Isolation of mRNA from specific tissues of Drosophila by mRNA tagging (original) (raw)
Abstract
To study the function of specific cells or tissues using genomic tools like microarray analyses, it is highly desirable to obtain mRNA from a homogeneous source. However, this is particularly challenging for small organisms, like Caenorhabditis elegans and Drosophila melanogaster. We have optimized and applied a new technique, mRNA tagging, to isolate mRNA from specific tissues of D.melanogaster. A FLAG-tagged poly(A)-binding protein (PABP) is expressed in a specific tissue and mRNA from that tissue is thus tagged by the recombinant PABP and separated from mRNA in other tissues by co-immunoprecipitation with a FLAG-tag specific antibody. The fractionated mRNA is then amplified and used as probe in microarray experiments. As a test system, we employed the procedures to identify genes expressed in Drosophila photoreceptor cells. We found that most known photoreceptor cell-specific mRNAs were identified by mRNA tagging. Furthermore, at least 11 novel genes have been identified as enriched in photoreceptor cells. mRNA tagging is a powerful general method for profiling gene expression in specific tissues and for identifying tissue-specific genes.
INTRODUCTION
Eukaryotic organisms have thousands of genes whose patterns of expression underlie biological function at levels from molecule to organismal behavior. To understand biological complexity, it is necessary to elucidate how these genes are expressed and how individual expression patterns influence one another. With the advent of genomic techniques like microarray analyses (1) and serial analysis of gene expression (2), it is now feasible to monitor the expression pattern of most, if not all, of the genes of an organism simultaneously. However, gene expression in multicellular eukaryotes is regulated in the dimensions of both time and space, so for these genomic techniques to yield maximum information, it is most useful to begin with a homogenous population of cells synchronized to a specific developmental time. The use of heterogeneous tissue or asynchronous cells as starting material contaminates the gene expression profiles for the cells of interest and reduces the power to detect changes in the target cells. Moderate or even dramatic changes of gene expression in one cell type may remain undetected using mRNA from complex organs or body parts.
Several methods have been used for isolating mRNAs from specific types of tissues or cells. Some involve physical separation of cells or tissues prior to RNA isolation; some involve methodology for separating mRNAs after homogenization of complex tissue by using RNA-binding proteins. Physical separation methods include the removal of specific tissue types by physical dissection; the difficulties in dissecting tissues make this problematic in many small model organisms. Physical separation methods also include laser capture microdissection (3), or separation of cell types based on an intrinsic property such as the fluorescence conferred by transfected green fluorescent protein (4). Recently, a new functional genomics approach, termed as ribonomics, was developed to fractionate subpopulations of mRNA contained in cellular messenger ribonucleoprotein complexes from tissue culture cells (5–7). This method takes advantage of the in vivo interaction of RNA-binding protein and mRNA. It has further evolved into a procedure referred to as mRNA tagging (8), to isolate mRNA from specific tissues of small organisms. RNA-binding proteins, such as poly(A)-binding protein (PABP) (9,10), can be epitope- tagged and expressed within the cells or tissues of interest using specific promoters. The mRNA from these tissues can then be separated from the mRNA of other tissues or cells by using an epitope-specific antibody to co-immunoprecipitate the desired mRNAs. The mRNA tagging method was successfully used to identify muscle-specific and ciliated sensory neuron-expressed genes in Caenohabditis elegans (8,11).
Here, we report the application and optimization of this technique for tissue-specific gene profiling of Drosophila melanogaster. The mRNA tagging method offers enormous analytical power to complement the superb genetics of this organism for studying a wide variety of biological problems, and obviates the difficulties for microarray studies posed by the small size of the organism. We have directed the expression of either D.melanogaster PABP (dPABP) or human PABP (hPABP) to all neurons, mushroom body neurons, or photoreceptor cells using the GAL4/UAS system (12). We demonstrate that the recombinant PABP can bind cellular mRNAs in vivo and these mRNAs can be retrieved and employed as probes for microarray studies. We applied this method to isolate mRNA from photoreceptor cells R1–R6 and followed this with microarray analyses, thus obtaining the gene expression profile of these cells. Consistent with our expectations, the mRNA level of most known photoreceptor cell-specific genes in the photoreceptor cell-specific mRNA population was >2-fold higher than in the mRNA population from whole heads. Furthermore, we identified at least 11 novel photoreceptor cell-enriched genes that may function in fly phototransduction or retinal degeneration.
MATERIALS AND METHODS
Generation of transgenic flies that express recombinant PABP
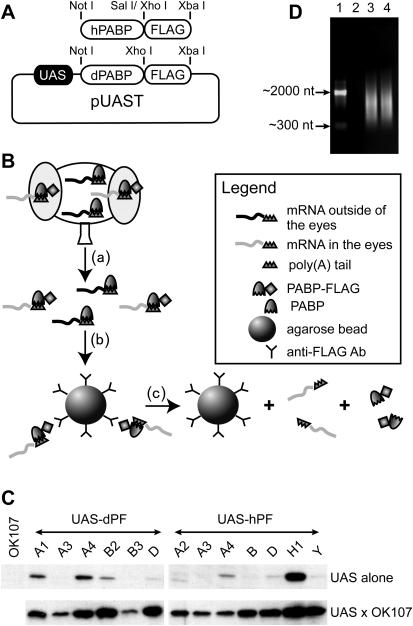
Two complementary oligonucleotides that contain the amino acid coding sequence of the FLAG tag (DYKDDDDK), FLAG1, 5′-TCGAGGATTACAAGGATGACGACGATAAGTAAT-3′, and FLAG2, 5′-CTAGATTACTTATCGTCGTCATCCTTGTAATCC-3′, were annealed together and cloned into the Xho1 and Xba1 sites of the vector pUAST (12); the resultant recombinant clone was named pP{UAST-FLAG}. The coding sequences of Drosophila dPABP and hPABP were amplified from the fly expressed sequence tag cDNA clone SD22319 and the human I.M.A.G.E. clone 3940309, respectively. These coding sequences were then cloned in-frame upstream of the FLAG coding sequence in pP{UAST-FLAG}. The resulting clones, pP{UAS-dPABP-FLAG} (abbreviated as pP{UAS-dPF}) and pP{UAS-hPABP-FLAG} (abbreviated as pP{UAS-hPF}) (Figure 1A), were used to transform Cantonized-_w_1118 flies following standard procedures. We recovered six and eight transformant lines, respectively, for the two constructs.
