Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-α responses (original) (raw)
Abstract
Lipopolysaccharide (LPS) stimulation of the innate immune response requires the activation of signaling cascades that culminate in the synthesis and secretion of proinflammatory cytokines. Given the inhibitory effects of phosphodiesterase (PDE) inhibitors on LPS-induced cytokine production, we have investigated LPS responses in mice deficient in PDE4 (type 4 cAMP-specific PDE)-B and PDE4D. LPS stimulation of mouse peripheral leukocytes induced PDE4B mRNA accumulation and increased PDE4 activity. This response was completely absent in mice deficient in PDE4B but not PDE4D. LPS induction of tumor necrosis factor-α secretion by circulating leukocytes was decreased by approximately 90% in mice deficient in PDE4B but not in mice lacking PDE4D. The impaired LPS response was evident regardless of the LPS dose used for stimulation and was associated with a more than 90% decrease in tumor necrosis factor-α mRNA accumulation. A decreased responsiveness to LPS was also present in other inflammatory cells, including peritoneal and lung macrophages. These findings demonstrate that PDE4B gene activation by LPS constitutes a feedback regulation essential for an efficient immune response.
Innate and acquired immunity are defense mechanisms by which a host protects itself from microbial infection. Microbial pathogens activate the innate immune response via recognition of conserved motifs termed pathogen-associated molecular patterns (PAMPs) by specific receptors expressed in the host cells (1). On PAMPs binding, these receptors activate signaling pathways involved in inflammatory responses and phagocytosis. The receptors activated by lipopolysaccharide (LPS), a PAMP component of the Gram-negative bacterial cell wall, belong to the family of Toll-like receptors (TLRs) similar to the Toll receptors described in Drosophila (for review, see refs. 2 and 3). Activation of TLRs by LPS is facilitated by additional recognition receptor/binding proteins, including the serum LPS-binding protein (LBP) and CD14. Although TLRs are sufficient for LPS binding and activation, a prior interaction of LPS with LBP and CD14 augments the inflammatory responses mediated by TLRs (4, 5). The LPS–TLR complex in turn activates several signaling pathways, including the nuclear factor-κB (NF-κB)- and the Jun/Fos-mediated transcriptional regulation, both of which control the expression of cytokine genes (2).
In addition to its protective effects, innate immunity may cause harmful responses, the best characterized being the LPS-induced shock that follows bacterial sepsis. LPS-induced production of proinflammatory cytokines, in particular tumor necrosis factor-α (TNF-α), is responsible for these detrimental effects (6). Numerous reports point to a key role of TNF-α in orchestrating the complex events involved in the development of the immune system and in a variety of inflammatory disease states (7–10).
Monocytes and macrophages are a major source of TNF-α. Production of this cytokine is subjected to both transcriptional and posttranscriptional controls (11, 12). LPS stimulation of macrophages induces a large increase in TNF-α mRNA by increasing gene transcription via activation of the transcriptional factors NF-κB (13–15), c-jun, and c-fos (16). In addition, an endotoxin-responsive element with an AU-rich sequence has been identified in the 3′-untranslated region of mRNAs of various cytokines and proto-oncogenes (17). This element is involved in both the control of mRNA degradation and its translation (18–20). A study investigating the control of TNF-α synthesis by using reporter constructs has demonstrated that this AU-rich element and the flanking sequences are sufficient to mediate a more than 200-fold increase in the LPS-dependent synthesis of the reporter protein. This increase is not due to a change in cytoplasmic mRNA concentration but rather to a reversal of the translation repression in response to LPS (21).
Several mediators attenuate TNF-α production in mononuclear cells, including cAMP-elevating agents such as PGE2 (22–24), and their effects are reproduced by dibutyryl cAMP (25, 26) or by inhibitors of cyclic nucleotide phosphodiesterases (PDE) (24, 25, 27–34). Among the PDE inhibitors tested, those specific for type 4 cAMP-specific PDE (PDE4) exhibit profound antiinflammatory effects both in vivo and in vitro (35). In human circulating monocytes, inhibition of PDE4 by rolipram markedly suppresses TNF-α synthesis and release in response to LPS; however, this inhibitory action is considerably less evident or absent when PDE1, PDE3, or PDE5 inhibitors are administered, indicating that monocytes express predominantly PDE4s (24, 27–34). The mechanism(s) by which the selective inhibition of PDE4 blocks TNF-α synthesis and release remains largely unknown. The PDE4 family consists of four distinct genes termed PDE4A, PDE4B, PDE4C, and PDE4D, and each gene encodes multiple variants generated by alternate splicing and/or from different transcriptional units (36, 37). Although numerous reports have indicated that these variants differ in their regulation, subcellular localization, and protein–protein interaction, the exact physiological role of the four genes remains to be determined.
In the present study, we investigated whether LPS-mediated responses are affected by inactivation of the PDE4B and PDE4D genes in the mouse. Our data demonstrated that the LPS treatment of leukocytes regulates PDE4B expression and the interruption of this regulatory loop causes a major decrease in the TNF-α response.
Materials and Methods
Construction of the PDE4B Targeting Vector and Production of PDE4B-Deficient Mice.
