The γ-secretase-generated intracellular domain of β-amyloid precursor protein binds Numb and inhibits Notch signaling (original) (raw)
Abstract
The β-amyloid precursor protein (APP) and the Notch receptor undergo intramembranous proteolysis by the Presenilin-dependent γ-secretase. The cleavage of APP by γ-secretase releases amyloid-β peptides, which have been implicated in the pathogenesis of Alzheimer's disease, and the APP intracellular domain (AID), for which the function is not yet well understood. A similar γ-secretase-mediated cleavage of the Notch receptor liberates the Notch intracellular domain (NICD). NICD translocates to the nucleus and activates the transcription of genes that regulate the generation, differentiation, and survival of neuronal cells. Hence, some of the effects of APP signaling and Alzheimer's disease pathology may be mediated by the interaction of APP and Notch. Here, we show that membrane-tethered APP binds to the cytosolic Notch inhibitors Numb and Numb-like in mouse brain lysates. AID also binds Numb and Numb-like, and represses Notch activity when released by APP. Thus, γ-secretase may have opposing effects on Notch signaling; positive by cleaving Notch and generating NICD, and negative by processing APP and generating AID, which inhibits the function of NICD.
Alzheimer's disease (AD) is linked to increased β-amyloid precursor protein (APP) processing and is characterized by neurite dystrophy, synapse loss, and neuronal degeneration (1–4). APP normally undergoes a series of endoproteolytic cleavages: twice within the extracellular domain (α- and β-sites), mediated by tumor necrosis factor-converting enzyme and βAPP-cleaving enzyme, respectively, and once within the transmembrane domain (γ-site) (1–4). The cleavage of APP by the Presenilin-dependent γ-secretase liberates amyloid-β (Aβ) peptides (2–4) and the APP intracellular domain (AID) (5–14). Two Aβ species of either 42 or 40 residues (Aβ42 and Aβ40) are the major component of amyloid plaques in the brain of AD patients (1, 2). The corresponding AID peptides of either 57 or 59 aa have been detected only recently in human brain (5), likely because they are rapidly degraded (11). Although the biological function of APP is unclear, recent data indicate that AID can lower the cellular threshold to apoptosis (5, 10). Furthermore, AID was recently shown to form a multimeric complex with the nuclear adaptor protein Fe65 and the histone acetyltransferase Tip60, which possesses potent transcriptional activity (13). Therefore, AID may participate in signaling by modulating the function of intracellular proteins with which it interacts (13–17). Although the role of APP and Aβ in AD are widely studied, signaling by AID is not well understood.
Like APP, the Notch receptor undergoes a series of proteolytic cleavages that are a prerequisite for its activation and subsequent signal transduction (18). In response to binding the Delta-Serrate-Lag2 family of ligands, Notch undergoes regulated proteolysis in two sequential steps, first on the extracellular side by tumor necrosis factor-converting enzyme (3, 4), followed by an intramembranous cleavage mediated by the Presenilin-dependent γ-secretase that releases the Notch intracellular domain (NICD) (19–22). The functionally active NICD translocates to the nucleus where it interacts with the CBF1-SuH-Lag1 (CSL ) family of transcription factors and activates the transcription of genes that regulate the ability of cells to respond to various proliferation, differentiation, or apoptotic cues (18, 23). In the nervous system, NICD has been implicated in a variety of processes, including inhibition of neuronal differentiation and neurite growth, and regulation of neuronal cell death (23–28). The apparent similarities in proteolytic activation and signal transduction of APP and Notch raises the possibility that signals from these two receptors interact.
The present study was designed to examine the possibility that crosstalk exists between APP and the Notch signaling pathway. Here, we show that AID binds to the cytosolic adaptor proteins Numb and Numb-like (Nbl), known inhibitors of Notch signaling, and inhibits NICD when released by γ-secretase from membrane-tethered APP.
Materials and Methods
DNA Cloning and Constructs.
APP, APPCT-Gal4, AID, NICD, NΔE, CBF1-luciferase, and TP1-luciferase have been described (5, 13, 16, 27, 29–31). Mouse Nbl and Numb (e.g., p65, p66, p71, and p72) constructs (32) were cloned into Flag-tagged pcDNA3.1 (Invitrogen), pECFP-N1, or pEYFP-N1 (CLONTECH). Deletion mutants of Nbl and AID were generated by using PCR.
Cell Lines.
HEK 293, HeLa, and APP-HeLa Tet-on cells were maintained in RPMI medium 1640 (GIBCO) with 10% (vol/vol) FBS (Biofluids, Rockville, MD). N2a cells were maintained in DMEM (GIBCO) with 10% (vol/vol) FBS. Transfections were performed in six-well plates with Fugene 6 (Roche Molecular Biochemicals) by using 3 μl/μg DNA. DAPT30 (stock solution was 100 mM in DMSO; ref. 33) was synthesized as described in patent applications WO9822441-A2 and WO9822494-A2 filed by Athena Neurosciences (Elan Pharmaceuticals, San Francisco) and Eli Lilly.