Figure 1.
mRNA tagging in Drosophila. (A) Diagram of recombinant pP{UAS-dPF} and pP{UAS-hPF} vectors for the expression of FLAG-tagged dPABP and hPABP. (B) Flowchart of the methodology for mRNA tagging. The figure illustrates the expression of FLAG-tagged PABP in the Drosophila photoreceptor cells. In step (a), fly heads were fixed with formaldehyde to crosslink poly(A)+ RNA with PABP, and the heads were then homogenized. In step (b), FLAG-tagged PABP bound mRNA from the photoreceptor cells was immunoprecipitated using an anti-FLAG-specific antibody, thus fractionating the photoreceptor cell mRNAs from mRNAs from other tissues. In step (c), the mRNA–PABP complex was dissociated by treatment of the complex with SDS at 65°C. (C) Basal and GAL4-OK107 driven expression of FLAG-tagged dPABP or hPABP in the P{UAS-dPF} and P{UAS-hPF} transgenic flies. The western blot was labeled by anti-FLAG M2 antibody (Sigma). (D) dPABP-FLAG and hPABP-FLAG bind mRNA in vivo. The transgenes P{UAS-dPF}D and P{UAS-hPF}B were combined with GAL4-OK107 and RNA fractionated by mRNA tagging as described above. The selected RNA was amplified in vitro (22) and fractionated on a formaldehyde agarose gel. The lanes show the amplified RNA from GAL4-OK107 (Lane 2), GAL4-OK107/P{UAS-dPF}D (Lane 3) and GAL4-OK107/P{UAS-hPF}B (Lane 4). Total fly RNA (lane 1) was included as marker. The major visible band represents the rRNAs. No amplified product was detectable from the RNA selected from GAL4-OK107 flies. The RNA selected from flies carrying P{UAS-dPF}D or P{UAS-hPF}B produced amplified products with a similar size distribution.
mRNA tagging
Our procedures (Figure 1B) were modified from those described previously (6–8). About 200 fly heads were fixed in 1 ml of 1× phosphate-buffered saline (PBS) (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl and 2.7 mM KCl)containing 1% formaldehyde and 0.5% NP40 for 30 min at 4°C. After fixation, 140 µl of 2 M glycine was added and the sample incubated at 4°C for 5 min. The heads were rinsed three times with 1× PBS and homogenized in 0.8 ml of homogenization buffer. To prepare homogenization buffer (HB), a solution of 150 mM NaCl, 50 mM HEPES buffer at pH 7.6, 1 mM EGTA, 15 mM EDTA and 10% glycerol was treated with diethyl pyrocarbonate. Immediately before use, vanadyl ribonucleoside complex (Sigma), SUPERase-In (Ambion), and a protease inhibitor cocktail tablet (Roche) were added to the solution to final concentrations of 8 mM, 50 U/ml and 1 tablet/50 ml, respectively. The homogenate was sonicated and cleared by centrifugation at 13 000 g for 10 min. RNA bound to the FLAG-tagged PABP was recovered by co-immunoprecipitation using anti-FLAG-M2 affinity agarose beads (Sigma). Before use, the agarose beads were washed four times with HB at 4°C. For co-immunoprecipitation, we gently mixed 0.8 ml of homogenate with anti-FLAG-M2 affinity agarose beads from 100 µl of bead suspension for 2 h at 4°C. The beads were then washed four times with HB at 4°C. The RNA::PABP crosslink was then reversed by incubating the beads in 100 µl of elution buffer (50 mM Tris–HCl, pH 7.0, 10 mM EDTA, 1.3% SDS and 50 U/ml SUPERase-In) at 65°C for 30 min. To isolate RNA, 100 µl of eluant was mixed with 400 µl of Trizol (Invitrogen) and then mixed with 100 µl of chloroform. The mixture was incubated at room temperature for 10 min and then centrifuged at 12 000 g for 10 min at 4°C. The aqueous supernatant was transferred to a new tube and mixed with 250 µl of isopropanol, chilled at −80°C overnight, and centrifuged at 12 000 g for 10 min at 4°C. The RNA pellet was rinsed once with 0.5 ml of 70% ethanol and then dissolved in 20 µl of RNase free water.
Microarrays and data analysis
RNA was processed following standard Affymetrix protocols (GeneChip® Expression Analysis Technical Manual, Santa Clara, CA; Affymetrix, 2001). We started with 10 µg of total RNA from each of the pools from whole heads, and from the total amount of tagged RNA available from those pools. Biotinylated cRNA was synthesized in vitro from the double-stranded cDNA using the ENZO BioArray High Yield RNA Transcript Labeling Kit (ENZO Diagnostics, Inc., Farmingdale, NY). Yields of labeled RNA from whole heads were high, and 15 µg of fragmented, biotinylated cRNA was mixed into 300 µl of hybridization cocktail, of which 200 µl was used for each hybridization. Yields from the tagged RNA were lower (5–19 µg), so in two cases 10 µg (replicates 1 and 3) and in two cases 5 µg (replicates 2 and 4) of biotinylated cRNA were mixed into 200 µl of hybridization mix. Hybridization was for 17 h at 42°C. Washing, staining and scanning were carried out according to standard protocols.
