of these studies is complicated by evidence that a portion of the endogenous Cdc6 population remains bound to nuclear structures throughout the cell cycle (Mendez and Stillman, 2000; Alexandrow and Hamlin, 2004), exporting the unbound population may still be important in the block to rereplication. Demonstrating such a role, however, has been difficult, in part because multiple layers of rereplication control make most individual mechanisms dispensable. In budding yeast, simultaneous disruption of multiple mechanisms has established a role for regulated Mcm localization in the block to rereplication (Nguyen et al., 2001; Mimura et al., 2004; Wilmes et al., 2004), but the limited rereplication that is induced makes it difficult to assess the relative significance of each individual mechanism.
In this study, we show that phosphomimic mutations of the CDK consensus sites at the Mcm3 NLS-NES segment promote the premature net export of Mcm2-7 from G1 nuclei. Even though this net export does not result in complete nuclear exclusion of the complex, it impairs the initiation of DNA replication and delays cell division. Presumably, the more effective nuclear exclusion mediated by bona fide CDK phosphorylation is sufficient to block reinitiation of DNA replication from most, if not all, origins. Thus, our data provide the first evidence that CDK regulation of the localization of a replication initiation factor may play a significant role in the inhibition of rereplication in eukaryotes. Analogous regulation of other replication initiation proteins in other eukaryotes may have a similarly important impact on the block to rereplication.
Supplementary Material
[Supplemental Materials]
Acknowledgments
We thank Anita Sil, David Toczyski, David Morgan, Hiten Madhani, Erin O'Shea, and members of the Li laboratory for critical reading of the manuscript. We thank Minoru Yoshida for the generous gift of Leptomycin B and Karsten Weis for the crm1 strains and plasmids. This work was supported by grants to J.J.L. from the American Cancer Society (RPG-99-169-01-CCG), National Institutes of Health (NIH; RO1 GM59704), and Brook Byers Award. M.E.L. was support by a Ford Foundation Predoctoral Fellowship and NIH predoctoral fellow (1 F31 CA110268-01). V.Q.N. was funded by an NIH Training Grant (T32 CA09270) and a UCSF Chancellor's Fellowship. A.W.R. was funded by National Institute of General Medical Sciences grant (1 R25 GM56847).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0412) on August 10, 2005.
References
- Alexandrow, M. G., and Hamlin, J. L. (2004). Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 24, 1614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault, V., Ikui, A. E., Drapkin, B. J., and Cross, F. R. (2005). Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 25, 6707-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Delmolino, L. M., Saha, P., and Dutta, A. (2001). Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276, 26947-26954. [DOI] [PubMed] [Google Scholar]
- Deng, W., Lin, B. Y., Jin, G., Wheeler, C .G., Ma, T., Harper, J. W., Broker, T. R., and Chow, L. T. (2004). Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J. Virol. 78, 13954-13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler, C. S., and Li, J. J. (1998). Ectopic induction of Clb2 in early G1 phase is sufficient to block prereplicative complex formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95, 2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F. (2004). Regulation of early events in chromosome replication. Curr. Biol. 14, R778-R786. [DOI] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (1997). The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U., Huber, J., Boelens, W. C., Mattaj, I. W., and Luhrmann, R. (1995). The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82, 475-483. [DOI] [PubMed] [Google Scholar]
- Foiani, M., Marini, F., Gamba, D., Lucchini, G., and Plevani, P. (1994). The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14, 923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- Geymonat, M., Spanos, A., Wells, G. P., Smerdon, S. J., and Sedgwick, S. G. (2004). Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell. Biol. 24, 2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, S. I., Surosky, R. T., and Tye, B. K. (1990). The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol. Cell. Biol. 10, 5707-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. M., and Li, J. J. (2005). Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 16, 421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. (eds.) (1990). Guide to Yeast Genetics and Molecular Biology, New York: Academic Press.