Genomic clones corresponding to the PDE4B gene were isolated from a mouse 129svj lambda FIXII genomic library (Stratagene). Following the strategy used for the inactivation of the PDE4D gene (38), a 1.65-kb _Hin_dIII fragment 5′ to exon 8 and a 6.5-kb _Sac_I fragment 3′ to exon 10 of the mouse PDE4B locus were isolated and inserted in a targeting plasmid. The positive selection marker (PGK-hprt) and the negative selection marker (pMC1-tk) were in the opposite orientation to the PDE4B genomic sequence. In the constructs thus created, the hprt gene substituted for a 3-kb genomic fragment that contains exons 8–10 of the PDE4B gene (38). The targeting vector was linearized with _Not_I and electroporated into E14TG2a embryonic stem (ES) cells (39). Homologous recombinant ES clones were screened by Southern blot analysis. ES cells from positive clones were microinjected into C57BL/6 blastocysts and implanted into pseudopregnant females. The resulting chimeras were mated to C57BL/6 mice to produce heterozygous mice (PDE4B+/−) that were subsequently mated to C57BL/6 mice again (one run of backcrossing). Further crosses between the PDE4B+/− mice yielded homozygous mutants (PDE4B−/−) with a mixed 129/Ola (25%) and C57BL/6 (75%) background. Genotyping of animals was performed by Southern blot analysis on DNA recovered from tail biopsies as described for inactivation of the PDE4D gene (38). The expected sizes for wild-type and mutant PDE4B are 2.1 kb and 4.75 kb, respectively (data not shown).
Cell Culture.
THP-1, a human leukemic monocytic cell line, was obtained from American Type Culture Collection. THP-1 cells were incubated in the absence or presence of 1 μg/ml of LPS and/or 100 μM of dibutyryl cAMP for 3 h. The cells were pelleted and stored at −80°C until use.
Whole Blood Cultures.
Blood was collected from mouse tails in a sterile heparin-containing tube and was immediately diluted with one volume of RPMI medium 1640 supplemented with 1% each of penicillin and streptomycin and 2.5% heat-inactivated FBS. The diluted blood was then aliquoted (150–200 μl) into 96-well cell culture plates. To induce TNF-α and IL-6 production, bacterial LPS (from Escherichia coli, serotype O55:B5, Sigma) was added at 100 ng/ml final concentration, and cultures were incubated at 37°C in 5% CO2 for the indicated times. In dose–response studies, the blood culture was stimulated with LPS at various final concentrations (0–1,000 ng/ml) for 8 h. After incubation, the culture was centrifuged at 100 × g for 10 min and the supernatant was collected and stored at −20°C until assayed.
Preparation of Mouse Blood Leukocytes.
Mouse blood was collected and diluted as described above. The diluted blood was incubated in the absence or presence of 100 ng/ml of LPS for 3 h. After centrifugation at 100 × g for 10 min, the cell pellet was suspended in 4 ml of PharM Lyse solution (PharMingen) to lyse red blood cells. The remaining blood leukocytes were recovered by centrifugation and washed once with PBS. The cell pellets were stored at −80°C until RNA extraction.
Preparation of Thioglycolate-Elicited Peritoneal Macrophages (TEPM).
Mice were injected i.p. with 1.5 ml of thioglycolate medium (Sigma) to recruit macrophages into the peritoneal cavity. Four days after injection, the mice were killed and the cells were collected by washing the peritoneum with cold HBSS. Cells were washed once with DMEM supplemented with 10% heat-inactivated FBS and then cultured at a density of 1 × 106 cells per ml. Two hours later, plates were washed with medium to remove nonadherent cells. The remaining cells with at least 95% macrophages were cultured overnight in DMEM supplement with 1% each of penicillin and streptomycin and 0.5% FBS before LPS stimulation.
Preparation of Bronchoalveolar Lavage (BAL) Monocytes/Macrophages.
Mice were anaesthetized by i.p. injection with xylazine (4.4 mg/kg) and ketamine (65 mg/kg). The trachea was cannulated, and the lung was then lavaged with 0.7 ml of PBS three times and the fluid pooled. Cells in the lavage fluid were counted using a hemocytometer, and BAL cell differentials were determined on Cytospin slides stained with May-Grunwald and Giemsa stain (Sigma). At least 95% of BAL cells were monocytes and macrophages. Cells were washed once with RPMI medium 1640 supplemented with 1% each of penicillin and streptomycin and 2.5% heat-inactivated FBS, dispensed into 96-well plates at 1 × 104 cells per well, and then incubated in the absence or presence of 100 ng/ml of LPS.
Northern Blot Hybridization.
Total RNA was prepared from the THP-1 cells by the guanidine thiocyanate/cesium chloride method (40). For Northern analysis, total RNA was denatured in glyoxal and dimethyl sulfoxide, size-fractionated by electrophoresis on 1% agarose gel, and transferred to nylon membrane (ICN). The blot was hybridized at 37°C for 48 h with a [α-32P]dATP-labeled oligonucleotide probe corresponding to human PDE4B2-specific sequence from base 772–736, 5′-CCTTCATTATTTGCACGCTGGCTCCTTCCTTCCAGCT-3′ (GenBank accession no. L20971). After hybridization, the blot was washed with 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) at 52°C for 20 min. To determine whether similar amounts of RNA were loaded in each lane, the blot was stripped of the hybridized probe and rehybridized with a human β-actin cDNA probe.
Reverse Transcription (RT)-PCR and Southern Blot Analysis.