The intensity signals obtained from the microarray chips stored in CEL files were extracted and normalized by the DNA-Chip Analyzer (dChip) program (13). The RNA expression levels of each probe set were calculated as model-based expression indexes (MBEI) by the dChip program and exported to the R program (14) for statistical analyses. These results were exported to Microsoft Excel for further analyses such as sorting and filtering. The fold change was calculated by dividing the average MBEI for each gene in the mRNA tagged-mRNA population with the corresponding average MBEI in the whole head mRNA population. The annotation of individual genes was obtained from http://apps1.niaid.nih.gov/David (15).
RT–PCR and real-time PCR
mRNA was transcribed into first-strand cDNA using the Superscript II RT kit (Invitrogen) following the manufacture's instructions. Real-time PCR was performed using Applied Biosystem's Taqman PCR reagent kit, gene-specific primers and Taqman probes. Real-time PCR was performed using the ABI PRISM® 7700 Sequence Detection System. Relative quantification of the mRNA level was achieved using the comparative CT method as described in User Bulletin #2, ABI PRISM 7700 Sequence Detection System. The mRNA of constitutively expressed ribosomal protein gene rp49 was used as an internal control.
In situ hybridization
Gene-specific cDNA fragments were amplified by RT–PCR using gene-specific primers. The cDNA fragments were then used as templates to synthesize digoxigenin-labeled RNA probes using the digoxigenin SP6/T7 RNA labeling kit (Roche) following the manufacture's suggestions. In situ hybridization was performed essentially as described (16). After the chromogenic reaction with nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate, the samples were counterstained with Nuclear Fast Red (Vector Labs) for 5 min. Slides were then mounted in VectaMount (Vector Labs) and imaged with the Zeiss Axioplan 2 Imaging System.
RESULTS
Generation of P{UAS-dPF} and P{UAS-hPF} transgenic flies
We prepared transgenic flies that express FLAG-tagged Drosophila PABP dPABP or hPABP, in a tissue-specific manner. A FLAG coding sequence was fused in-frame to the 3′ end of the dPABP and hPABP coding sequences, and the resulting cassettes were inserted into an UAS vector pUAST (Figure 1A) so that each PABP could be expressed using the GAL4/UAS system. We recovered six and eight transgenic lines, respectively, for the pP{UAS-dPF} and pP{UAS-hPF} constructs.
Many transgenes built with P{UAST} exhibit significant levels of expression without GAL4 owing to position effects that promote expression. We screened the transgenic lines, selecting lines that exhibited no or very low basal expression of recombinant PABP without a GAL4 driver and significant expression when combined with the driver. The transgenic lines were crossed with the GAL4 line, P{GawB]OK107, which expresses GAL4 at high levels within the mushroom bodies (17). Heads from the progeny were used for western analyses. Among the P{UAS-PABP-FLAG} lines, the transgenes P{UAS-dPF}D and P{UAS-hPF}B were found to express the FLAG epitope at minimal levels in the absence of GAL4 and at robust levels after combining with GAL4 (Figure 1C). P{UAS-dPF}D and P{UAS-hPF}B were therefore used for subsequent studies unless otherwise noted.
Expression of dPABP-FLAG produces lethality with some GAL4 drivers
PABP plays important roles in translation initiation and mRNA stabilization/degradation (18). It binds to poly(A) tails of eukaryotic mRNAs (∼1 protomer/27 nt) (19). It can also interact with other components of the translation machinery, such as eIF4G and PAIP-1 (20). Because of these important cellular functions, we reasoned that the overexpression of PABP might potentially perturb the normal regulation of translation in the tissues where it is expressed. We tested this possibility by combining hPABP (P{UAS-hPF}) or Drosophila PABP (P{UAS-dPF}) transgenes with different GAL4 drivers and scoring the progeny for viability (Table 1 and data not shown).
Table 1.
Viability of flies carrying P{UAS-dPF} and P{UAS-hPF} transgenes with GAL4 drivers
| Cross | Non-Cy progeny | Cy progeny | Expected ratioa | Observed ratiob |
|---|---|---|---|---|
| P{UAS-dPF}A3 × Act5C | 0 | 165 | 0.5 | 0 |
| P{UAS-dPF}B2 × Act5C | 0 | 113 | 1 | 0 |
| P{UAS-dPF}D × Act5C | 0 | 162 | 1 | 0 |
| P{UAS-hPF}B × Act5C | 79 | 125 | 1 | 0.63 |
| P{UAS-hPF}Y × Act5C | 71 | 118 | 0.5 | 0.6 |
| P{UAS-dPF}A3 × GMR | 0 | 47 | 1 | 0 |
| P{UAS-dPF}B2 × GMR | 93c | NA | NA | |
| P{UAS-dPF}D × GMR | 52c | NA | NA | |
| P{UAS-hPF}B × GMR | 95 | NA | NA | |
| P{UAS-hPF}D × GMR | 30 | NA | NA | |
| P{UAS-hPF}Y × GMR | 61 | 39 | 1 | 1.56 |
We used the following GAL4 lines: P{Act5C-GAL4}25FO1, P{GawB}OK107, P{GawB}elav[C155], P{w[+mC]=GAL4-ninaE.GMR}12, Rh1-GAL4 and P{GawB}201Y. The Act5C-GAL4 provides broad expression in most if not all tissues. The ELAV-GAL4 transgene drives expression in all neurons of the organism. The transgenes GAL4-OK107 and GAL4-201Y provide preferential expression in the mushroom bodies, the transgene GMR-GAL4 provides specific expression in the eye, and Rh1-GAL4 provides specific expression in photoreceptor cells R1–R6. In addition to these expression patterns in the adult fly, the transgenes may also express GAL4 during development in other tissues. FLAG-tagged Drosophila PABP expressed from Act5C-GAL4, GMR-GAL4 and GAL4-201Y produced lethality at different developmental stages depending on the specific GAL4 line used. For instance, Act 5C-GAL4/PUAS-dPF} animals died at early larval instar, while GMR-GAL4/P{UAS-dPF} and GAL4-201Y/P{UAS-dPF} animals died at the pupal stage (Table 1 and data not shown). Rearing the animals at 18°C, which reduces GAL4 activity, failed to lessen the observed lethality (data not shown). In contrast, the GAL4 lines GAL4-OK107, ELAV-GAL4 and Rh1-GAL4 were viable in combination with P{UAS-dPF} and all GAL4 lines produced viable and healthy adult offspring in combination with P{UAS-hPF} (Table 1 and data not shown). Thus, the hPABP offers an alternative to the Drosophila PABP for cases in which the overexpression of dPABP is toxic. Alternatively, the use of modified GAL4 systems that offer experimenter control over the induction of GAL4 activity (21) may also provide a method for avoiding toxicity of dPABP overexpression during development.