Immunoprecipitation and Immunoblot Analysis.
Lysates from transfected cells or mouse brain were immunoprecipitated as described (29) with the following antibodies: α-Flag-agarose (Sigma), α−LC (CLONTECH), α-APP (1736; a kind gift of D. Selkoe, Harvard Medical School, Boston, MA), α-Nbl (34) or rabbit α-mouse IgG (ICN), and protein A/G agarose beads (Pierce). APP was detected with the 22C11 antibody (Chemicon). Proteins were detected by using the Supersignal West Pic chemiluminescent system (Pierce).
Fluorescence Resonance Energy Transfer (FRET) Analysis.
HEK 293T cells were cotransfected with the enhanced cyan fluorescent protein (ECFP) and enhanced yellow fluorescent protein (EYFP) fusion proteins by using 12 μl of Fugene 6 and a total of 2 μg of each DNA and were harvested 18–24 h later. A MoFlo Multi Laser Sorter (Cytomation, Fort Collins, CO) was configured as described (16). The threshold for a positive FRET cell was set by using the standard cytometry method of comparing each sample to negative controls. Additionally, if a sample did not have a cotransfection profile matching its control it was rejected from our analysis. Data acquisition and analysis was done with the SUMMIT version 3.0 software package (Cytomation).
Neuronal Cultures.
Primary cultures of cortical neurons were prepared as described (27) and maintained in serum free Neurobasal medium with B27 supplement (GIBCO). After 5 days neurons were immunostained or transfected by using the calcium-phosphate method. Neurons were incubated in DMEM supplemented with 10 μM 6-cyano-7-ni-troquioxaline-2, 3dione, pH 7.5, for 30 min. Calcium phosphate/DNA precipitates were prepared by adding a mix of DNA and 250 mM CaCl2 dropwise to 2× HBS while vortexing gently, and then added to the cells. After 30 min, the medium was replaced with conditioned medium.
Immunofluorescence.
The following antibodies were: rabbit α-APP, C-terminal (Sigma); mouse α-Numb (Transduction Laboratories, Lexington, KY); and goat α-Notch1 (C20, Santa Cruz Biotechnology). Immunofluorescence was performed as described (27). Images were collected on a LSM 510 confocal microscope (Carl Zeiss).
Luciferase Assays.
Transfections of HeLa, HEK293, and N2a cells were performed by using 3 μl of Fugene and a total of 1 μg of DNA. Each transfection included 100 ng of 4xwt-CBF1-luciferase, 300 ng of NICD or NΔE, and 500 ng of AID-EYFP (unless indicated otherwise; see Fig. 4F) or AID-EYFP mutants. Expression of APP in HeLa Tet-on cells was induced by treating the cells for at least 24 h before transfections with 1 mg/ml of Doxycycline (CLONTECH). Cells were harvested 14 h after transfection.
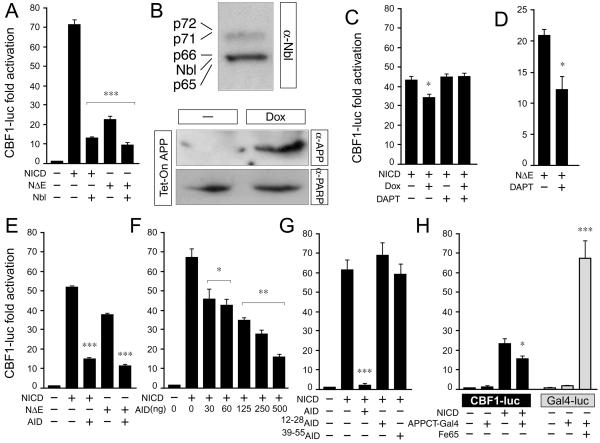
Figure 4.
