Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells (original) (raw)
Abstract
A high degree of aneuploidy characterizes the majority of human tumors. Aneuploid status can arise through mitotic or cleavage failure coupled with failure of tetraploid G1 checkpoint control, or through deregulation of centrosome number, thus altering the number of mitotic spindle poles. p53 and the RB pocket proteins are important to the control of G1 progression, and p53 has previously been suggested as important to the control of centrosome duplication. We demonstrate here that neither suppression of p53 nor of the RB pocket protein family directly generates altered centrosome numbers in any of several mammalian primary cell lines. Instead, amplification of centrosome number occurs in two steps. The first step is failure to arrest at a G1 tetraploidy checkpoint after failure to segregate the genome in mitosis, and the second step is clustering of centrosomes at a single spindle pole in subsequent tetraploid or aneuploid mitosis. The trigger for these events is mitotic or cleavage failure that is independent of p53 or RB status. Finally, we find that mouse embryo fibroblasts spontaneously enter tetraploid G1, explaining the previous demonstration of centrosome amplification by p53 abrogation alone in these cells.
Keywords: aneuploidy‖G1 phase‖pRB
Aneuploidy and chromosomal instability (CIN) are characteristic of the great majority of human tumors (1) and are linked to the progressive development of high-grade, invasive tumors (2–4). Additionally, a high degree of aneuploidy is correlated with poor prognosis (5–7). Recent evidence suggests that aneuploidy may be a necessary intermediate in the formation of many solid human tumors (8).
Aneuploidy can arise through two principle mechanisms. Cells either can proceed through a tetraploid intermediate to a multipolar mitosis that creates random chromosome distribution or can proceed directly to aneuploidy through failure of a critical control of euploidy. Although aneuploidy can arise directly, tetraploidy seems to be a frequent intermediate step toward aneuploidy. In many human carcinomas, cells with tetraploid DNA content arise as an early step in tumorigenesis, preceding the formation of aneuploid cells (9, 10). Human tumors such as esophageal adenocarcinoma (3, 11), cervical carcinoma (12), and rodent tumor model systems (13, 14) proceed through tetraploid intermediates.
Tetraploidy can arise either through disruption of chromosome segregation during mitosis (15–18) or through failure of cytokinesis (19). Progression to aneuploidy then occurs in the absence of a p53-dependent checkpoint control that normally acts in G1 to arrest tetraploid cells.
Alternatively, aneuploidy can arise from mechanisms independent of a tetraploid intermediate. For example, loss of control of centrosome duplication, leading to abnormal centrosome amplification, will create multipolar spindles and aneuploidy. Centrosome duplication normally occurs during S phase (20, 21) through a cdk2 dependent mechanism (22–25), and is under a system of constraint that ensures there is one and only one duplication event during interphase. As a result of fidelity in duplication, a nontransformed cell has two centrosomes at mitosis, which dictate the formation of two spindle poles. If more than one duplication event occurs in interphase, a multipolar spindle could result, and the genome would segregate in an aneuploid manner.
It has been reported that p53 (26) and aurora A (27) are involved in the regulation of centrosome duplication during interphase, as centrosome number abnormalities can result from disruption of either protein's function. However, neither p53 nor aurora A has convincingly been shown to influence centrosome duplication during a single cell cycle. Indeed, Meraldi et al. (28) have recently presented evidence that neither aurora A nor p53 directly controls centrosome number during the course of S phase.
In accord with Meraldi et al. (28), we demonstrate here that p53 status has no direct influence on centrosome duplication in each of several mammalian cell lines. Instead, suppression of p53 causes centrosome amplification through an obligate failure of a G1 tetraploidy checkpoint. Importantly, we further establish that the effect of p53 suppression on centrosome amplification is indistinguishable from results obtained by suppression of the RB pocket proteins. As the RB pocket protein pathway appears to be compromised in virtually all tumors (29–31), our result suggests that the G1 tetraploidy checkpoint is routinely suppressed in tumors, making them vulnerable to both aneuploidy progression and centrosome amplification. In this pathway, we show that centrosome clustering in the tetraploid mitosis that follows escape from G1 tetraploidy is critical to the centrosome amplification process.
Finally, our results offer an explanation for previous findings that appeared to establish a direct link between p53 status and centrosome amplification in mouse embryo fibroblasts (MEFs; ref. 26). We show that p53-competent MEF cells, the model system used in that study, rapidly cease cycling in vitro and become partially tetraploid. However, when either p53 or RB pocket proteins are suppressed, MEFs exhibit the same behavior as we describe for other cells, undergoing centrosome amplification as a result of progression past tetraploid G1 followed by centrosome clustering in mitosis.
Materials and Methods
Generation of p53−/− MEF.
C57B1/6J mice heterozygotic for p53 were purchased from The Jackson Laboratory. Mice were mated and at 13-days gestation, the female was killed. Embryos were washed in PBS, the head and internal organs were removed, and the remainder was minced and then passed through a 1-ml syringe with an 18 gauge needle to disperse cells. Cells were plated on a 10-cm plate coated with 0.1% gelatin and grown in DMEM containing pen-strep and 15% (vol/vol) FBS (HyClone), as described by Harvey et al. (32). Cells were genotyped by PCR according to instructions from The Jackson Laboratory.