For semiquantitative measurements of LPS-induced expression of PDE4B and TNF-α, total RNA was extracted from the cell pellets of mouse blood leukocytes. After treatment with RNase-free DNase I (0.1 units per μg total RNA, Boehringer Mannheim), first-strand cDNA was synthesized from 1 μg of total RNA in the presence of oligo dT primer or random primer according to the manufacturer's protocol (cDNA Cycle kit, Invitrogen). For PCR, 2 μl of aliquots of the resulting 20 μl of cDNA were amplified in a 50-μl reaction volume containing 1× PCR buffer, 0.2 mM dNTP mix, 1 μM of each specific primer, and 1 unit of _Taq_DNA polymerase. Oligonucleotide primers were as follows: PDE4B, 5′-TGGAAATCCTGGCTGCCAT-3′ and 5′-TCCACAGAAGCTGTGTGCT-3′ (from base 1473–1491 and base 2007–1989, respectively; accession no. AF208023), defining a 535-bp product encompassing exons 7–10 of the PDE4B gene; TNF-α, 5′-GTGACAAGCCTGTAGCCCA-3′ and 5′-AAAGTAGACCTGCCCGGAC-3′ (from base 419–437 and base 846–828, respectively; accession no. X02611), yielding a 428-bp product; glyceraldehyde-3-phosphate dehydrogenase (GAPDH, CLONTECH), 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′ and 5′-CATGTAGGCCATGAGGTCCACCAC-3′, yielding a 983-bp product. Conditions for the amplification were as follows: PDE4B, annealing temperature of 57°C for 27 cycles; TNF-α and GAPDH, annealing temperature of 60°C for 20 cycles.
The PCR products were fractionated by agarose gel electrophoresis and then transferred to nylon membrane (ICN). Blots were hybridized with [γ-32P]ATP-labeled oligonucleotide probes corresponding to nucleotide sequences nested between the specific PCR primers. The sequences of the oligonucleotide probes were as follows: PDE4B, 5′-GTTAGCAACTGATATGTC-3′ (from base 1714–1731; accession no. AF208023), TNF-α, 5′-CATACCAGGGTTTGAGCT-3′ (from base 736–719; accession no. X02611), and GAPDH, 5′-CAGTGGCAAAGTGGAGATT-3′ (from base 112–130; accession no. M32599). The blots were then washed in 1.5× SSC/0.1% SDS at 48°C, followed by autoradiography.
TNF-α and IL-6 ELISA.
Levels of TNF-α and IL-6 in the whole blood or macrophage cell culture supernatants were measured with commercially available ELISA kits (BioSource International, Camarillo, CA). The sensitivities of the assays were 19.5 pg/ml and 7.8 pg/ml, respectively.
PDE Assay.
Following incubation with 100 ng/ml of LPS for 4 h, TEPM were harvested for PDE activity measurement. Cells were scraped in PBS (1 ml per dish) with a rubber policeman, centrifuged, washed with PBS, and then incubated on ice for 15 min in a buffer containing 0.5 mg/ml digitonin, 250 mM sucrose, 10 mM Tris⋅Cl (pH 7.4), 50 mM sodium fluoride, 1 mM EDTA, 10 mM benzamidine, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 2 μg/ml aprotinin, 100 μg/ml soybean trypsin inhibitor, and 0.5 mM phenylmethylsulfonyl fluoride. After sonication with 25 bursts, aliquots of the cell extracts were assayed for total PDE activity, and rolipram-insensitive PDE activity in the presence of 10 μM rolipram. The PDE assay was performed according to the method of Thompson and Appleman (41) as detailed (38). The rolipram-sensitive activity (i.e., PDE4 activity) was obtained by subtracting the rolipram-insensitive activity from the total activity. Protein concentration was determined using the Bradford method (42).
Results
Targeted Disruption of the PDE4B Gene and General Characteristics of PDE4B−/− Mice.
The PDE4B gene was disrupted following a strategy similar to that described for the inactivation of the PDE4D gene (38). Because multiple promoters control transcription of the exons that encode divergent amino termini of the PDE4B proteins, exons 8–10 coding for the common catalytic domain at the carboxyl terminus were targeted by insertion of a PGK-hprt cassette. This strategy does not disrupt the start of transcription or translation, leaving open the possibility of generating truncated mRNAs and proteins (see below). Although a PDE4B mRNA was readily detected in the wild-type mice, RT-PCR followed by Southern blot analysis showed that no mRNA coding for the catalytic domain is present in the PDE4B−/− mice (see below). However, truncated mRNAs containing 5′ exons were detected in these mice (data not shown). The loss of the full-length PDE4B mRNA was reflected in the complete absence of PDE4B protein expression in the cerebellum. Western blot analysis demonstrated that an immunoreactive PDE4B protein could not be detected using antibodies that recognize the carboxyl terminus of the protein (data not shown).
The PDE4B−/− mice appeared normal and exhibited no overt morphological abnormalities. Litter size, body weight, and growth rate of these mice were similar to those of wild-type littermates. Total and differential white cell counts in peripheral blood revealed no significant difference between the wild-type and PDE4B−/− mice.
Induction of PDE4B by LPS in Mouse Circulating Leukocytes.