Processing of APP and release of AID inhibits Notch signaling. (A) HeLa cells were transfected with 4xCBF1-luciferase (CBF-luc) along with the indicated Notch1 and Nbl constructs. Nbl inhibits the transactivation of CBF-luc by both NICD and NΔE. (B) Western blotting (WB) with the α-Nbl antiserum shows expression of Nbl and/or Numb proteins in HeLa cells (Top). WB with α-APP (22C11) shows induction of APP by Dox in APP-HeLa Tet-on cells (Middle). An α-PARP antibody (2C10) was used to normalize for protein loading (Bottom). (C and D) APP-HeLa Tet-on cells were transfected with CBF-luc along with (C) NICD or (D) NΔE. Some samples were treated with Dox to induce APP expression and/or with the γ-secretase inhibitor DAPT (100 nM). The decrease in NICD activity by APP induction is inhibited by DAPT (C). DAPT treatment significantly reduces NΔE activity in HeLa Tet-on cells (D). (E_–_H) HeLa cells were transfected with CBF-luc along NICD, NΔE, AID, or mutant forms of AID as indicated. AID inhibits the activation of CBF-luc by both NICD and NΔE (E). The inhibition is dose-dependent (F) and correlates with the ability of AID to interact with Nbl (G). (H) HeLa cells were transfected with either CBF-luc or GAL4-luciferase (GAL4-luc) reporter genes along with APPCT-Gal4 in the presence or absence of cotransfected Fe65. APPCT-Gal4 reduces NICD activity but enhances Fe65 transactivation. Significance was determined by using a two-tailed Student's t test (*, P < 0.05; **, P ≤ 0 01; ***, P ≤ 0.001).
Neuronal cultures were transfected with 250 ng of the reporter plasmids CBF-luciferase or TP1-luciferase in the presence or absence of cotransfected NICD (500 ng), Nbl (1 μg), APP (1 μg), or AID (0.25–1 μg). The total amount of DNA per well was equalized by the addition of pcDNA3. Cells were harvested 18–20 h after transfection and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega). The total amount of DNA was adjusted to 1 μg by the addition of pcDNA3.1 and/or pEYFP-N1. All luciferase measurements were normalized for transfection efficiency by using TK-renilla luciferase or β-galactosidase. Data are plotted as the means where each point was assayed in triplicate (n ≥ 3 trials).
Results and Discussion
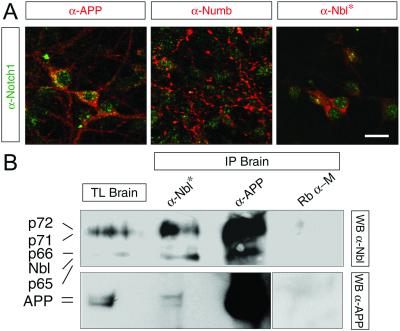
Both APP and Notch undergo a series of sequential proteolytic events resulting in the release of their intracellular domains, AID and NICD, respectively. Intracellular modulators of Notch, such as Nbl and the four Numb isoforms (32, 34, 35) (p65, p66, p71, and p72, are henceforth referred to collectively as Numbs) are potential molecular links between AID and NICD. Like other APP-interacting proteins, Nbl and Numbs contain a phosphotyrosine binding (PTB) domain (32, 34–35) that could bind the Y682ENPTY687 motif present in the cytoplasmic tail of APP (15–17). Notch1, APP, and Numbs/Nbl are present in the mouse brain and are coexpressed by cortical neurons (Fig. 1A). To determine whether endogenous APP, Nbl, and Numbs associate in vivo, mouse brain homogenates were immunoprecipitated with either an α-APP antiserum or an α-Nbl polyclonal antibody, which crossreacts with all four Numb isoforms (34). APP was immunoprecipitated with both α-APP and α-Nbl antibodies (Fig. 1B). Interaction between APP, Nbl, and/or Numbs was confirmed by the reverse experiment, that is, Nbl and Numbs were immunoprecipitated by the α-APP and α-Nbl antibodies but not by the nonspecific rabbit α-mouse IgG antibody (Fig. 1B). Thus, APP interacts with Nbl and/or Numb proteins in mouse brain lysates.
Figure 1.
APP and Numbs/Numb-like are coexpressed with Notch1 in cortical neurons and are coimmunoprecipitated from adult mouse brain. (A) Representative confocal images (1 μm thick) of cultured cortical neurons immunostained for APP, Numb, Numb-like (all red), and Notch1 (green). (Bar = 15 μm.) (B) Coimmunoprecipitation (IP) of APP and Nbl from total lysates (TL) of adult mouse brain was done by using the α-APP (no. 1736) and α-Nbl antibodies. *, α-Nbl polyclonal antibody crossreacts with all four Numb isoforms. Western blotting (WB) was performed with either the α-Nbl antiserum or the α-APP monoclonal antibody 22C11. The rabbit α-mouse IgG (Rb α-M) antibody was used as a negative control.