Recombinant dPABP-FLAG and hPABP-FLAG bind mRNA in vivo
To test whether the recombinant and transgenically-supplied PABP could bind mRNA in vivo, we crossed P{UAS-dPF} and P{UAS-hPF} to GAL4-OK107. We isolated RNA from the heads of progeny flies by mRNA tagging (Materials and Methods and Figure 1B). RNA was then used as template for RNA amplification (22). As shown in Figure 1D, no amplified RNA product was detected using RNA template prepared from the control GAL4-OK107 flies, while significant amounts of amplified RNA were detected from RNA templates prepared from GAL4-OK107/P{UAS-dPF} and GAL4-OK107/P{UAS-hPF} flies. The size of the amplified RNA ranged from ∼300 to ∼2000 nt, similar to the size distribution of the mRNA population of adult flies; the size distribution was similar for both P{UAS-dPF} and P{UAS-hPF}. These results indicated that the recombinant dPABP-FLAG and hPABP-FLAG are able to bind to mRNA in vivo.
Effect of formaldehyde fixation on mRNA tagging
The mRNA tagging method is based upon formaldehyde crosslinking of RNA-binding protein and mRNA, which allows co-immunoprecipitation with antibodies to the FLAG epitope. One critical concern is the degree of formaldehyde crosslinking, since excessive crosslinking may result in irreversible crosslinks and subsequent loss of RNA, or in the reduction of antigen available for antibody binding (23). However, under-fixation may result in poor selection. Therefore, we explored the effect of formaldehyde fixation on the efficiency of mRNA tagging.
We constructed flies Rh1-GAL4/P{UAS-hPF} and GAL4-201Y/P{UAS-hPF} that express hPABP-FLAG in photoreceptor cells and in mushroom body neurons, respectively. The heads of these flies were fixed with 1% formaldehyde for 0, 30 or 60 min, and real-time PCR was then used to estimate the enrichment or depletion of inaD mRNA in the photoreceptor cell- and mushroom body neuron-enriched mRNA populations relative to whole heads. Since inaD mRNA is exclusively found in photoreceptor cells (24), we expected that inaD mRNA would be enriched in the photoreceptor cell-enriched mRNA sample and depleted in the mRNA sample selected from mushroom body neurons. As illustrated in Table 2, inaD mRNA was enriched ∼10-fold in the mRNA samples selected from photoreceptor cells. Formaldehyde fixation for 0, 30 or 60 min was without effect. In contrast, the relative abundance of inaD mRNA in mRNA samples selected from mushroom body neurons without fixation was reduced to ∼1/10 of that in whole head mRNA control. It was reduced further when fly heads were treated with formaldehyde for 30 and 60 min, respectively. Formaldehyde treatment was used in the mRNA tagging protocols of two previous studies of C.elegans (8,11). However, it has been reported that no mixing of PABP and mRNA occurs between cells in cell extracts (25), and thus formaldehyde fixation prior to mRNA tagging may not be necessary. Our results suggest that mRNA tagging without prior formaldehyde treatment can effectively enrich mRNA from tissues where recombinant PABP is expressed and deplete mRNA from other tissues. However, crosslinking with formaldehyde helps to reduce the contamination of mRNA from other tissues.
Table 2.
Effect of formaldehyde fixation on enrichment of inaD mRNA by mRNA tagging
| Fixation time | Fold change after mRNA tagging selection | ||
|---|---|---|---|
| 0 min | 30 min | 60 min | |
| Rh1 GAL4/UAS-hPABP-FLAG | 11.8 ± 4.2 | 10.1 ± 0.6 | 8.4 ± 0.3 |
| 201Y GAL4/UAS-hPABP-FLAG | 1/(11.1 ± 3.8) | 1/(59.8 ± 19.7) | 1/(33.8 ± 15.8) |
Gene expression profiling in Drosophila photoreceptor cells
Although visual transduction in Drosophila is among the best understood signal transduction pathways (26,27), there remain many outstanding questions. Forward genetic screening for visually defective mutants is at saturation so reverse genetic tools, such as RNAi (28) and mRNA tagging, offer new methodologies to define the function of novel photoreceptor cell-expressed genes. We performed microarray analysis of mRNA co-immunoprecipitated from fly photoreceptor cells with dPABP as well as mRNA from whole fly heads, to gain insights into the profile of genes expressed in these cells and to identify novel, photoreceptor cell-enriched genes.
