Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins (original) (raw)
Abstract
We have observed a common sequence motif in membrane proteins, which we call a glycine zipper. Glycine zipper motifs are strongly overrepresented and conserved in membrane protein sequences, and mutations in glycine zipper motifs are deleterious to function in many cases. The glycine zipper has a significant structural impact, engendering a strong driving force for right-handed packing against a neighboring helix. Thus, the presence of a glycine zipper motif leads directly to testable structural hypotheses, particularly for a subclass of glycine zipper proteins that form channels. For example, we suggest that the membrane pores formed by the amyloid-β peptide in vitro are constructed by glycine zipper packing and find that mutations in the glycine zipper motif block channel formation. Our findings highlight an important structural motif in a wide variety of normal and pathological processes.
Keywords: amyloid-β, membrane channel, membrane protein structure, prion, transmembrane helix
More than 13 structures a day are currently being deposited in the Protein Data Bank (1), and structural genomics centers have been created to obtain structures even faster [such as the National Institute of General Medical Sciences (NIGMS) Protein Structure Initiative, www.nigms.nih.gov/psi]. In this assault on protein structure, however, technical challenges have left membrane proteins far behind. Membrane protein structures are currently being solved at ≈0.2% of the pace of soluble proteins (2). Thus, membrane protein biochemists are relatively starved for structural insight into these key proteins. In the absence of dramatic technical improvements, alternatives to experimental structure determination are needed.
Here, we describe a transmembrane (TM) sequence motif, the glycine zipper, that can lead directly to structural models for many membrane proteins. The most significant glycine zipper sequence patterns are (G,A,S)XXXGXXXG and GXXXGXXX(G,S,T). These patterns contain a GXXXG motif, which is known to be important in TM helix homodimers where the Gly faces are in direct contact (3-5). The GXXXG sequence pattern is statistically overrepresented in membrane proteins in general, not just in TM homodimers (4). Nevertheless, the structural role of the GXXXG pattern in other types of TM helix packing interactions has not been elucidated. We find that the addition of an appropriately spaced small residue, as found in the glycine zipper, leads to a distinct preference for right-handed packing against a heterologous helix surface. Thus, the presence of a glycine zipper generates a strong helix packing prediction, particularly for homooligomeric channel proteins, providing a structural foundation for hypothesis-driven investigations.
Methods
Glycine Zipper Motif Search. We started with Swiss-Prot release 41.15 containing 129,996 proteins (6). All sequences <50 residues in length were removed, leaving 125,887 proteins. Helical membrane proteins were identified by using the Eisenberg hydrophobicity scale and a window length of 21 residues (7). A protein was flagged as a membrane protein if the most hydrophobic segment had an average hydrophobicity of >0.68 or if two segments had a summed average hydrophobicity of >1.1. By these criteria, 29,416 proteins (≈23.4%) were identified as helical membrane proteins. TM regions within the membrane proteins were identified by using an average hydrophobicity of 0.42. A nonredundant membrane protein data set was made by eliminating sequences with >90% sequence identity. The final data set contained 23,102 proteins. We then searched for proteins containing the GXXXGXXXG motif in the TM segments and found 1,772 proteins and 2,250 motifs. After removing proteins with low complexity G repeats (more than four G residues in a row), 1,726 proteins with the glycine zipper motif remained. The same method was applied to identify proteins with the other strong glycine zipper motifs.
Relative Conservation Score (RCS). The program conseq (http://conseq.bioinfo.tau.ac.il) (8) was used to determine the sequence conservation reflected in multiple sequence alignments of glycine zipper proteins. This server compares the sequence of a reference protein with proteins deposited in Swiss-Prot (6) to find homologs. The number of psi-blast iterations and the E value cutoff used were 1 and 0.001, respectively. All of the sequences that were found to be evolutionarily related with the glycine zipper proteins in the data set were used in conservation scoring. Briefly, the conseq server assigns a conservation score to each residue, taking into account the evolutionary relationships among the family of homologs. The scores are normalized such that the average score is zero, and negative and positive deviations represent the degrees of conservation and variation, respectively. RCS values were calculated by taking normalized ratio between glycines in glycine zippers and other glycines in TMs. RCS = |_CS_Gly_GZ - _CS_TM|/|_CS_Gly_Ran - _CS_TM|, where _CS_Gly_GZ, _CS_Gly_Ran, and _CS_TM are the conservation scores for Gly in glycine zippers, random glycines, and all TM residues, respectively.