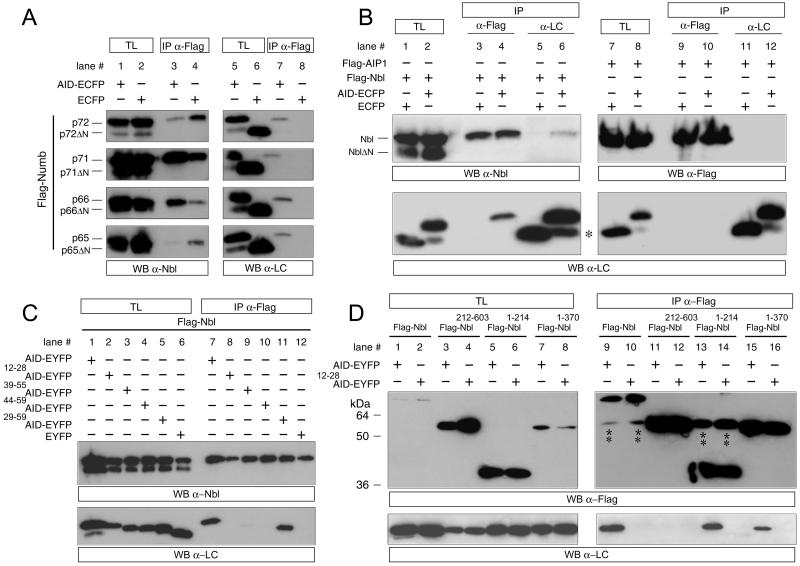
However, given the lack of specificity of the α-Nbl antibody and the difficulty in discriminating the Nbl and Numb proteins by size, this experiment does not resolve whether Nbl and all four Numbs associate with APP. To test directly if APP binds to Nbl and/or all four Numb proteins, HEK293T cells were cotransfected with constructs expressing either ECFP or AID-ECFP along with Flag-Numbs (p65, p66, p71, p72) (Fig. 2A), Flag-Nbl (Fig. 2B), or the unrelated protein Flag-AIP1 (Fig. 2B). Cell lysates were immunoprecipitated with a monoclonal antibody directed against the Flag epitope and were analyzed by Western blot by using the anti-Living Colors (α-LC) antibody, which recognizes ECFP and EYFP. Our results indicated that the four Numb isoforms (Fig. 2A, lane 7, all panels) interact with AID-ECFP but not with ECFP alone (Fig. 2A, lane 8, all panels). Furthermore, by using the same method of immunoprecipitation we found that Nbl (Fig. 2B, lane 4, Lower), but not the unrelated protein AIP1 (Fig. 2B, lane 10, Lower), interacts with AID-ECFP but not with ECFP alone (Fig. 2B, lane 3, Lower). The ability of Nbl to interact with AID-ECFP but not ECFP was confirmed by performing the reverse experiment by using α-LC for the immunoprecipitation and blotting back with an α-Nbl antibody (Fig. 2B, compare lanes 5 and 6, Upper). Therefore, AID-ECFP specifically interacts with all four Numb isoforms and Nbl.
Figure 2.
AID interacts with Nbl and Numbs. (A) HEK293 cells were transfected with either AID-ECFP or ECFP along with Flag-Numb p72 (p72, Upper), Flag-Numb p71 (p71, Upper middle), Flag-Numb p66 (p66, Lower middle), or Flag-Numb p65 (p65, Lower). Cell lysates were coimmunoprecipitated (IP) with either α-Flag or α-Living Colors (α-LC), which recognizes ECFP- and EYFP-tagged proteins. Expression of transfected constructs in total lysates (TL) and immunoprecipitates (IP) also were analyzed by Western blot (WB) with the α-Nbl or α-LC antibodies. p72ΔN, p71ΔN, p66ΔN, and p65ΔN are recognized by the α-Nbl antisera, which is specific for the COOH-terminal region of Numb/Nbl, but are not immunoprecipitated by the α-Flag antibody, which is specific for the Flag epitope fused at the NH2 terminus of these proteins. Therefore, they represent degradation products lacking the NH2-terminal portion and part of the PTB domain, which do not interact with AID-ECFP. (B) HEK293 cells were transfected with AID-ECFP or ECFP along with either Flag-Nbl (lanes 1–6) or the unrelated protein Flag-AIP (lanes 7–12). Lysates were immunoprecipitated with either the α-Flag or α-LC antibodies as indicated. Lysates and immunoprecipitates were analyzed by WB with either the α-Flag, α-LC, or α-Nbl antibodies as indicated. NblΔN is a degradation product of Nbl lacking the NH2-terminal PTB domain and does not interact with AID-ECFP (see above for explanation). * indicates degradation products of AID fusion proteins containing ECFP alone, which does not bind Nbl. (C) HEK293 cells were transfected with Flag-Nbl along with EYFP-tagged deletion construction of AID as indicated. Lysates were immunoprecipitated with α-Flag and analyzed by WB with either α-Nbl or α-LC as indicated. AID mutant numbering is based on the AID59 peptide sequence. (D) HEK 293 cells were transfected with either AID-EYFP or 12–28AID-EYFP along with Flag tagged deletion constructs of Nbl as indicated. Lysates were immunoprecipitated with α-Flag and analyzed by WB with either α-Flag or α-LC as indicated. ** indicates the signal from the α-Flag heavy chain. The difference in molecular mass between AID-EYFP and mutant forms of AID-EYFP, which can be appreciated in (C), is not apparent in (D), because the proteins were separated less extensively by SDS/PAGE.