We utilized the photoreceptor cell-specific GAL4 driver, Rh1-GAL4, combined with P{UAS-dPF}D, to drive the expression of dPABP-FLAG in fly photoreceptor cells R1–R6. To identify photoreceptor-expressed genes, we prepared two groups of RNA samples, each with four replicates. In group I, total RNA was isolated from whole fly heads with Trizol reagent (Invitrogen); in group II, mRNA associated with dPABP-FLAG in photoreceptor cells was selected by mRNA tagging (Materials and Methods). These RNA samples were amplified and used to probe Affymetrix Drosophila whole genome array chips, which contain 14010 probe sets for ∼13 600 genes. Microarray data obtained with groups I and II therefore represent the transcriptome of whole fly heads and the mRNA population associated with PABP in photoreceptor cells 1–6, respectively. For the convenience of discussion, we refer to the latter subpopulation as the transcriptome of photoreceptor cells, although in reality, group II does not represent all photoreceptor cell mRNAs but only those capable of interacting with PABP. The complete microarray experimental data can be found at the website of the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo) with the accession number GSE1790.
The data for the four microarray replicates from group I samples were highly correlated with an inter se correlation coefficient >0.99. However, the correlation coefficients for replicate 4 with the three other replicates of group II samples were 0.82, 0.89 and 0.86, respectively, while the inter se correlation coefficients among replicates 1–3 were all >0.95.We also found that the average coefficient of variance for the microarray data generated from group I RNA samples was 9.2%; while the average coefficient of variance of data obtained using group II RNA samples was 20.0%. This variance decreased to 14.3% if replicate 4 was excluded from analysis. Therefore, we excluded replicate 4 of group II for further data analysis.
The distributions of transcripts according to expression level in the transcriptome of whole head versus photoreceptor cells were similar. In the transcriptome of whole head, ∼20% of the genes were expressed at a level of relative abundance <0.001% (MBEI < 73), 68% of the genes were expressed between 0.001 and 0.01% (73 < MBEI < 730), 11% of the genes between 0.01 and 0.1% (730 < MBEI ≤ 7300), and only 1.2% of the genes were expressed at levels >0.1% (MBEI > 7300). This compares with 20, 67, 12 and 0.9% in the transcriptome of photoreceptor cells.
We selected genes with mRNAs of >0.001% relative abundance in whole head transcriptome for further analysis. Of the 11 212 genes analyzed, we found that the mRNAs of 1161 of the genes were underrepresented in the transcriptome of photoreceptor cell by >2-fold when compared with the transcriptome of whole head (Supplementary Table 1); Of these 98% were statistically significant at the 5% level of significance. In contrast, the mRNAs of 743 genes were enriched >2-fold in the transcriptome of photoreceptor cells (Supplementary Table 2); Of these 85% were significantly different at the 5% level.
Twenty-two genes are currently known to be involved in phototransduction or retinal degeneration (29). The enrichment of these genes in the photoreceptor cells as determined by mRNA tagging is illustrated in the left column of Table 3. Four of these genes (Rh3, Rh4, Rh5 and Rh6) are expressed outside of the photoreceptor cells R1–R6, and may therefore not be highly enriched after mRNA selection. However, the mRNAs for Rh3, Rh4 and Rh6 were modestly enriched by PABP selection. As illustrated in Table 3, the mRNAs for 13 of the remaining 18 genes are enriched >2-fold after mRNA tagging. The genes listed on the right side of Table 3 are also known to be eye-enriched but have no known function in phototransduction or retinal degeneration (29). The transcripts of nine of these genes were enriched >2-fold after mRNA tagging (Table 3). Taken together, the mRNAs for >70% of the known eye-enriched genes were enriched >2-fold after mRNA tagging. These data, therefore, offer support for a significant enrichment of tissue-specific mRNAs after mRNA tagging.
Table 3.
Enrichment of known eye-specific genes by mRNA tagging
| Gene | Expression level | Fold change | Gene | Expression level | Fold change | ||
|---|---|---|---|---|---|---|---|
| Whole head | Photoreceptor cell | Whole head | Photoreceptor cell | ||||
| inaF | 144 ± 30 | 1396 ± 527 | 9.72 | hdc | 198 ± 28 | 1473 ± 140 | 7.43 |
| ninaA | 407 ± 31 | 3070 ± 191 | 7.54 | eyc | 343 ± 26 | 2310 ± 836 | 6.73 |
| trpl | 757 ± 75 | 5649 ± 462 | 7.46 | dlg | 518 ± 40 | 3463 ± 1685 | 6.68 |
| Ga76C | 895 ± 142 | 6620 ± 1692 | 7.39 | boss | 563 ± 19 | 3036 ± 960 | 5.40 |
| ninaC | 967 ± 113 | 6723 ± 241 | 6.95 | cpn | 764 ± 58 | 3829 ± 59 | 5.01 |
| trp | 1135 ± 86 | 7859 ± 929 | 6.92 | glass | 244 ± 28 | 1188 ± 238 | 4.88 |
| inaD | 598 ± 45 | 3879 ± 284 | 6.49 | chp | 2328 ± 194 | 9723 ± 1644 | 4.18 |
| norpA | 750 ± 124 | 4710 ± 1935 | 6.28 | so | 2401 ± 11 | 695 ± 101 | 2.89 |
| inaC | 695 ± 17 | 3139 ± 1177 | 4.51 | lqf | 563 ± 43 | 1171 ± 419 | 2.08 |
| Rh2 | 814 ± 145 | 2834 ± 839 | 3.48 | st | 434 ± 58 | 587 ± 33 | 1.35 |
| Gα30A | 1187 ± 128 | 3760 ± 1779 | 3.17 | pdh | 10 757 ± 470 | 10 246 ± 841 | 0.95 |
| Gα49B | 161 ± 32 | 492 ± 225 | 3.06 | Sh | 184 ± 28 | 94 ± 13 | 0.51 |
| ninaE | 9843 ± 650 | 20 532 ± 3411 | 2.09 | Cry | 1241 ± 136 | 509 ± 79 | 0.41 |
| arr2 | 9119 ± 228 | 15331 ± 726 | 1.68 | ||||
| arr1 | 5312 ± 273 | 8444 ± 1287 | 1.59 | ||||
| cds | 2880 ± 388 | 3292 ± 436 | 1.14 | ||||
| rdgC | 378 ± 41 | 262 ± 145 | 0.69 | ||||
| rdgA | 378 ± 51 | 257 ± 48 | 0.68 | ||||
| Rh3 | 5343 ± 570 | 8275 ± 751 | 1.55 | ||||
| Rh4 | 3967 ± 351 | 7499 ± 1201 | 1.89 | ||||
| Rh5 | 2482 ± 312 | 2035 ± 40 | 0.82 | ||||
| Rh6 | 4292 ± 655 | 7249 ± 1997 | 1.69 |
In addition, the mRNAs of genes that are known to be enriched in other tissues were found to be underrepresented in the transcriptome of photoreceptor cells. For instance, the mRNA level of the muscle-specific myosin genes Mhc, Mlc1, Mlc2 in the transcriptome of photoreceptor cells was reduced to <20% of the value in the transcriptome of whole heads. Similarly, the mRNAs of 10 odorant-binding proteins were underrepresented >2-fold and only 1 odorant-binding protein mRNA was enriched after the mRNA tagging procedure (Supplementary Tables 1 and 2). This is consistent with the finding that odorant-binding protein genes are specifically expressed in gustatory and olfactory sensilla (30).