Ion Channel Current Measurements. The methods for measuring channel currents were reported in ref. 9. Briefly, planar bilayers were formed by spreading a 50:50 (weight:weight) mixture of 1-palmitoyl-2-oleoyl phosphatidylethanolamine and phosphatidyl serine (Avanti Polar Lipids) over a hole in a Teflon barrier between two chambers. The chamber compartments were filled with symmetrical KCl solution (140 mM KCl/1 mM CaCl/10 mM Hepes, pH7.4). Measurements of mutant channel activity were facilitated by the addition 0.15 nM SDS, which had no effect in the absence of the peptides (10). Ion channel currents were recorded with an Axopatch amplifier (Axon Instruments, Union City, CA) with a 1-kHz low-pass filter during data acquisition. After the formation of the membrane, we verified that there was no channel-like activity before the addition of the amyloid-β (Aβ) 1-42 peptides to the chamber. The peptides were dissolved in 10 mM NaOH at a concentration of 0.42 mM. Aβ1-42 peptide incorporation into acidic phospholipid planar bilayer membranes was achieved by adding peptide solutions into the chamber to make a final peptide concentration of 3 μg/ml. The Aβ1-42 peptides were synthesized and purified to >95% homogeneity by CS Bio (Menlo Park, CA).
Neuronal Cell Viability Assay. Mouse Neuro-2a (N2A) neuroblastoma cells (American Type Culture Collection) were grown on cell culture plates by using standard methods in DMEM. The medium contained high glucose, glutamine, pyridoxine, 110 mg/liter sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated FBS (Invitrogen). Aβ42 peptides were pre-dissolved in 10 mM sodium hydroxide solution at a concentration of 480 μM. The solutions, along with a control containing no peptide, were diluted 4-fold with Dulbecco's PBS (D-PBS) buffer and immediately added to the cells at final concentrations up to 5 μM. Cell viability was assayed by using the live/dead assay kit from Molecular Probes according to the supplied instructions. Cells were washed with a 1,000× volume of D-PBS, and 200 μl of a solution containing 2 μM ethidium homodimer and 1 μM CalceinAM was added. After incubation for 30-45 min at room temperature, the cells were viewed and counted by using an inverted fluorescence microscope at 580 nm for CalceinAM and 617 nm for ethidium bromide.
Results
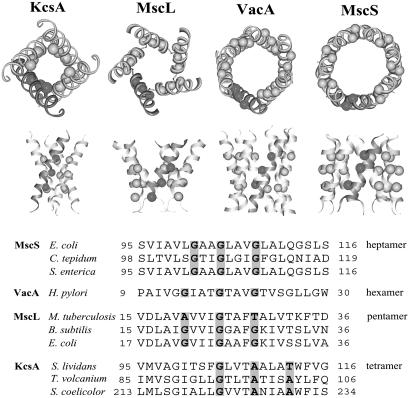
Glycine Zipper Motifs Mediate TM Helix Packing Interactions in Channel Proteins. We first noticed glycine zipper packing in homooligomeric channel proteins. The unrelated channel proteins, shown in Fig. 1 [KcsA (potassium channel), MscL (mechanosensitive channel of large conductance), VacA (vacuolating toxin A), and MscS (mechanosensitive channel of small conductance)], all use a stripe of small residues in the packing interface (11-14). Although Gly, Ala, and Thr are all observed in these structures, glycine is the most common interfacial residue. Thus, we call this packing motif a glycine zipper, because it is reminiscent of another common oligomerization motif found in soluble proteins, the leucine zipper (15). Like the leucine zipper, the glycine zipper can lead to a variety of different oligomerization states. The channels shown in Fig. 1 range from tetramers to heptamers. The fact that such a large fraction of the relatively few known membrane channel structures use the glycine zipper suggests that the sequence motif plays a special role in forming these homooligomeric bundles. As discussed below, the glycine zipper is also used in asymmetric helix packings as well.
Fig. 1.
Homooligomeric glycine zipper channel structures. Shown are KcsA, potassium channel pore-lining helices (Protein Data Bank ID code 1BL8) (11); MscL, mechanosensitive channel of large conductance (PDB ID code 1MSL) (12); VacA, vacuolating toxin A anion selective channel model (PDB ID code 1SEW) (13); and MscS, mechanosensitive channel of small conductance (PDB ID code 1MXM) (14). The balls highlight the Cα carbon positions of the glycine zipper packing residues. The glycine zipper packing residues are also highlighted on the TM sequences shown below. Small residues (Gly, Ala, or Thr) were overrepresented at the highlighted positions on the homologs.