To characterize the interaction between these proteins further, we transfected deletion mutants of either AID (Fig. 2 C and D) or Nbl (Fig. 2D) into HEK293T cells performing coimmunoprecipitation studies. As for AID, a full-length construct (amino acids 1–59, Fig. 2C, lane 7, Lower), as well as a mutant coding for the COOH-terminal 31 aa (i.e., AID29–59), which includes the YENPTY motif (Fig. 2C, lane 11, Lower), bound Nbl efficiently. Two other mutants encompassing the YENPTY motif plus NH2- and COOH-terminal flanking regions of different lengths (i.e., AID39–55 and AID44–59) bound Nbl with greatly diminished efficiency, such that the interaction was only visible after a longer exposure (Fig. 2C, lanes 9 and 10, Lower; data not shown). The AID12–28 mutant, coding for a portion of AID NH2-terminal to the YENPTY motif, did not interact with Nbl at detectable levels, as expected (Fig. 2C, lane 8, Lower). With respect to Nbl, deletion of the NH2-terminal region comprising the PTB domain and flanking amino-terminal amino acids (Nbl212–603 abolished interaction with full-length AID (Fig. 2D, lane 11, Lower). Conversely, Nbl constructs containing the PTB domain (Nbl1–370 and Nbl1–214 were sufficient to coprecipitate full-length AID (Fig. 2D, lanes 13 and 15, Lower), but not AID12–28, which lacks the YENPTY motif (Fig. 2D, lanes 14 and 16, Lower), demonstrating the specificity of this interaction.
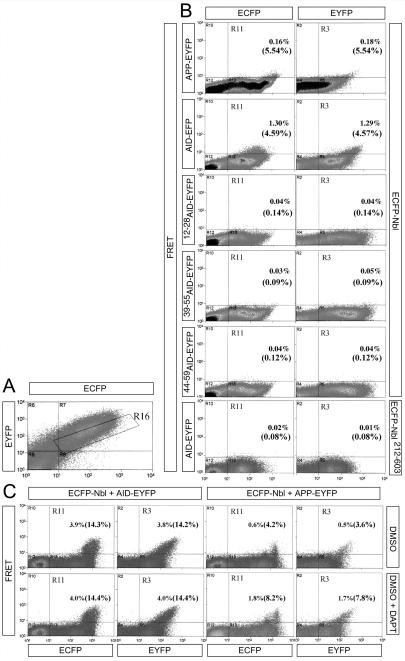
To determine whether AID and Nbl interact in living HEK293T cells, we used FRET. FRET is observed in living cells by fluorescence-activated cell sorting only if an ECFP-fusion protein and an EYFP-fusion protein are in close proximity. When Nbl-ECFP was coexpressed with AID-EYFP, FRET was detected (Fig. 3B). Consistent with the immunoprecipitation data, Nbl-ECFP did not FRET with AID39–55-EYFP, AID44–59-EYFP, or AID12–28-EYFP (i.e., the AID mutants lacking the YENPTY motif), and conversely Nbl212–603-EYFP (i.e., the Nbl mutant lacking the PTB domain) did not FRET with AID-ECFP (Fig. 3B). Furthermore, Nbl-EYFP showed positive FRET when cotransfected with APP-ECFP, suggesting that Nbl associates with membrane-bound APP in vivo (Fig. 3B). Alternatively, the observed FRET may reflect an interaction between Nbl and AID molecules released by the intramembranous cleavage of APP, because EYFP is fused to the COOH terminus of APP and γ-secretase cleavage would result in the release of an AID-EYFP fusion protein. To distinguish between these two possibilities, we used the specific γ-secretase inhibitor _N_-[_N_-(3,5-difluorophenacetyl)-L-alanyl]-_S_-phenylglycine _t_-butyl ester (DAPT) to block production of AID (33). As expected, DAPT did not affect FRET between Nbl and AID (Fig. 3C). However, the number of Nbl/APP-transfected cells showing significant FRET was doubled by treatment with the inhibitor (Fig. 3C), which indicates that Nbl interacts with membrane-associated APP and that a fraction of APP/Nbl complexes are cleaved by γ-secretase, releasing short-lived AID/Nbl complexes. The transitory nature of the AID/Nbl interaction may permit accurate regulation and reversibility of signaling pathways affected by this complex. Nbl and Numbs inhibit Notch activity, at least to some extent, by direct physical interaction with NICD (34). AID may therefore associate with NICD and modulate its activity indirectly through Nbl-Numbs.
Figure 3.