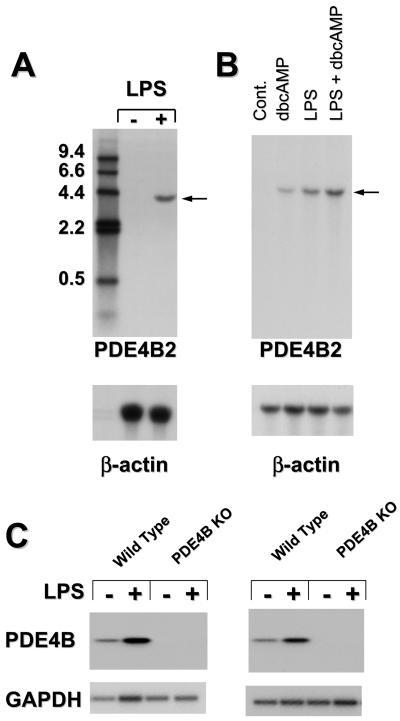
Given the findings that PDE4 inhibitors have profound effects on monocyte activation and that PDE4B is the predominant subtype expressed in human monocytes (35, 43), we investigated whether the PDE pattern of expression and the function of monocytes were affected in the PDE4B-deficient mice. A human monocytic cell line, THP-1, was used as a control. Northern blot analysis of RNA from THP-1 cells revealed that PDE4B mRNA is undetectable under basal conditions, whereas treatment with LPS produced a major increase in PDE4B mRNA with a molecular size of ≈4 kb (Fig. 1A). As previously reported by us and others, cAMP also induced PDE4B mRNA (43–46), and this induction was synergistic with LPS (Fig. 1B). To confirm that a similar regulation of PDE4B mRNA is operating in mouse circulating mononuclear cells, leukocytes from wild-type and PDE4B−/− mice were treated with 100 ng/ml of LPS for 3 h, and the RNA extracted and analyzed by RT-PCR followed by Southern blot. When PCR primers specific to the catalytic domain of mouse PDE4B were used in the amplification, mouse circulating leukocytes exhibited a basal expression of PDE4B mRNA (Fig. 1C). As demonstrated for human THP-1 cells, the level of PDE4B mRNA was markedly increased after LPS treatment, indicating an increase in mRNA transcription and/or stabilization. As expected, PDE4B mRNA was not detected in the cells of PDE4B−/− mice under basal or LPS-stimulated conditions even after 35 cycles of amplification (Fig. 1C). The induction of PDE4B in blood leukocytes probably reflects the expression of PDE4B2, a short splicing variant of the PDE4B gene, because the induced RNA could be detected when a PDE4B2-specific oligonucleotide probe was used (Fig. 1 A and B). This finding is consistent with the recent observation that LPS specifically increases PDE4B transcription in human monocytes (43).
Figure 1.
LPS effects on PDE4B mRNA steady state. (A and B) THP-1 cells were incubated in the absence or presence of 1 μg/ml LPS and/or 100 μM of dibutyryl cAMP (dbcAMP) for 3 h. Extraction of total RNA and Northern blot analysis were performed as described in Materials and Methods. Twenty-five micrograms of total RNA were loaded onto each lane. Numbers on the left are sizes of DNA molecular weight markers in kb. (C) Circulating leukocytes from wild-type and PDE4B−/− mice were cultured in the absence or presence of 100 ng/ml of LPS for 3 h. Extraction of total RNA from leukocytes (pooled from four animals per group), RT-PCR using primers corresponding to mouse PDE4B cDNA sequence, and Southern blot analysis were performed as described in Materials and Methods. Results derived from cDNAs prepared with random primers (Left) and oligo dT primers (Right) are reported. Amplification of a GAPDH fragment was included to monitor the amount of RNA in each sample.
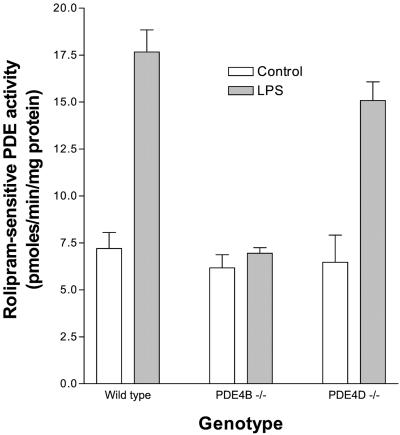
Because of the technical limitation in isolating sufficient monocytes from mouse blood, TEPM from the wild-type and PDE4B−/− mice were used to study the induction of PDE4B protein. In wild-type TEPM, LPS produced a 2.5-fold increase in rolipram-sensitive PDE4 activity within 4 h of incubation (Fig. 2). A similar increase was present in macrophages from PDE4D−/− mice; it was, however, completely absent in macrophages from the PDE4B−/− mice (Fig. 2). The basal PDE4 activity was comparable in TEPM from wild-type, PDE4D−/−, and PDE4B−/− mice, indicating minimal or no expression of PDE4B or PDE4D in the untreated TEPM. In addition, the activity of rolipram-insensitive PDEs was not affected by the LPS treatment (pmol/min/mg protein; wild type, 22.8 ± 1.97 vs. 23.8 ± 1.39; PDE4B−/−, 28.1 ± 3.45 vs. 26.3 ± 3.21, n = 4). Similar results were obtained with macrophages not exposed to thioglycolate (data not shown). Thus, these data demonstrated that inactivation of a single PDE4B gene completely prevented the LPS-induced PDE activation, confirming that PDE4B is the predominant subtype induced by LPS in mouse monocytes and macrophages.
Figure 2.
Absence of LPS-induced PDE4 activity in thioglycolate-elicited peritoneal macrophages (TEPM) from PDE4B−/− mice. TEPM from wild-type, PDE4B−/−, and PDE4D−/− mice were incubated in the absence or presence of 100 ng/ml LPS for 4 h. Cells were harvested, permeabilized with digitonin, and sonicated as described in Materials and Methods. Total and rolipram-insensitive PDE activities in the cell extracts were assayed in the absence or presence of 10 μM rolipram. Values reported are the rolipram-sensitive PDE activity (i.e., PDE4 activity). Data are the mean ± SEM (3–8 mice per genotype). Significant difference in the LPS-stimulated PDE4 activity between wild-type and PDE4B−/− mice was determined by t test (P < 0.0001).
LPS-Stimulated TNF-α Production in PDE4B−/− Mice.