The Glycine Zipper Motif Is Unusually Common. The glycine zipper packing mode is distinct from the well known GXXXG dimerization motif found in glycophorin A and other symmetric dimers that involve direct packing between the Gly faces (16). In glycine zipper packing, the glycine zipper packs against a different face of the associated helix. Thus, the glycine zipper may be more promiscuous in the nature of the helix face that it packs against, and it seemed possible that the glycine zipper could be used in a more catholic fashion throughout membrane protein structures, not just in symmetric oligomers like the channel proteins.
A method to discover sequence motifs that are important for defining membrane protein structure was introduced by Engelman and coworkers (4), who looked for sequence patterns that were significantly more common than expected by chance. Indeed, Senes et al. (4) found that a perfect glycine zipper motif, GXXXGXXXG, with Gly residues spaced every four positions is one of the most overrepresented triplets in predicted TM helices from all membrane proteins, without restriction to a specific class (odds ratio = 1.92; P = 1.38 × 10-13). Although the heptameric packing in the MscS pore has a slightly different spacing (see Fig. 1), a four-residue spacing is strongly preferred over other possible Gly patterns, reinforcing the significance of the GXXXGXXXG sequence pattern. Nevertheless, other spacings could lead to glycine zipper packing if the Gly residues are placed on the same face of the helix as in MscS.
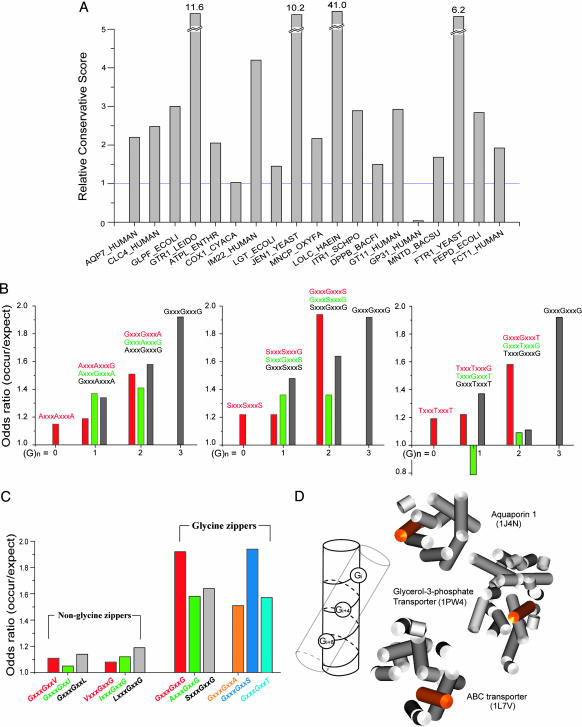
Glycine Zippers Are Conserved. As another measure of the importance of the glycine zipper motif, we find that glycine zipper sequences are strongly conserved. Nineteen proteins with perfect glycine zippers and their homologs were examined. As shown in Fig. 2_A_, in all but two of the proteins, Gly residues in glycine zippers are more strongly conserved than random Gly residues. These results again indicate that the glycine zipper motif plays a particularly important role in membrane protein structure.
Fig. 2.
Amino acid preferences and conservation of glycine zippers. (A) Glys in glycine zipper motifs are more conserved than random Gly residues. A histogram of the RCS for a set of 19 glycine zipper motif proteins is plotted. A ratio >1 indicates that Gly residues are more conserved in the glycine zipper motif than they are elsewhere in the structure. Conservation scores and protein codes are given in Table 2. (B) Odds ratios for Ala, Ser, and Thr substitutions. (C) Comparison of odds ratios for the strong glycine zipper motifs with single substitutions of Val, Leu, or Ile. Many triplet motifs with the small residues (Gly, Ala, Ser, or Thr) spaced four apart are at least somewhat overrepresented in TM helices. Single substitutions with Val, Leu, or Ile are not overrepresented, indicating that they are not generally good substitutes for Gly. (D) A glycine zipper drives a preference for right-handed helix packings. Glycine residues located at every fourth position, represented as balls on cylindrical model helices, create a polar stripe of small residues by exposing the backbone carbonyl and amide atoms that may nucleate helix interactions in the apolar membrane environments. The right-handed packing found in the glycine zipper helices of aquaporin 1 (PDB ID code 1J4N, residues 212-231), glycerol-3-phospate transporter (PDB ID code 1PW4, residues 380-404), and ABC transporter (PDB ID code 1L7V, residues 92-107) are highlighted.