Nbl interacts with AID and membrane-tethered APP in vivo. (A) Representative plot of ECFP vs. EYFP. The R16 gate, containing cells with nearly equal ECFP and EYFP intensity, was used to calculate the percentage of cotransfected cells exhibiting FRET. (B) HEK293 cells were transfected with the indicated EYFP-tagged APP/AID constructs (Left) along with ECFP-Nbl constructs (Right). Samples are plotted based on the intensity of the FRET signal (y axis) versus its ECFP or EYFP emission (x axis, left and right column, respectively). Cells in the R11 (ECFP column) or R3 (EYFP column) gates were scored as positive. Percentages are calculated as the number of positive cells divided by the total number of cells, or, in parentheses, the number of cotransfected cells (gate R16). Although both APP and AID interact with Nbl (as indicated by positive FRET), the AID mutants 12–28AID-EYFP, 39–55AID-EYFP, and 44–59AID-EYFP do not exhibit FRET when coexpressed with ECFP-Nbl. In addition, the Nbl deletion mutant ECFP-Nbl212–603, which lacks the PTB domain, does not interact with AID-EYFP to detectable levels. (C) Effect of the γ-secretase inhibitor DAPT (100 nM) on FRET between Nbl and AID or APP. HEK293 cells were transfected with ECFP-Nbl along with either EYFP-AID or EYFP-APP. Inhibition of γ-secretase activity by DAPT doubles the number of ECFP-Nbl/EYFP-APP transfected cells exhibiting FRET. As expected, DAPT does not affect FRET in ECFP-Nbl/EYFP-AID transfected cells.
Upon cleavage, NICD interacts with the CSL family of transcription factors and translocates to the nucleus where it regulates transcription (18). As shown, Nbl and Numbs inhibit the transactivation of a CBF1-dependent luciferase reporter construct by NICD and NΔE (Fig. 4A) (22, 24, 27). NΔE contains the membrane-spanning region of Notch1, and the release of NICD from NΔE depends on γ-secretase cleavage (29). To determine whether APP and its processing can regulate NICD-dependent transactivation, we used HeLa cells stably expressing APP under the control of a tetracycline-inducible promoter (Fig. 4B). Induction of APP expression significantly reduced (≈25%) NICD-dependent CBF1-luciferase activity (Fig. 4C). If this inhibition were caused by AID peptides derived from APP cleavage, then the γ-secretase inhibitor DAPT should counteract it (33). Treatment with DAPT fully reconstituted the NICD-dependent CBF1-luciferase activity in HeLa cells expressing APP (Fig. 4C). As expected, DAPT did not influence CBF1-luciferase activity driven by NICD in uninduced cells (Fig. 4C), but significantly inhibited (45%) the transactivation of CBF-luciferase by NΔE (Fig. 4D), as the release of NICD from NΔE is a γ-secretase-dependent event. These results indicate that AID molecules released by intramembranous cleavage of APP attenuate NICD function. To test directly for the ability of AID to inhibit NICD function, we transfected AID in HEK293, N2a neuroblastoma, and HeLa cell lines, all of which express endogenous Nbl and/or Numb proteins (Fig. 4 E–G and data not shown). We found that AID significantly inhibited transactivation of the CBF1-luciferase reporter construct by either NICD (Fig. 4 E–G and data not shown) or NΔE (Fig. 4E) in all cell lines tested. The inhibition of CBF1-luciferase by AID was dose-dependent (Fig. 4F) and correlated with the ability of AID molecules to interact with Nbl, such that AID39–55 and AID12–28 did not inhibit CBF1-luciferase activation by NICD (Fig. 4G). To determine whether the inhibitory function of AID on Notch transactivation was specific, we used a construct in which the cytoplasmic tail of APP was fused to the Gal4 DNA-binding domain (i.e., APPCT-Gal4; ref. 13). As described, this construct can promote transcriptional activation of a Gal4-dependent luciferase reporter when coexpressed with the APP-binding protein Fe65 (ref. 19; Fig. 4H, gray bars). Conversely, the presence of APPCT-Gal4 significantly inhibited the transactivation of CBF-luciferase by NICD (Fig. 4H, black bars), indicating that AID either promotes or represses transcription in a signaling-pathway-specific manner.