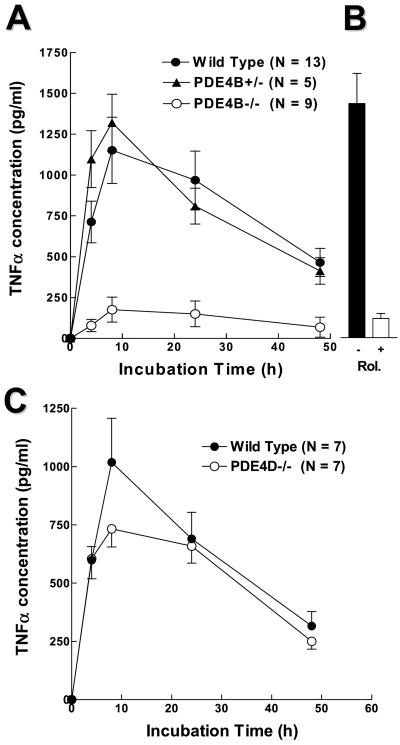
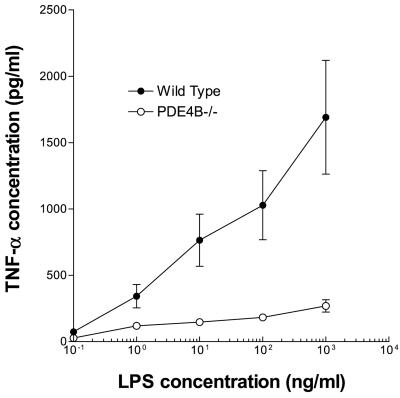
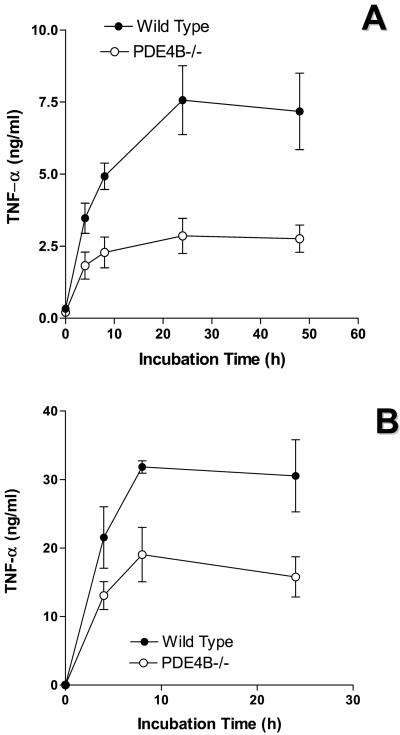
To determine the physiological significance of this PDE4B regulation, we investigated whether cytokine production induced by LPS is affected in PDE4B−/− mice. The levels of TNF-α in leukocytes cultured ex vivo were measured at different times after LPS stimulation. As shown in Fig. 3A, cells from both wild-type and PDE4B−/− mice produced TNF-α at the limit of detection under basal conditions. In the presence of 100 ng/ml of LPS, TNF-α release was induced at least 50-fold in leukocytes from the wild-type mice. Stimulation reached a maximum in 8 h and declined thereafter. TNF-α production in response to LPS was decreased by approximately 90% in circulating leukocytes derived from the PDE4B−/− mice. Conversely, the TNF-α response of PDE4B+/− heterozygous cells was identical to the wild type, thus lessening the possibility that disruption of the PDE4B locus generates truncated proteins with dominant negative effects. Circulating leukocytes from the PDE4D−/− mice responded to LPS in a manner similar to the wild-type cells (Fig. 3C). Identical results were obtained whether whole blood or leukocytes were used in the incubation (data not shown). The decrease in TNF-α production in the PDE4B null mice was in the same range as that obtained by treating the wild-type leukocytes with rolipram (Fig. 3B). Conversely, ablation of PDE4B or an acute PDE4 inhibition with rolipram had no effect on the LPS-induced IL-6 production (pg/ml IL-6 at 4 h incubation with 100 ng/ml LPS: PDE4B+/+ 235.2 ± 43.9, n = 14; PDE4B+/+ plus 50 μM Rolipram 244.7 ± 67.6, n = 5; PDE4B−/− 287.7 ± 58.7, n = 12). This dichotomy in the effect on TNF-α and IL-6 production suggests that the different signaling pathways emanating from the TLRs are not equally sensitive to the PDE4B feedback regulation. The large decrease in LPS response was not overcome by increasing the concentration of LPS (Fig. 4). When enriched macrophage preparations from bronchoalveolar lavage (Fig. 5A) or TEPM (Fig. 5B) were used, a significant decrease in TNF-α production in response to LPS was also present; however, this decrease was not as dramatic as that observed with peripheral leukocytes. TNF-α production by PDE4B−/− macrophages was impaired also when a different stimulus—i.e., phorbol myristate acetate—was used (data not shown).
Figure 3.
LPS-induced TNF-α production in circulating leukocytes ex vivo. Circulating leukocytes from wild-type, PDE4B+/−, and PDE4B−/− mice (A) and from wild-type and PDE4D−/− mice (C) were incubated in the absence or presence of 100 ng/ml LPS for indicated times. In B, wild-type leukocytes were pretreated with 50 μM Rolipram for 30 min before the LPS stimulation for 8 h. Data are the mean ± SEM. The number of mice tested individually in each group is reported on the right of the graph.
Figure 4.
Concentration dependence of the LPS-induced TNF-α accumulation. Circulating leukocytes from wild-type and PDE4B−/− mice were treated with increasing concentrations of LPS for 8 h. Data are the mean ± SEM (n = 4 mice per group).
Figure 5.
LPS-induced TNF-α production in bronchoalveolar monocyte/macrophages (A) and thioglycolate-elicited peritoneal macrophages (B). Cells from the bronchoalveolar lavage fluid of unprimed mice or TEPM from wild-type and PDE4B−/− mice were stimulated in vitro with 100 ng/ml LPS for indicated times. Data are the mean ± SEM (n = 3–4 mice per group).