Small Residues Can Substitute in Glycine Zipper Motifs. Because different small residues were found to substitute for Gly in the channel proteins discussed above, we next examined whether other possible sequence patterns, consistent with the glycine zipper spacing, were overrepresented in the TMSTAT database of Engelman and coworkers (4). Senes et al. (4) previously presented the effects on pattern abundance when either the first or third glycine in the GXXXGXXXG motif was replaced with a small residue. We expanded this analysis to examine the abundance of sequence patterns when the perfect glycine zipper motif was adulterated with one, two, or three small residues at any position in the zipper (Ala, Ser, or Thr). As shown in Fig. 2_B_, the strongest glycine zipper sequence motifs, with odds ratios >1.5, were GXXXGXXXG, AXXXGXXXG, GXXXGXXXA, SXXXGXXXG, GXXXGXXXS, and GXXXGXXXT. These six motifs contain two or more Gly residues, and all maintain the central Gly residue in the pattern. Not only are these sequence motifs overrepresented in TM helices in general, but Russ and Engelman (5) found that many of these zipper patterns are found in homooligomerizing TM sequences discovered experimentally by using the TOX-CAT assay. Indeed, the GXXXGXXXT sequence pattern was particularly common in these TM helices (5).
Although the pattern GXXXG by itself is overrepresented in TM helices (4), the presence of a GXXXG sequence alone is not necessarily significant in the triplet patterns. In Fig. 2_C_, the odds ratios for the strong glycine zipper motifs are compared with GXXXG containing triplets with Val, Ile, or Leu in place of Ala, Ser, or Thr [also see Senes _et al._ (4)]. None of these triplets is significantly overrepresented. Thus, there is apparently special significance associated with the strong glycine zipper sequence patterns, and glycine zippers are apparently part of the increased abundance of the GXXXG subpattern.
Glycine Zippers Strongly Influence TM Helix Packing. The presence of a glycine zipper motif has a profound effect on protein structure. We searched for the preferred glycine zipper motifs in membrane proteins of known structure and found 13. All of these glycine zipper motifs are directly involved in helix packing (Fig. 5, which is published as supporting information on the PNAS web site). Thus, the glycine zipper face apparently provides a magnet for helix packing. Moreover, the glycine zipper motif strongly drives right-handed helix packing, as found in the structures shown in Fig. 1. On average, <30% of helix packings in membrane proteins are right-handed (17). Of the glycine zipper helices in proteins of known structure, however, this preference is completely reversed. Ten of 13 (77%) are involved in right-handed helix packing. As shown in Fig. 2_D_, a four-residue spacing creates an ≈20° angle with respect to the helix axis. Small residues are favored in TM helix packing sites (18), and a right-handed helix packing maximizes contact with the small residues of the glycine zipper motif.
Glycine Zippers Are Common in Membrane Proteins. We searched for the preferred glycine zipper motifs in a database of predicted TM helical proteins from the Swiss-Prot database (6). Sequences with >90% sequence identity were removed from the TM protein database, leaving a total of 23,102 protein sequences. Of this set, we found that 1,726 proteins (7.5%) contain the perfect glycine zipper motif, GXXXGXXXG, and 5,684 (25%) contain one of the preferred motifs (see Table 2, which is published as supporting information on the PNAS web site). Thus, approximately one-quarter of all membrane proteins contain one of the strong glycine zipper motifs, and many others likely use weaker glycine zipper motifs. For example, MscS, which is known to use glycine zipper packing (Fig. 1), would not be counted because of the slightly different spacing of the pattern. The analysis of known protein structures discussed above suggests that virtually all of these glycine zipper motifs are involved in helix packing and that the vast majority of these packings are right-handed.
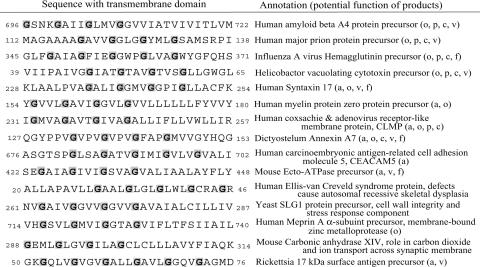
Prediction of Right-Handed Homooligomeric Bundles. Our finding that a glycine zipper creates a strong driving force for right-handed helix packing suggests that the presence of a glycine zipper motif in a single-pass protein is likely to drive the formation of right-handed homooligomeric bundles like those shown in Fig. 1. To look for proteins that are highly likely to form these homooligomeric bundle structures, we searched for single-pass proteins with extended glycine zipper motifs containing four Gly residues (GXXXGXXXGXXXG). The proteins retrieved in this search are listed in Table 1. We predict that these proteins form right-handed helix bundles to deliver their physiological or pathological function.
Table 1. List of single-pass TM proteins containing extended glycine zipper motifs.