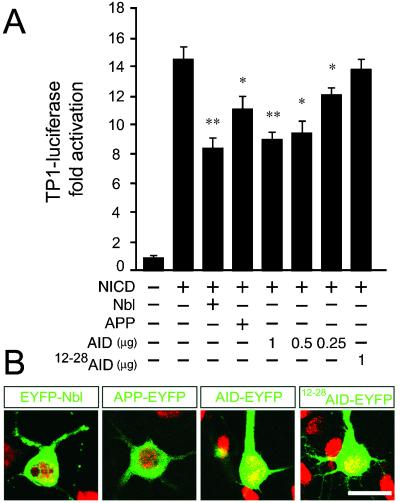
To assess the effect of APP and AID on NICD signaling in neurons, primary neuronal cultures were cotransfected with CSL-dependent luciferase reporters, NICD, and APP/AID constructs. In addition to the CBF1-luciferase reporter construct (30), we also tested the activity of another CSL-dependent regulatory element, the TP1 promoter (31), which regularly elicits much higher transactivation by Notch in neurons (N.S. and B.E.B., unpublished data). As expected, NICD caused an activation of both the CBF1- and TP1-luciferase reporter constructs (Fig. 5A and data not shown). The transactivation of TP1 by NICD was significantly inhibited by coexpression of Nbl (Fig. 5A). APP and AID coexpression also significantly reduced NICD-dependent transactivation of both luciferase reporter constructs (Fig. 5A and data not shown). Furthermore, the inhibition by AID was dose-dependent and correlated with the ability of AID molecules to interact with Nbl, because AID12–28, which does not bind Nbl, did not inhibit the transactivation of TP1-luciferase (Fig. 5A).
Figure 5.
AID inhibits NICD signaling in primary cortical neurons. (A) Cultured cortical neurons (5 to 6 days old) were transfected with TP1-luciferase along with NICD, Nbl, APP, AID, or AID12–28 as indicated. The activity of NICD was inhibited by Nbl, APP, and AID in a dose-dependent manner (0.25, 0.5, and 1 μg/well). No significant change in NICD-dependent transactivation of TP1-luciferase was observed when NICD was cotransfected with AID12–28. Significance was determined by using a two-tailed Student's t test (*, P < 0.05; **, P ≤ 0.01). (B) Representative confocal images (1 μm thick) of cultured cortical neurons transiently transfected with AID-EYFP, AID12–28-EYFP, APP-EYFP, and EYFP-Nbl fusion constructs (shown in green). Neuronal DNA was stained with propidium iodide (shown in red). (Bar = 20 μm.)
Our data indicate that APP processing may physiologically regulate Notch activity. Considering the presence of APP-Nbl/Numb complexes in the brain, it is plausible that AID-Nbl/Numb complexes released as a result of APP processing might bind NICD and antagonize the role of Notch in the generation, differentiation, and survival of neuronal cells. Analysis of the intracellular distribution of EYFP fusion proteins by using confocal microscopy revealed that APP-EYFP and EYFP-Nbl were localized throughout the cytoplasm, including the dendritic and axonal processes, but not the nuclei of cultured neurons (Fig. 5B). In contrast, AID-EYFP and AID12–28-EYFP were found in both the cytoplasm and nuclei of cultured neurons (Fig. 5B), indicating that AID gains access to neuronal nuclei. Although our studies suggest that the inhibition of Notch by AID is mediated by Numbs and/or Nbl, formal proof of this hypothesis will require studies performed in cells deficient for both Nbl and all four Numb isoforms. Furthermore, it remains to be tested whether AID interacts with NICD by a trimeric complex with Numb/Nbl, because Numb and Nbl interact with the intracellular domains of both APP and Notch. Nevertheless, this study suggests that γ-secretase has opposing effects on Notch signaling depending on the proteolytic substrate and the intracellular peptide released: positive by cleavage of Notch, negative by cleavage of APP. Inhibition of Notch function by AID might accelerate the neurodegenerative process of AD by enhancing synapse loss, neurite dystrophy, and neuronal degeneration. Some Presenilin FAD mutants form γ-secretase complexes that produce more AID57 and Aβ42 peptides, whereas they cleave Notch with reduced efficiency and decrease its activity (3, 4). These mutations may thereby have a dual inhibitory effect on Notch signaling suggesting that AD might involve divergent changes in γ-secretase activity (i.e., enhanced on APP and reduced on Notch). According to this view, γ-secretase inhibitors may be detrimental AD drugs not only because of adverse effects due to inhibition of other γ-secretase substrates, either known (2–4) or unknown, but also because Notch inhibition might exacerbate AD pathology. Interfering with the function of AID may prove advantageous in the development of therapeutics to treat and prevent AD.
Acknowledgments
We thank Spyros Artavanis-Tsakonas, Todd Golde, Diane Hayward, Tasuku Honjo, Raphael Kopan, Dennis Selkoe, Thomas Südhof, and Weimin Zhong for generously providing reagents. This work was supported by funds from the Irene Diamond Foundation (to L.D.) and National Institutes of Health Grants NS14841 and AG19394 (to P.R.) and DA15014-01 (to O.M.).