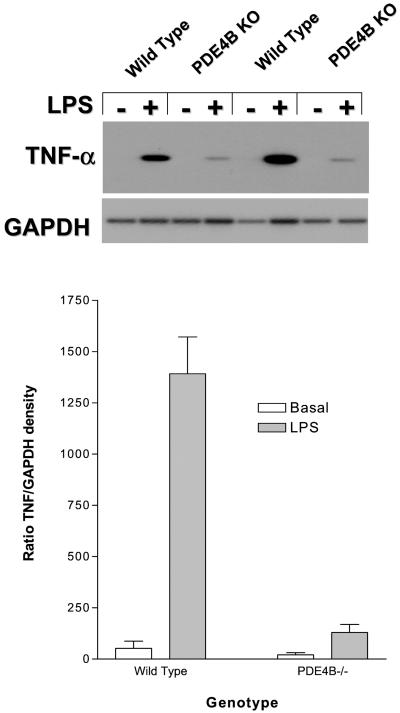
To determine whether the decrease in TNF-α production is due to altered transcription or mRNA stabilization, the levels of TNF-α mRNA derived from circulating leukocytes were measured by a semiquantitative PCR. No amplified fragment for TNF-α was detected in cells from the wild-type or PDE4B−/− mice unless cells were treated with LPS (Fig. 6). The induction was about 10 times higher in blood leukocytes from the wild-type mice than in cells from PDE4B−/− mice (Fig. 6).
Figure 6.
Effects of LPS on TNF-α mRNA accumulation in circulating leukocytes from the wild-type and PDE4B−/− mice. Treatment of circulating leukocytes with LPS, RNA extraction, RT-PCR, and Southern blot analysis were performed as described in Fig. 1C and Materials and Methods. The left four lanes present results derived from cDNAs prepared from oligo dT primers and the right four lanes from random primers. (Lower) The ratios of integrated densities of the TNF-α and GAPDH.
Discussion
Using mice deficient in PDE4 genes, we report here that LPS-elicited innate immune response is associated with the induction of PDE4B, but not PDE4D, in monocytes and macrophages. Exposure to LPS causes an increase in PDE4B2 mRNA and protein synthesis, which leads to an increase in the PDE activity in monocytes and macrophages. We propose that induction of this PDE constitutes a positive feedback loop essential for cytokine production. Ablation of PDE4B, but not PDE4D, inactivates this regulatory loop and prevents the LPS-stimulated TNF-α mRNA and protein accumulation.
Our data show that LPS induces an increase in the PDE4B mRNA levels in both human monocytic THP-1 cells and circulating leukocytes of the wild-type mice. This finding is consistent with observations in human circulating monocytes that PDE4B gene expression is selectively induced by LPS, and that PDE4B2 is the PDE4B variant being up-regulated (43, 47). Although no basal expression of PDE4B was observed in THP-1 cells, PDE4B mRNA was clearly detected in the absence of LPS in circulating leukocytes. This finding may be due to the heterogeneity of the leukocyte population used. It is possible that the basal level of the RNA detected in the wild-type mouse leukocytes is due to constitutive expression of PDE4B in neutrophils, whereas the LPS-stimulated PDE4B mRNA induction occurs primarily in monocytes. Wang et al. have demonstrated that in human circulating neutrophils the PDE4B gene is expressed constitutively and this expression is not affected by LPS (47).
The induction of the PDE4B mRNA is followed by an increase in PDE4 activity. Even though a direct measurement of the enzyme in circulating monocytes was precluded by the low number of cells that can be isolated in the mouse, the analysis of the PDE activity in macrophages from the wild-type, PDE4B−/−, and PDE4D−/− mice is consistent with this conclusion. The basal PDE4 activity was unaffected after disruption of the PDE4B and PDE4D genes, suggesting that PDE4A is the predominant subtype expressed in the unstimulated macrophages. On LPS stimulation, the induction of PDE4 activity was absent in PDE4B−/− cells, but not significantly affected in PDE4D−/− cells, indicating a selective activation of PDE4B by LPS. Thus, these data from mice deficient in different PDE4s are consistent with the conclusion that PDE4B is the major PDE4 form inducible by LPS.
It is well documented that PDE4-selective inhibitors produce profound inhibitory effects on LPS-stimulated TNF-α production in circulating monocytes (24, 27–34). However, with this widely used pharmacological approach it is not possible to assess the role of PDE4B induction on LPS-mediated responses, nor to distinguish the contribution of each PDE4 to these processes. Available PDE4 inhibitors are nonselective and therefore block the activity of all PDE4s expressed in these cells. In addition, they inhibit both the basal and LPS-induced PDE4 activity, further confounding the interpretation of the results. By using mice genetically altered in two PDE4 genes and TNF-α accumulation as a readout, we have been able to determine the exact role of PDE4B induction on the LPS-induced responses. Whereas the TNF-α response to LPS stimulation was normal in PDE4D-deficient circulating leukocytes, this response was decreased 80–90% in leukocytes from the PDE4B−/− mice. This large reduction in TNF-α induction was confirmed by the finding of a more than 90% reduction in TNF-α mRNA accumulation in these cells. The major decrease in responsiveness is not due to decreased sensitivity of the PDE4B−/− cells to LPS because a reduction was present at all LPS concentrations tested. In addition, the IL-6 response was unaffected, thus ruling out a generalized disruption of the mononuclear cell function. It should be pointed out that other PDE4s present in circulating leukocytes are apparently not up-regulated in the PDE4B−/− mice, as determined by measuring total and rolipram-sensitive PDE activity. Therefore, we can conclude that a major decrease in the LPS-induced TNF-α response is due solely to the absence of PDE4B induction.