For many of the extended glycine zipper proteins, there is already experimental evidence for the importance of the glycine zipper motif in their function. One of the known pore-forming proteins is VacA, a toxin secreted by Helicobacter pylori, the etiologic agent in roughly half of all stomach ulcers (19). VacA is known to form a hexameric anion selective channel, and mutations in the glycine zipper motif residues G47 and G51 abolish the channel activity and cytotoxicity of VacA (20). Influenza hemagglutinin, syntaxin 17, annexin A7, and ecto-ATPase are involved in membrane fusion, a process that involves formation of a fusion pore (21-23). In the best studied of these proteins, influenza hemagglutinin, the glycine positions are highly conserved, and mutations in the glycine residues or alterations of the glycine spacing in the glycine zipper motif block viral fusion (24). A number of the extended glycine zipper proteins are involved in cell adhesion. Of these extended glycine zipper proteins, myelin protein zero is known to be a tetramer, and glycine mutations at position G163 and G167 in the TM domain are linked to Charcot-Marie-Tooth disease and Dejerine-Sottas syndrome (25, 26).
Extended glycine zippers are also found in tight junction (TJ) proteins such as CLMP that act as gated intercellular pores regulating the exchange of small molecules, ions, and water between cells (27). Moreover, other major TJ proteins, such as the occludins, also have extended glycine zipper motifs but are not included in Table 1 because they contain multiple TM domains (see Table 2). It is therefore tempting to speculate that glycine zipper pore structures mediate the passage of molecules through TJs.
Two glycine zipper proteins, Aβ and prion protein (PrP), are involved in the neurodegenerative diseases Alzheimer's disease and spongiform encephalopathies. The glycine residues in these TM domains are completely conserved, which suggests that the glycine zipper motifs are playing an important role in their normal function. Cytotoxic fragments of Aβ and PrP include the glycine zipper motifs and are also known to form pores in artificial membranes in vitro that could play a role in disease etiology (28-33). The fact that these peptides contain glycine zipper motifs suggests that in vitro channel formation by these peptides could be driven by glycine zipper packing (see below).
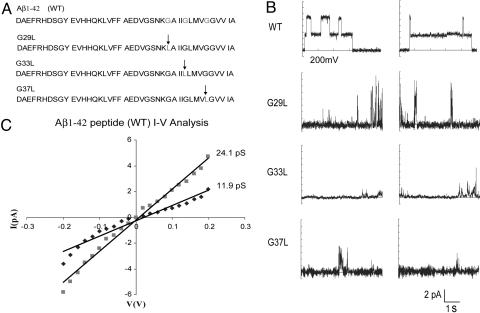
Experimental Test of a Glycine Zipper Packing Prediction. To test whether the glycine zipper motif in the Aβ42 peptide is important for channel formation in vitro, we made three mutant Aβ42 peptides in which Gly-29, Gly-33, and Gly-37 were substituted individually with Leu, disrupting the glycine zipper packing interface. Fig. 3_B_ shows some representative current traces across a polar lipid bilayer membrane exposed to 3 μg/ml Aβ1-42 peptides. Consistent with earlier studies (31-33), we found that the WT Aβ1-42 readily forms channels. Analysis of the current distributions and current-voltage (_I_-V) plots shown in Fig. 3_C_ demonstrate that the current increased in quantized units with a conductance of ≈12 pS. These results confirm that Aβ42 can indeed form unique, well organized channel structures under our measurement conditions. In contrast, none of the Gly-to-Leu mutants showed organized channel behavior. All of the mutants induced variable current spikes, indicating that they can disrupt the membrane to some extent, but they are apparently unable to maintain stable, organized channels (see Fig. 6, which is published as supporting information on the PNAS web site). Although overall delivery to the membrane may be altered by peptide solubility, the character of the current spikes induced by the peptides is clearly altered with the mutants. Moreover, we saw no evidence for insoluble fibril formation in our peptide samples as judged by Congo red staining (not shown). As shown in Fig. 4, the ability to disrupt membranes is correlated with cytotoxicity (Fig. 4). These results suggest that the Aβ42 pore formation depends on the glycine zipper motif. Although we cannot be certain that this dependence is due to direct helix packing involving the glycine zipper motif, the fact that all glycine zippers in known structures are in packing interfaces favors a glycine zipper packing hypothesis. In this manner, the identification of a simple sequence motif defines a limited set of structural templates for hypothesis-driven experiments.
Fig. 3.