Identification of previously uncharacterized photoreceptor-enriched genes
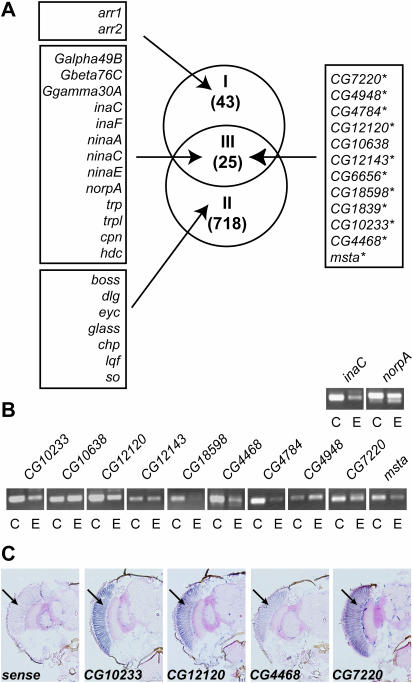
A fortuitous observation enabled us to identify 11 novel photoreceptor-enriched genes. We compared the transcriptomes of whole heads of Rh1-GAL4/P{UAS-dPF} flies and Canton-S flies. We found that there were 78 genes whose mRNA level was >2-fold higher in Canton-S flies than in Rh1-GAL4/P{UAS-dPF} flies. As discussed below, we believe that this may be owing to a squelching effect that GAL4 has on transcription when overexpressed (31). The mRNAs of 25 of these 78 genes were also enriched >2-fold by mRNA tagging from photoreceptor cells (Figure 2A, Group III). Surprisingly, all 13 genes previously characterized to be eye-specific were found among these 25 genes. Of these 13 genes (all except cpn and hdc), 11 are known to be involved in phototransduction or retinal degeneration (Figure 2A, left column of Group III). Therefore, we hypothesized that the remaining 12 unknown genes of this group genes are probably to be enriched in the eye as well. To test this hypothesis, we designed specific primers for 10 out of the 12 uncharacterized genes (Figure 2A, right column) and performed RT–PCR to measure their mRNA level in the heads of Canton-S flies and that in the heads of eye absent (eya) mutant flies, which are devoid of normal compound eyes. As shown in Figure 2B, at least 7 (CG10233, CG12120, CG18598, CG4468, CG4784, CG7220 and msta) of the 10 genes tested exhibited obviously higher levels of amplification from Canton-S fly heads compared with eya fly heads. This strongly suggests that these seven functionally unannotated genes are also photoreceptor cell-enriched genes. In situ hybridization experiments confirmed the photoreceptor cell-restricted expression pattern of these genes (Figure 2C and data not shown). In addition, a recent study (29) showed that three more genes (CG12143, CG4948 and CG6656) of the group are also eye-enriched genes, although our RT–PCR results on two of these (CG12143 and CG4948) were inconclusive (Figure 2B). Overall, 24 out of the 25 genes in group III shown in Figure 2A exhibit enriched expression in photoreceptor cells.
Figure 2.
Identification of novel photoreceptor-enriched genes. (A) Classification of photoreceptor-enriched genes based on the ratio of their mRNA level in Canton-S wild-type flies to that in Rh1-GAL4/P{UAS-dPF} flies, and on the ratio of mRNA level after mRNA tagging compared with whole head mRNA. Group I contains genes whose mRNA level in Canton-S heads was >2-fold higher than in Rh1-GAL4/P{UAS-dPF} heads. Group II contains genes whose mRNA level after mRNA tagging was >2-fold greater than before selection. Group III represents the overlap of the two previous groups. Numbers in brackets indicate the number of genes in each group. All members of Group III and the known eye-enriched genes involved in phototransduction and retinal degeneration are listed in the left column. Previously uncharacterized genes are listed on the right. Genes labeled with an asterisk were shown to be photoreceptor cell-enriched in this study and eye-enriched in a related study (29). (B) RT–PCR detection of mRNA of putative photoreceptor cell-enriched genes in Canton-S flies and eye absent mutant flies. The mRNA prepared from heads of Canton-S flies (C) and eye absent mutant flies (E) was reverse transcribed, and the quantity of RT products was normalized to rp49 mRNA (data not shown). Same amount of RT products were then used as template for PCR using primers specific to genes as indicated. (C) Spatial expression of previously unknown photoreceptor-enriched genes revealed by in situ hybridization. Antisense RNA probes, except for the CG12120 sense control (sense), were hybridized to frontal sections of fly heads. For all antisense probes, enhanced signal was observed in the eye (arrows).