Abbreviations
AD
Alzheimer's disease
APP
β-amyloid precursor protein
AID
APP intracellular domain
Nbl
Numb-like
NICD
Notch intracellular domain
Aβ
amyloid-β peptide
α-LC
anti-living colors
FRET
fluorescence resonance energy transfer
PTB
phosphotyrosine binding
CSL
CBF1-SuH-Lag1
DAPT
_N_-[_N_-(3,5-difluorophenacetyl)-l-alanyl]-_5_-phenylglycine _t_-butyl ester
ECFP
enhanced cyan fluorescent protein
EYFP
enhanced yellow fluorescent protein
References
- 1.Price D L, Tanzi R E, Borchelt D R, Sisodia S S. Annu Rev Genet. 1998;32:461–493. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D J. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 3.Esler W P, Wolfe M S. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- 4.Haass C, De Strooper B. Science. 2001;286:916–919. doi: 10.1126/science.286.5441.916. [DOI] [PubMed] [Google Scholar]
- 5.Passer B, Pellegrini L, Russo C, Siegel R M, Lenardo M J, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimer Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 6.Pinnix I, Musunuru U, Tun H, Sridharan A, Golde T, Eckman C, Ziani-Cherif C, Onstead L, Sambamurti K. J Biol Chem. 2001;276:481–487. doi: 10.1074/jbc.M005968200. [DOI] [PubMed] [Google Scholar]
- 7.Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron M M, Teplow D B, Haass C. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y. J Biol Chem. 2001;276:35235–35238. doi: 10.1074/jbc.C100357200. [DOI] [PubMed] [Google Scholar]
- 9.Yu C, Kim S H, Ikeuchi T, Xu H, Gasparini L, Wang R, Sisodia S S. J Biol Chem. 2001;276:43756–43760. doi: 10.1074/jbc.C100410200. [DOI] [PubMed] [Google Scholar]
- 10.D'Adamio L. In: Notch from Neurodevelopment to Neurodegeneration: Keeping the Fate. Israel A, De Strooper B, Checler F, Christen Y, editors. Berlin: Springer; 2002. [Google Scholar]
- 11.Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B. J Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 12.Kimberly W T, Zheng J B, Guenette S Y, Selkoe D J. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 13.Cao X, Sudhof T C. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Pimplikar S W. Proc Natl Acad Sci USA. 2001;98:14979–14784. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minopoli G, de Candia P, Bonetti A, Faraonio R, Zambrano N, Russo T. J Biol Chem. 2001;276:6545–6550. doi: 10.1074/jbc.M007340200. [DOI] [PubMed] [Google Scholar]
- 16.Scheinfeld M H, Roncarati R, Vito P, Lopez P A, Abdallah M, D'Adamio L. J Biol Chem. 2002;277:3767–3775. doi: 10.1074/jbc.M108357200. [DOI] [PubMed] [Google Scholar]
- 17.Van Gassen G, Annaert W, Van Broeckhoven C. Neurobiol Dis. 2000;7:135–151. doi: 10.1006/nbdi.2000.0306. [DOI] [PubMed] [Google Scholar]
- 18.Artavanis-Tsakonas S, Rand M, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 19.Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner B A. Nat Cell Biol. 2000;2:463–465. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- 21.Struhl G, Greenwald I. Proc Natl Acad Sci USA. 2001;98:229–234. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berechid B E, Kitzmann M, Foltz D R, Roach A H, Seiffert D, Thompson L A, Olson R E, Bernstein A, Donoviel D B, Nye J S. J Biol Chem. 2002;227:8154–8165. doi: 10.1074/jbc.M108238200. [DOI] [PubMed] [Google Scholar]
- 23.Sestan N, Rakic P. In: Notch from Neurodevelopment to Neurodegeneration: Keeping the Fate. Israel A, De Strooper B, Checler F, Christen Y, editors. Berlin: Springer; 2002. [Google Scholar]
- 24.Berezovska O, McLean P, Knowles R, Frosh M, Lu F M, Lux S E, Hyman B T. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- 25.Franklin J L, Berechid B E, Cutting F B, Presente A, Chambers C B, Foltz D R, Ferreira A, Nye J S. Curr Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- 26.Qi H, Rand M D, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 27.Sestan N, Artavanis-Tsakonas S, Rakic P. Science. 1999;286:41–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 28.Redmond L, Oh S R, Hicks C, Weinmaster G, Ghosh A. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 29.Kopan R, Schroeter E H, Weintraub H, Nye J S. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S, Fujimuro M, Hsieh J J, Chen L, Miyamoto A, Weinmaster G, Hayward S D. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Development (Cambridge, UK) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 32.Dho S E, French M B, Woods S A, McGlade C J. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 33.Dovey H F, John V, Anderson J P, Chen L Z, de Saint Andrieu P, Fang L Y, Freedman S B, Folmer B, Goldbach E, Holsztynska E J, et al. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhong W, Jiang M M, Weinmaster G, Jan L Y. Development (Cambridge, UK) 1997;124:1887–1997. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 35.Verdi J M, Bashirullah A, Goldhawk D E, Kubu C J, Jamali M, Meakin S O, Lipshitz H D. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]