Channel formation of Aβ42 WT peptide and mutants. (A) Sequences of the Aβ42 peptide variants. The position of the mutation in the glycine zipper motif is indicated by the arrow. (B) Channel activities of Aβ42 peptides recorded in the symmetrical K+ solutions (140 mM KCl/1 mM CaCl/10 mM Hepes, pH7.4) at a TM potential of -200mV (see Methods for details). Multiple conductance levels were found with the WT Aβ42 peptides, indicating the presence of multiple channels in the membrane. (C) The current-voltage relationships for the WT Aβ42 peptides. A single channel conductance of ≈12 pS was calculated from the difference in slopes of the regression line drawn to fit the Aβ42 channel current (▪, 24.1 pS) and the baseline current from the lipid bilayer (♦, 11.9 pS).
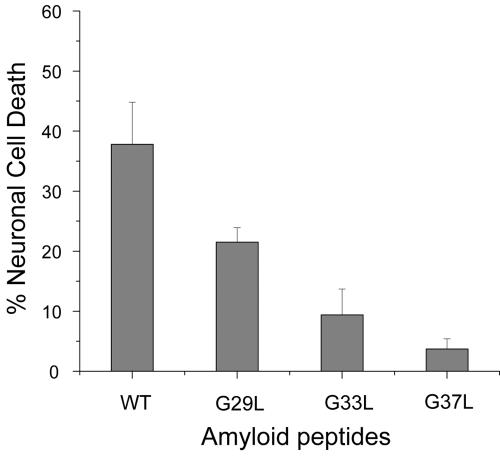
Fig. 4.
Neuronal cell viability after treatment of Aβ42 WT and mutants. The percentage of dead Neuro-2a cells was measured after exposure of the culture to 5 μMAβ42 peptide for 5 h. With the WT Aβ42 peptide, 37.8 ± 7.0% of the cells were dead after 5 h. With G29L, G33L, and G37L, only 21.5 ± 2.4%, 9.4 ± 4.3%, and 3.7 ± 1.7% were dead, respectively. Thus, all of the mutants were less toxic than WT. The order of increasing toxicity was WT > G29L > G33L > G37L. This order correlates well with the mutants' ability to disrupt membranes, suggesting that toxicity in vitro may depended on membrane permeabilization.
Discussion
We have described a common TM packing motif in membrane proteins, the glycine zipper. The importance of the motif in membrane protein structure is implied by its statistically significant overrepresentation in TM helices, its unusual conservation in membrane protein families, and its clear influence on helix packing seen in proteins of known structure. Thus, glycine zippers likely play a significant structural role in the many membrane proteins where they are found.
The glycine zipper involves a row of small residues on one face of a helix. The preference for small residues in TM helix packing interactions is well known (4, 5), but the confluence of three small residues spaced by four appears to have special significance. In particular, TM glycines in a glycine zipper motif are much more strongly conserved than random TM glycines. Moreover, the GXXXG pattern by itself is not necessarily significant unless it is associated with additional sequence constraints (34, 35) as are found in the glycine zipper.
The most favorable glycine zippers have at least two glycines, and glycine occupies the central position. The source of the glycine preference may be manifold. One possibility is the minimal entropy cost required to bury a small residue (36). The exposure of backbone atoms could also facilitate weakly polar interactions such as the CH···O hydrogen bonds found in the glycophorin A dimer (37, 38). In addition, small residues can allow closer approach of the helices, perhaps maximizing packing interactions (37-39).
The finding of glycine zipper motifs in Aβ and PrP, which are associated with Alzheimer's and prion diseases, suggests the possibility of a common pathological role for the motif. Although there are many similarities in these diseases, to our knowledge, no other obvious protein sequence or structural connections have been identified. A hallmark of both Alzheimer's disease and spongiform encephalopathies is the formation of fibrillar deposits in the brain (40). Although these fibrillar aggregates can be an obvious symptom of the diseases, their presence is not well correlated with disease progression, and there is growing evidence that smaller, prefibrillar aggregates are significantly more cytotoxic than mature fibrils (41-44). Both toxic Aβ peptides and PrP peptides (residues 106-126) have been found to form ion channels, suggesting a channel hypothesis of pathogenesis in which a loss of ion hemeostasis ultimately leads to neuronal cell death (30-33). Moreover, pore-like structures have been observed by electron and atomic force microscopy (28, 33). In view of this hypothesis, it is an interesting coincidence that both Aβ and PrP contain glycine zipper motifs, which are so common in channel proteins. It is also notable that diseases involving the extended glycine zipper proteins we identified (VacA, Rickettsia surface protein, Aβ, and PrP) all induce extensive vacuolation in the affected cells. Thus, despite the complete lack of any other apparent relationship among these proteins, the associated diseases show a remarkable histological similarity, suggesting that glycine zipper motifs may impart a common vacuolating channel function to all of these proteins. The many commonalities between Alzheimer's disease and the spongiform encephalopathies, and the absence of any other apparent sequence relationships, suggest that the involvement of the glycine zippers in disease etiology deserves further scrutiny.