- Harreman, M. T., Kline, T. M., Milford, H. G., Harben, M. B., Hodel, A. E., and Corbett, A. H. (2004). Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem. 279, 20613-20621. [DOI] [PubMed] [Google Scholar]
- Hennessy, K. M., Lee, A., Chen, E., and Botstein, D. (1991). A group of interacting yeast DNA replication genes. Genes Dev. 5, 958-969. [DOI] [PubMed] [Google Scholar]
- Ishimi, Y., Komamura, Y., You, Z., and Kimura, H. (1998). Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 273, 8369-8375. [DOI] [PubMed] [Google Scholar]
- Jacobson, M. D., Munoz, C. X., Knox, K. S., Williams, B. E., Lu, L. L., Cross, F. R., and Vallen, E. A. (2001). Mutations in SID2, a novel gene in Saccharomyces cerevisiae, cause synthetic lethality with sic1 deletion and may cause a defect during S phase. Genetics 159, 17-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans, D. A., Xiao, C. Y., and Lam, M. H. (2000). Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22, 532-544. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Wells, N. J., and Hunter, T. (1999). Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96, 6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman, A., and O'Shea, E. K. (1999). Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15, 291-339. [DOI] [PubMed] [Google Scholar]
- Komeili, A., and O'Shea, E. K. (1999). Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284, 977-980. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E. P., Wolff, B., Yoshida, M., and Horinouchi, S. (1999). Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96, 9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Diffley, J.F.X., and Kearsey, S. E. (1999). G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1, 415-422. [DOI] [PubMed] [Google Scholar]
- Labib, K., Kearsey, S. E., and Diffley, J. F. (2001). MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell 12, 3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Tercero, J. A., and Diffley, J. F. (2000). Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288, 1643-1647. [DOI] [PubMed] [Google Scholar]
- Lee, J. K., and Hurwitz, J. (2001). Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98, 54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, D. M., Zhang, W., Timney, B. L., Chait, B. T., Rout, M. P., Wozniak, R. W., and Aitchison, J. D. (2004). Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24, 8487-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, E., Li, X., Yan, F., Zhao, Q., and Wu, X. (2004). Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 279, 17283-17288. [DOI] [PubMed] [Google Scholar]
- Loog, M., and Morgan, D. O. (2005). Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434, 104-108. [DOI] [PubMed] [Google Scholar]
- Maridor, G., Gallant, P., Golsteyn, R., and Nigg, E. A. (1993). Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J. Cell Sci. 106(Pt 2), 535-544. [DOI] [PubMed] [Google Scholar]
- Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004). Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165, 473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, J., and Stillman, B. (2000). Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, A. M., Kawasaki, Y., Chen, Y., Lei, M., and Tye, B. K. (1997). A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L., and Zhang, H. (2002). Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22, 1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, S., Seki, T., Tanaka, S., and Diffley, J. F. (2004). Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431, 1118-1123. [DOI] [PubMed] [Google Scholar]
- Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743-758. [DOI] [PubMed] [Google Scholar]
- Neville, M., and Rosbash, M. (1999). The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18, 3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., Irie, K., and Li, J. J. (2000). Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10, 195-205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., and Li, J. J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- Nigg, E. A. (1993). Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 3, 296-301. [DOI] [PubMed] [Google Scholar]
- Nishi, K., Yoshida, M., Fujiwara, D., Nishikawa, M., Horinouchi, S., and Beppu, T. (1994). Leptomycin B. targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269, 6320-6324. [PubMed] [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997). Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278, 141-144. [DOI] [PubMed] [Google Scholar]
- Pasion, S. G., and Forsburg, S. L. (1999). Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell 10, 4043-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizon, C., Madine, M. A., Romanowski, P., and Laskey, R. A. (2000). Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 14, 2526-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., Knop, M., and Schiebel, E. (1998). Spc98p directs the yeast gamma-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell 9, 775-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, B. O., Lukas, J., Sorensen, C. S., Bartek, J., and Helin, K. (1999). Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18, 396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, S., Lengauer, C., and Nasmyth, K. (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a `reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, P., Chen, J., Thome, K. C., Lawlis, S. J., Hou, Z. H., Hendricks, M., Parvin, J. D., and Dutta, A. (1998). Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18, 2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova, J. M., Mikesell, G. E., and Breeden, L. L. (1995). Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 6, 1641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., Ford, C. S., Guthrie, C., and Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041-1050. [DOI] [PubMed] [Google Scholar]
- Subramaniam, P. S., and Johnson, H. M. (2004). The IFNAR1 subunit of the type I IFN receptor complex contains a functional nuclear localization sequence. FEBS Lett. 578, 207-210. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H., and Fujita, M. (2004). Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279, 19691-19697. [DOI] [PubMed] [Google Scholar]
- Thomer, M., May, N. R., Aggarwal, B. D., Kwok, G., and Calvi, B. R. (2004). Drosophila double-parked is sufficient to induce re-replication during development and is regulated by cyclin E/CDK2. Development 131, 4807-4818. [DOI] [PubMed] [Google Scholar]
- Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859-864. [DOI] [PubMed] [Google Scholar]
- Weis, K. (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441-451. [DOI] [PubMed] [Google Scholar]
- Wen, W., Meinkoth, J. L., Tsien, R. Y., and Taylor, S. S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463-473. [DOI] [PubMed] [Google Scholar]
- Wendler, P., Lehmann, A., Janek, K., Baumgart, S., and Enenkel, C. (2004). The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alphabeta and proteasome functions. J. Biol. Chem. 279, 37751-37762. [DOI] [PubMed] [Google Scholar]
- Wilmes, G. M., Archambault, V., Austin, R. J., Jacobson, M. D., Bell, S. P., and Cross, F. R. (2004). Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18, 981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M. R., Suzuki, K., Yan, H., Gibson, S., and Tye, B. K. (1997). Nuclear accumulation of Saccharomyces cerevisiae Mcm3 is dependent on its nuclear localization sequence. Genes Cells 2, 631-643. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Chen, Y., and Dutta, A. (2004). Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24, 7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]