DISCUSSION
These results demonstrate the utility of mRNA tagging using PABP as a tag for mRNA isolation from specific cell types in Drosophila prior to microarray analyses. With larger model organisms, one can often obtain sufficient quantities of tissue or cell samples by physical dissection that are pure enough for analytical procedures such as microarray experiments. The small size of Drosophila generally prohibits the use of physical dissection for obtaining large enough quantities of relatively pure tissue for such procedures. However, mRNA tagging requires no special instruments and offers the ability to purify mRNA from virtually any cell type in Drosophila at any developmental stage, given the large supply of GAL4 lines that are available. Moreover, starting material should no longer be limiting since, in principle, tens to thousands of flies, larvae or embryos can be used as starting material.
However, the mRNA recovered by mRNA tagging is affected by at least four factors and these need to be considered. The first factor is the mRNA transcriptome of the tissue of interest. This establishes the identity of mRNA that is available for binding to PABP. The second factor is established by the population of mRNAs that can bind PABP. Not all mRNAs are capable of binding, and these are therefore excluded by PABP-mediated selection. The third factor is the physiological status of the tissue or cell type used for selection. PABP has well-established functions in translation initiation, termination and the maintenance of mRNA stability (32). Therefore, the binding of PABP to mRNA is probably influenced by the status of translation and mRNA metabolism in the cells of interest. Thus, the mRNA associated with PABP is not necessarily equivalent to the transcriptome of the same cell, because of these potential regulatory processes. The net effect of this, though, is that PABP-mediated RNA selection may not be ideal for elucidating tissue or cell transcriptomes. However, this also means that the PABP-associated mRNA population resembles a cell's actively translated mRNA population or proteome, and these may provide more biological insights than the transcriptome. By comparing the transcriptome of whole fly heads with the PABP-selected mRNA population from fly photoreceptor cells, the mRNAs of 743 genes of 11 212 examined were found to be enriched >2-fold in the latter population. In contrast, a recent and comprehensive microarray analysis of the eye transcriptome (29) found 128 genes to be preferentially expressed. One possible explanation for the numerical discrepancy is that the PABP-selected mRNA population reflects those actively translated genes rather than the mRNA transcriptome. Moreover, results similar to those presented here were obtained in a recent study (25), which compared the PABP-associated mRNA with the transcriptome of the same murine endothelial PY4.1 cells. It was found that when comparisons were made after global normalization, the mRNAs of 8% of 333 genes examined were enriched >2-fold and the mRNAs of 12% of the genes reduced to <1/2 in the PABP-associated mRNA population. The final factor that affects the mRNA selection is experimental errors. These could include nonspecific binding of mRNA to antibodies or agarose beads used in immunoprecipitation, and leaky expression of tagged PABP in tissues other than those of interest.
Because of these factors, we cannot accurately estimate how many of the 743 identified genes actually have elevated mRNA abundance levels in photoreceptor neurons R1–R6. However, genes of this type must represent a significant fraction of photoreceptor enriched genes must be included in this population. Pilot experiments for the enrichment of inaD mRNA using Rh1 GAL4 and its depletion using 201Y GAL4 establish that the overall procedure can identify genes enriched in expression at the mRNA level. By extension this suggests that a significant fraction of the 743 genes identified in our large-scale experiment must have higher mRNA abundance levels in photoreceptor cells compared with other cells of the head. Also, genes known to be preferentially expressed at the RNA level in other tissues, including known muscle genes and odorant-binding protein genes, were underrepresented in the RNA fraction selected from photoreceptor cells. Moreover, the 743 genes identified include genes known to have elevated mRNA abundance in photoreceptor cells. Indeed, the mRNA tagging procedure identified 70% (22/31) of the genes currently known to be preferentially expressed in photoreceptor cells R1–R6 (Table 3). Random chance alone would predict the identification of only 7% of the photoreceptor-enriched mRNAs (743/11 212 genes screened). Finally, we identified 11 novel genes with preferential mRNA expression in the eye based on RT–PCR and/or RNA in situ hybridization experiments, consistent with the selection of RNAs preferentially expressed at the mRNA level by the mRNA tagging protocol.
One practical concern when using mRNA tagging to study Drosophila gene expression is the toxicity of dPABP overexpression. Indeed, overexpression of dPABP produced lethality depending on the GAL4 driver used (Table 1). This lethality is probably more dependent on the expression pattern in time and in space than on the absolute level of expression. GMR-GAL4 and Rh1-GAL4 have similar levels of expression in adult flies, but GAL4 is expressed more broadly in GMR-GAL4 than Rh1-GAL4 (photoreceptor cells R1–R8 versus R1–R6), and the expression of GMR-GAL4 begins earlier than Rh1-GAL4, commencing in the larval stages versus late in pupariation (33). Although the development of GMR-GAL4/P{UAS-dPF} flies was arrested at the pupal stage, the Rh1-GAL4/P{UAS-dPF} flies developed into adulthood without any discernible morphological or viability difference from wild-type flies.