The following model of LPS mechanism of action reconciles the findings described above. As shown in several in vitro studies, cAMP, via activation of PKAs or cAMP-GEF (48), exerts a tonic negative constraint on LPS-induced signaling by functioning as a gating pathway (49). To overcome this tonic inhibition, LPS signaling requires the activation of PDE4B transcription and accumulation of PDE4B protein. The consequent increase in PDE activity leads to a decrease in cAMP, thus removing the cAMP constraint and allowing a full induction of TNF-α mRNA and protein synthesis. The induction of PDE4B should then be viewed as a positive feedback loop that contributes to a full activation of the cytokine responses. When this induction is ablated, as in the PDE4B-deficient cells, the cAMP-negative constraint is not removed and the LPS-induced responses are severely blunted. Such a regulatory loop involving PDE4s, as well as other PDEs, is probably functional in other inflammatory cells, including T cells. For instance, activation of T cells induces the expression of PDE7 and PDE8 (50, 51), which function by removing a cAMP constraint on T cell-receptor-mediated signals. Indeed, treatment with antisense oligonucleotides against PDE7 blocks T cell receptor activation of cell replication and IL-2 production (50).
It has been generally accepted that TNF-α production plays a critical role in a number of disease states, such as rheumatoid arthritis, Crohn's disease, and septic shock (52–54). Treatment of rheumatoid arthritis, Crohn's disease, and Jarisch-Herxheimer reaction with TNF-α antibodies has clearly demonstrated the central role of TNF-α in these diseases (7–10). Although there is uncertainty on exactly how the innate immune response and the TLR-dependent signaling pathway are involved in the pathogenesis of these TNF-α-mediated diseases, the requirement of PDE4B in LPS-induced TNF-α production in monocytes may be exploited for pharmacological intervention. The present study strongly indicates that inhibitors selective for PDE4B may be promising pharmacological agents for the treatment of diseases in which the release of TNF-α from monocytes is important for their pathogenesis.
Acknowledgments
We are indebted to Drs. Dale Umetsu and Peng Wang for helpful discussions. This study was supported by an award from the Sandler Program for Asthma Research (to M.C.), National Institutes of Health Grant HD20788, and a grant from Pfizer Inc.
Abbreviations
PDE
cyclic nucleotide phosphodiesterase
PDE4
type 4 cAMP-specific PDE
TNF-α
tumor necrosis factor-α
LPS
lipopolysaccharide
GAPDH
glyceraldehyde-3phosphate dehydrogenase
TEPM
thioglycolate-elicited peritoneal macrophages
RT
reverse transcription
TLR
Toll-like receptors
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Janeway C A, Jr, Medzhitov R. Semin Immunol. 1998;10:349–350. doi: 10.1006/smim.1998.0142. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A, Ulevitch R J. Nature (London) 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K V. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 4.Antal-Szalmas P. Eur J Clin Invest. 2000;30:167–179. doi: 10.1046/j.1365-2362.2000.00610.x. [DOI] [PubMed] [Google Scholar]
- 5.Hailman E, Lichenstein H S, Wurfel M M, Miller D S, Johnson D A, Kelley M, Busse L A, Zukowski M M, Wright S D. J Exp Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutler B A. J Rheumatol. 1999;26, Suppl. 57:16–21. [PubMed] [Google Scholar]
- 7.Elliott M J, Maini R N, Feldmann M, Kalden J R, Antoni C, Smolen J S, Leeb B, Breedveld F C, Macfarlane J D, Bijl H, et al. Lancet. 1994;344:1105–1110. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 8.Stack W A, Mann S D, Roy A J, Heath P, Sopwith M, Freeman J, Holmes G, Long R, Forbes A, Kamm M A. Lancet. 1997;349:521–524. doi: 10.1016/s0140-6736(97)80083-9. [DOI] [PubMed] [Google Scholar]
- 9.Fekade D, Knox K, Hussein K, Melka A, Lalloo D G, Coxon R E, Warrell D A. N Engl J Med. 1996;335:311–315. doi: 10.1056/NEJM199608013350503. [DOI] [PubMed] [Google Scholar]
- 10.Moreland L W, Baumgartner S W, Schiff M H, Tindall E A, Fleischmann R M, Weaver A L, Ettlinger R E, Cohen S, Koopman W J, Mohler K, et al. N Engl J Med. 1997;337:141–147. doi: 10.1056/NEJM199707173370301. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Thompson P, Beutler B. J Exp Med. 1990;172:391–394. doi: 10.1084/jem.172.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raabe T, Bukrinsky M, Currie R A. J Biol Chem. 1998;273:974–980. doi: 10.1074/jbc.273.2.974. [DOI] [PubMed] [Google Scholar]
- 13.Shakhov A N, Collart M A, Vassalli P, Nedospasov S A, Jongeneel C V. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collart M A, Baeuerle P, Vassalli P. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drouet C, Shakhov A N, Jongeneel C V. J Immunol. 1991;147:1694–1700. [PubMed] [Google Scholar]