The findings that all glycine zipper motifs in known structures are directly involved in helix interactions and that nearly 80% pack in a right-handed orientation make strong structural predictions. In particular, an important subset of glycine zipper TM helices create right-handed homooligomers that can line membrane pores. Thus, the presence of a glycine zipper motif presents simple and testable structural models for proteins that are often beyond the reach of current structure determination methods. For example, the Aβ pores, fusion pores, and TJ channels exist only transiently, impeding high-resolution structure determination. Thus, the identification of glycine zipper motifs can provide a critical structural foundation for the design of structure-based, hypothesis-driven experiments in the thousands of membrane proteins where they are found.
Supplementary Material
Supporting Information
Acknowledgments
We thank Alessandro Senes for helpful discussions and members of J.U.B.'s laboratory for discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM3919 and a National Science Foundation Integrative Graduate Education and Research Traineeship predoctoral award (to A.O.). J.U.B. is a Leukemia and Lymphoma Society Scholar.
Author contributions: S.K., T.-J.J., A.O., D.Y., J.J.S., and J.U.B. designed research; S.K., T.-J.J., A.O., and D.Y. performed research; T.-J.J., A.O., and D.Y. contributed new reagents/analytic tools; S.K., T.-J.J., A.O., D.Y., J.J.S., and J.U.B. analyzed data; and S.K., T.-J.J., J.J.S., and J.U.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TM, transmembrane; Aβ, amyloid-β; RCS, relative conservation score; PrP, prion protein; MscS, mechanosensitive channel of small conductance; VacA, vacuolating toxin A; TJ, tight junction.
References
- 1.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28**,** 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White, S. H. (2004) Protein Sci. 13**,** 1948-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon, M. A., Flanagan, J. M., Treutlein, H. R., Zhang, J. & Engelman, D. M. (1992) Biochemistry 31**,** 12719-12725. [DOI] [PubMed] [Google Scholar]
- 4.Senes, A., Gerstein, M. & Engelman, D. M. (2000) J. Mol. Biol. 296**,** 921-936. [DOI] [PubMed] [Google Scholar]
- 5.Russ, W. P. & Engelman, D. M. (2000) J. Mol. Biol. 25**,** 911-919. [DOI] [PubMed] [Google Scholar]
- 6.Bairoch, A., Boeckmann, B., Ferro, S. & Gasteiger, E. (2004) Brief. Bioinformatics 5**,** 39-55. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg, D., Schwarz, E., Komaromy, M. & Wall, R. (1984) J. Mol. Biol. 179**,** 125-142. [DOI] [PubMed] [Google Scholar]
- 8.Berezin, C., Glaser, F., Rosenberg, J., Paz, I., Pupko, T., Fariselli, P., Casadio, R. & Ben-Tal, N. (2004) Bioinformatics 20**,** 1322-1324. [DOI] [PubMed] [Google Scholar]
- 9.Arispe, N., Pollard, H. B. & Rojas, E. (1993) Proc. Natl. Acad. Sci. USA 90**,** 10573-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Micelli, S., Meleleo, D., Picciarelli, V., Stoico, M. G. & Gallucci, E. (2004) Biophys. J. 87**,** 1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle, D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T. & MacKinnon, R. (1998) Science 280**,** 69-77. [DOI] [PubMed] [Google Scholar]
- 12.Chang, G., Spencer, R. H., Lee, A. T., Barclay, M. T. & Rees, D. C. (1998) Science 282**,** 2220-2226. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S., Chamberlain, A. K. & Bowie, J. U. (2004) Proc. Natl. Acad. Sci. USA 101**,** 5988-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass, R. B., Strop, P., Barclay, M. & Rees, D. C. (2002) Science 298**,** 1582-1587. [DOI] [PubMed] [Google Scholar]
- 15.Landschulz, W. H., Johnson, P. F. & McKnight, S. L. (1988) Science 240**,** 1759-1764. [DOI] [PubMed] [Google Scholar]