Because of this potential lethality, we also fashioned affinity probes using hPABP, reasoning that divergent protein–protein interaction domains might make hPABP less toxic. PABP has two functional domains: the N-terminus consists of four highly conserved RNA recognition motifs that are responsible for PABP's binding to the poly(A) tail, and the less-conserved C-terminal region contains sites for interaction with other proteins, such as eIF4G and PIP1 (9). The N-terminal domain of hPABP is 71% identical in amino acid sequence with that of dPABP, while the identity between the C-terminal domains is only 30%. Therefore, we thought it probable that in Drosophila cells, hPABP would retain the ability to bind to the poly(A) tails of mRNAs but fail to interact with other translation-regulating proteins and thereby display less toxicity. With the option of using either dPABP and hPABP, the range of fly tissues that can be studied by mRNA tagging is broadened. Nevertheless, the mis-expression of either dPABP or hPABP might produce alterations in the physiological state of the expressing cells in unknown ways.
We serendipitously discovered that the mRNA level of most, if not all, genes involved in visual transduction in Rh1-GAL4/P{UAS-dPABP} flies is less than one half of that in Canton-S flies (Figure 2A). This led us to define 25 genes (Group III in Figure 2A), which are both underrepresented in Rh1-GAL4/P{UAS-dPF} heads compared with Canton-S heads and were enriched by mRNA tagging with Rh1-GAL4. Aside from 13 known eye-specific genes in this group, 11 of the other 12 previously uncharacterized genes were also proved to be photoreceptor cell-enriched genes (Figure 2) (29). We suggest that an unknown master transcription factor may regulate the transcription of the genes in this group, and this factor may be the target of the squelching effect of GAL4 that was first described in yeast (31). Interestingly, only the transcription of genes with direct function in visual function were subjected to the presumed squelching effect, while the mRNA level of other known eye-enriched genes (except hdc and cpn) were not affected. Thus, we speculate that the 11 new photoreceptor cell-enriched genes, identified together with the genes known to be involved in visual transduction are also probable to be involved in phototransduction or retinal degeneration. Indeed, one of these genes, CG12143, or Sunglasses, was shown recently to be involved in retinal degeneration (29). However, we cannot rule out the possibility that the GAL4 expression in photoreceptor cells leads to the misidentification of some genes as photoreceptor cell-expressed or photoreceptor cell-enriched. It is possible, for instance, that the presumed squelching effect of GAL4 in photoreceptor cells non-autonomously alters gene expression in cells other than photoreceptor neurons.
An alternative approach for isolating photoreceptor cell-enriched mRNAs is via a differential expression using mutant flies. A recent and comprehensive microarray comparison study (29) of mRNA from wild-type fly heads and mRNA from heads of an ommatidia devoid mutant (sine oculis) reported the identification of 93 new eye-enriched genes. This approach potentially identifies mRNAs enriched in the whole eye irrespective of cell type. The mRNA tagging approach described here, however, using Rh1-GAL4, specifically identifies PABP-associated mRNA in photoreceptor neurons R1–R6. Although both approaches are valuable, this comparison illustrates a major advantage of mRNA tagging: the ability to identify PABP-associated mRNAs in specific cell types even within one tissue.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
[Supplementary Material]
Acknowledgments
We thank R. E. Jerome and C. Zhu for technical expertise in the microarray experiments, Dr S. Pletcher for helping with microarray data analysis, Dr K. Choi for providing fly stocks and valuable comments and C. Wilson for assistance in histology. This research was supported by NIH grant U01AA13476 (Integrative Neuroscience Initiative in Alcoholism) to R.L.D. and U01AA13518 to H.J.E. Microarray studies were carried out in the Center for Medical Genomics at Indiana University School of Medicine, supported in part by the Indiana Genomics Initiative; INGEN® is partially supported by the Lilly Endowment, Inc. R.L.D. is the R. P. Doherty-Welch Chair in Science at the Baylor College of Medicine. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuaqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., Liotta L.A. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 4.Bryant Z., Subrahmanyan L., Tworoger M., LaTray L., Liu C.R., Li M.J., van den Engh G., Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl Acad. Sci. USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenenbaum S.A., Carson C.C., Lager P.J., Keene J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 7.Tenenbaum S.A., Lager P.J., Carson C.C., Keene J.D. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 8.Roy P.J., Stuart J.M., Lund J., Kim S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 9.Gorlach M., Burd C.G., Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 10.Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 11.Kunitomo H., Uesugi H., Kohara Y., Iino U. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 2005;6:R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 13.Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ihaka R., Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 15.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 16.Vosshall L.B., Amrein H., Morozov P.S., Rzhetsky A., Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 17.Lee T., Lee A., Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 18.Sachs A., Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 19.Baer B.W., Kornberg R.D. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc. Natl Acad. Sci. USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 21.McGuire S.E., Roman G., Davis R.L. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Van Gelder R.N., von Zastrow M.E., Yool A., Dement W.C., Barchas J.D., Eberwine J.H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 24.Shieh B.H., Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- 25.Penalva L.O., Burdick M.D., Lin S.M., Sutterluetyand H., Keene J.D. RNA-binding proteins to assess gene expression states of co-cultivated cells in response to tumor cells. Mol. Cancer. 2004;3:24. doi: 10.1186/1476-4598-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montell C. Visual transduction in Drosophila. Annu. Rev. Cell Dev. Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- 27.Hardie R.C., Raghu P. Visual transduction in Drosophila. Nature. 2001;413:186–193. doi: 10.1038/35093002. [DOI] [PubMed] [Google Scholar]
- 28.Kalidas S., Smith D.P. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- 29.Xu H., Lee S.J., Suzuki E., Dugan K.D., Stoddard A., Li H.S., Chodosh L.A., Montell C. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galindo K., Smith D.P. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 32.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtin K.D., Zhang Z., Wyman R.J. Gap junction proteins expressed during development are required for adult neural function in the Drosophila optic lamina. J. Neurosci. 2002;22:7088–7096. doi: 10.1523/JNEUROSCI.22-16-07088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]