- 16.Guha M, Mackman N. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 17.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 19.Wilson T, Treisman R. Nature (London) 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 20.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Brown T, Beutler B. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel S L, Spengler M, May M A, Spengler R, Larrick J, Remick D. J Biol Chem. 1988;263:5380–5384. [PubMed] [Google Scholar]
- 23.Schade F U, Schudt C. Eur J Pharmacol. 1993;230:9–14. doi: 10.1016/0014-2999(93)90403-5. [DOI] [PubMed] [Google Scholar]
- 24.Sinha B, Semmler J, Eisenhut T, Eigler A, Endres S. Eur J Immunol. 1995;25:147–153. doi: 10.1002/eji.1830250125. [DOI] [PubMed] [Google Scholar]
- 25.Strieter R M, Remick D G, Ward P A, Spengler R N, Lynch J P, III, Larrick J, Kunkel S L. Biochem Biophys Res Commun. 1988;155:1230–1236. doi: 10.1016/s0006-291x(88)81271-3. [DOI] [PubMed] [Google Scholar]
- 26.Spengler R N, Spengler M L, Lincoln P, Remick D G, Strieter R M, Kunkel S L. Infect Immun. 1989;57:2837–2841. doi: 10.1128/iai.57.9.2837-2841.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semmler J, Wachtel H, Endres S. Int J Immunopharmacol. 1993;15:409–413. doi: 10.1016/0192-0561(93)90052-z. [DOI] [PubMed] [Google Scholar]
- 28.Molnar-Kimber K, Yonno L, Heaslip R, Weichman B. Agents Actions. 1993;39:C77–C79. doi: 10.1007/BF01972726. [DOI] [PubMed] [Google Scholar]
- 29.Prabhakar U, Lipshutz D, Bartus J O, Slivjak M J, Smith E F, III, Lee J C, Esser K M. Int J Immunopharmacol. 1994;16:805–816. doi: 10.1016/0192-0561(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 30.Verghese M W, McConnell R T, Strickland A B, Gooding R C, Stimpson S A, Yarnall D P, Taylor J D, Furdon P J. J Pharmacol Exp Ther. 1995;272:1313–1320. [PubMed] [Google Scholar]
- 31.Souness J E, Griffin M, Maslen C, Ebsworth K, Scott L C, Pollock K, Palfreyman M N, Karlsson J A. Br J Pharmacol. 1996;118:649–658. doi: 10.1111/j.1476-5381.1996.tb15450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimura T, Kurita C, Nagao T, Usami E, Nakao T, Watanabe S, Kobayashi J, Yamazaki F, Tanaka H, Nagai H. Gen Pharmacol. 1997;29:633–638. doi: 10.1016/s0306-3623(96)00580-0. [DOI] [PubMed] [Google Scholar]
- 33.Eigler A, Siegmund B, Emmerich U, Baumann K H, Hartmann G, Endres S. J Leukocyte Biol. 1998;63:101–107. doi: 10.1002/jlb.63.1.101. [DOI] [PubMed] [Google Scholar]
- 34.Barnette M S, Christensen S B, Essayan D M, Grous M, Prabhakar U, Rush J A, Kagey-Sobotka A, Torphy T J. J Pharmacol Exp Ther. 1998;284:420–426. [PubMed] [Google Scholar]
- 35.Torphy T J. Am J Respir Crit Care Med. 1998;157:351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]
- 36.Houslay M D, Sullivan M, Bolger G B. Adv Pharmacol. 1998;44:225–342. doi: 10.1016/s1054-3589(08)60128-3. [DOI] [PubMed] [Google Scholar]
- 37.Conti M, Jin S L. Prog Nucleic Acid Res Mol Biol. 1999;63:1–38. doi: 10.1016/s0079-6603(08)60718-7. [DOI] [PubMed] [Google Scholar]
- 38.Jin S L, Richard F J, Kuo W P, D'Ercole A J, Conti M. Proc Natl Acad Sci USA. 1999;96:11998–12003. doi: 10.1073/pnas.96.21.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 40.Ullrich A, Shine J, Chirgwin J, Pictet R, Tischer E, Rutter W J, Goodman H M. Science. 1977;196:1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- 41.Thompson W J, Appleman M M. Biochemistry. 1971;10:311–316. [PubMed] [Google Scholar]
- 42.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 43.Ma D, Wu P, Egan R W, Billah M M, Wang P. Mol Pharmacol. 1999;55:50–57. doi: 10.1124/mol.55.1.50. [DOI] [PubMed] [Google Scholar]
- 44.Swinnen J V, Tsikalas K E, Conti M. J Biol Chem. 1991;266:18370–18377. [PubMed] [Google Scholar]
- 45.Torphy T J, Zhou H L, Foley J J, Sarau H M, Manning C D, Barnette M S. J Biol Chem. 1995;270:23598–23604. doi: 10.1074/jbc.270.40.23598. [DOI] [PubMed] [Google Scholar]
- 46.Verghese M W, McConnell R T, Lenhard J M, Hamacher L, Jin S L. Mol Pharmacol. 1995;47:1164–1171. [PubMed] [Google Scholar]
- 47.Wang P, Wu P, Ohleth K M, Egan R W, Billah M M. Mol Pharmacol. 1999;56:170–174. doi: 10.1124/mol.56.1.170. [DOI] [PubMed] [Google Scholar]
- 48.de Rooij J, Rehmann H, van Triest M, Cool R H, Wittinghofer A, Bos J L. J Biol Chem. 2000;275:20829–20836. doi: 10.1074/jbc.M001113200. [DOI] [PubMed] [Google Scholar]
- 49.Iyengar R. Science. 1996;271:461–463. doi: 10.1126/science.271.5248.461. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Yee C, Beavo J A. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 51.Glavas N A, Ostenson C, Schaefer J B, Vasta V, Beavo J A. Proc Natl Acad Sci USA. 2001;98:6319–6324. doi: 10.1073/pnas.101131098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maini R N, Taylor P C. Annu Rev Med. 2000;51:207–229. doi: 10.1146/annurev.med.51.1.207. [DOI] [PubMed] [Google Scholar]
- 53.Bell S, Kamm M A. Lancet. 2000;355:858–860. doi: 10.1016/S0140-6736(99)00442-0. [DOI] [PubMed] [Google Scholar]
- 54.Beutler B, Kruys V. J Cardiovasc Pharmacol. 1995;25:S1–S8. doi: 10.1097/00005344-199500252-00002. [DOI] [PubMed] [Google Scholar]