- 16.MacKenzie, K. R., Prestegard, J. H. & Engelman, D. M. (1997) Science 276**,** 131-133. [DOI] [PubMed] [Google Scholar]
- 17.Bowie, J. U. (1997) Nat. Struct. Biol. 4**,** 915-917. [DOI] [PubMed] [Google Scholar]
- 18.Javadpour, M. M., Eilers, M., Groesbeek, M. & Smith, S. O. (1999) Biophys. J. 77**,** 1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covacci, A., Telford, J. L., Del Giudice, G., Parsonnet, J. & Rappuoli, R. (1999) Science 284**,** 1328-1333. [DOI] [PubMed] [Google Scholar]
- 20.McClain, M. S., Iwamoto, H., Cao, P., Vinion-Dubiel, A. D., Li, Y., Szabo, G., Shao, Z. & Cover, T. L. (2003) J. Biol. Chem. 278**,** 12101-12108. [DOI] [PubMed] [Google Scholar]
- 21.Bonnafous, P. & Stegmann, T. (2000) J. Biol. Chem. 275**,** 6160-6166. [DOI] [PubMed] [Google Scholar]
- 22.Han, X., Wang, C. T., Bai, J., Chapman, E. R. & Jackson, M. B. (2004) Science 304**,** 289-292. [DOI] [PubMed] [Google Scholar]
- 23.Salzer, U., Hinterdorfer, P., Hunger, U., Borken, C. & Prohaska, R. (2002) Blood 99**,** 2569-2577. [DOI] [PubMed] [Google Scholar]
- 24.Cross, K. J., Wharton, S. A., Skehel, J. J., Wiley, D. C. & Steinhauer, D. A. (2001) EMBO J. 20**,** 4432-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro, L., Doyle, J. P., Hensley, P., Colman, D. R. & Hendrickson, W. A. (1996) Neuron 17**,** 435-449. [DOI] [PubMed] [Google Scholar]
- 26.Nelis, E., Haites, N. & Van Broeckhoven, C. (1999) Hum. Mutat. 13**,** 11-28. [DOI] [PubMed] [Google Scholar]
- 27.Raschperger, E., Engstrom, U., Pettersson, R. F. & Fuxe, J. (2004) J. Biol. Chem. 279**,** 796-804. [DOI] [PubMed] [Google Scholar]
- 28.Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T., Jr. (2002) Nature 418**,** 291-292. [DOI] [PubMed] [Google Scholar]
- 29.Stefani, M. & Dobson, C. M. (2003) J. Mol. Med. 81**,** 678-699. [DOI] [PubMed] [Google Scholar]
- 30.Kourie, J. I. & Culverson, A. (2000) J. Neurosci. Res. 62**,** 120-133. [DOI] [PubMed] [Google Scholar]
- 31.Arispe, N., Rojas, E. & Pollard, H. B. (1993) Proc. Natl. Acad. Sci. USA 90**,** 567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirakura, Y., Lin, M. C. & Kagan, B. L. (1999) J. Neurosci. Res. 57**,** 458-466. [DOI] [PubMed] [Google Scholar]
- 33.Lin, H., Bhatia, R. & Lal, R. (2001) FASEB J. 15**,** 2433-2444. [DOI] [PubMed] [Google Scholar]
- 34.Melnyk, R. A., Kim, S., Curran, A. R., Engelman, D. M., Bowie, J. U. & Deber, C. M. (2004) J. Biol. Chem. 279**,** 16591-16597. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, D. & Engelman, D. M. (2004) J. Mol. Biol. 343**,** 799-804. [DOI] [PubMed] [Google Scholar]
- 36.MacKenzie, K. R. & Engelman, D. M. (1998) Proc. Natl. Acad. Sci. USA 95**,** 3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senes, A., Ubarretxena-Belandia, I. & Engelman, D. M. (2001) Proc. Natl. Acad. Sci. USA 98**,** 9056-9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, S. O., Eilers, M., Song, D., Crocker, E., Ying, W., Groesbeek, M., Metz, G., Ziliox, M. & Aimoto, S. (2002) Biophys. J. 82**,** 2476-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faham, S., Yang, D., Bare, E., Yohannan, S., Whitelegge, J. P. & Bowie, J. U. (2004) J. Mol. Biol. 335**,** 297-305. [DOI] [PubMed] [Google Scholar]
- 40.Aguzzi, A. & Haass, C. (2003) Science 302**,** 814-818. [DOI] [PubMed] [Google Scholar]
- 41.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416**,** 507-511. [DOI] [PubMed] [Google Scholar]
- 42.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J. & Selkoe, D. J. (2002) Nature 416**,** 535-539. [DOI] [PubMed] [Google Scholar]
- 43.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300**,** 486-489. [DOI] [PubMed] [Google Scholar]
- 44.Kayed, R., Sokolov, Y., Edmonds, B., McIntire, T. M., Milton, S. C., Hall, J. E. & Glabe, C. G. (2004) J. Biol. Chem. 279**,** 46363-46366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information