Cell Culture.
Primary rat embryo fibroblasts (REF)-52 cells and their simian virus-40 large T antigen-transformed derivatives (TAG; ref. 33) were a kind gift of G. R. Stark (Cleveland Clinic, Cleveland, OH). IMR-90 and VA-13 cells were obtained from Coriell Cell Repositories (Camden, NJ). p53DD REF-52 were prepared by infecting REF-52 with murine retroviruses expressing the p53DD truncated mutant of p53, as described in Andreassen et al. (19). RB pocket protein triple knockout (TKO) MEF were a kind gift of Julien Sage and Tyler Jacks (Massachusetts Institute of Technology, Cambridge, MA) (34).
Cell doubling times at mid-log phase were 28 h for wild-type MEF, 24 h for REF-52, p53DD REF-52, IMR-90, p53−/− MEF and TKO MEF, and 20 h for TAG and VA-13. All cells were cultured as monolayers in DMEM (Invitrogen) supplemented with 10% (vol/vol) FCS (Biological Industries, Beit Haemek, Israel). Cells were maintained in a humid incubator at 37°C in a 5% CO2 environment.
Drug Treatment and Synchronization Protocols.
To arrest cells at the G1/S phase boundary, randomly cycling cells were incubated with 2 mM hydroxyurea for the indicated times. To induce cytokinesis failure, p53DD-expressing REF-52 cells were synchronized in mitosis and then incubated with 10 μM DCB for 5 hours as described (19) and then released in drug-free medium for the indicated times. Alternatively, p53−/− and TKO MEF were exposed to 10 μM DCB for 24 h and then released. Hydroxyurea, DCB, and nocodazole were obtained from Sigma.
Flow Cytometric Analysis.
To analyze cell-cycle profiles, cells were prepared for flow cytometry by using propidium iodide as a marker for DNA content, as described (35). Data were collected by using a FACScan flow cytometer, and results were analyzed by using CELLQUEST software (both from Becton Dickinson). For each sample, 10,000 events were collected, and aggregated cells were gated out.
Immunofluorescence Microscopy.
Cells prepared for immunofluorescence microscopy were grown on poly-d-lysine-coated glass coverslips. To assay for centrosome and spindle pole number, cells were fixed with 2% (wt/vol) paraformaldehyde in PBS at 37°C for 20 min and then permeabilized 3 min with 0.2% Triton X-100 in PBS. Cells were incubated with primary anti γ-tubulin antibodies (Clone GTU-88, Sigma) diluted 1/100 in PBS containing 0.05% Tween-20 and 3% BSA for 1 h at 37°C in a humid chamber. After three washes with PBS, cells were incubated for 30 min with 2.5 μg/ml FITC-conjugated goat anti-mouse IgG secondary antibody from The Jackson Laboratory. DNA was counterstained by the addition of 0.5 μg/ml propidium iodide for 5 min. Cells were observed with an Optiphot II microscope (Nikon) attached to an MRC-600 laser scanning confocal apparatus (Bio-Rad Microscience Division, Herts, England).
Results
Absence of p53 and RB Pocket Protein Family Function Does Not Influence Centrosome Number.
Compromised function of p53 has been linked to centrosome amplification in randomly cycling MEF cells, and suppression of p53 function is frequently associated with aneuploid status of tumors. Abnormal centrosome numbers will give rise to aneuploidy by altering the number of spindle poles. Thus, we first asked whether compromised p53 function had any influence per se on cell ploidy in continuous cell culture. p53 function could be suppressed either by expression of p53DD, a dominant negative form of p53, or by expression of SV40 large T-antigen in REF-52 cells, a nontransformed line of REFs.
As determined by FACScan analysis (Fig. 1), there is no gross change evident in DNA content in p53DD-expressing cells nor in TAG cells after long term culture, in comparison with control REF-52 cells expressing functional p53. As T-antigen suppresses the function of the RB pocket protein family as well as p53 (36), it is also evident that suppression of the RB family does not influence ploidy and, by inference, centrosome number.
Figure 1.
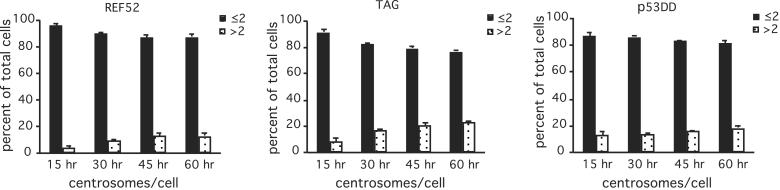
Absence of p53 and RB pocket protein family functions does not modify ploidy nor centrosome number in fibroblast cells. (A) DNA content of random cycling REF-52, p53DD REF-52, and TAG cells, assayed by flow cytometry. Centrosome (B) and spindle pole (C) numbers in the indicated cell lines, determined by microscopic analysis of random cycling populations stained with γ-tubulin and PI. (B) Interphase cells were scored as having a normal number of centrosomes (1 or 2 γ-tubulin spots; black bars) or numbers in excess of two (white bars). (C) Mitotic cells were counted as having either two (black bars) or more than two spindle poles (white bars). In both B and C, values are means of three counts of 100–200 cells each, ± SD.
The apparent retention of euploidy during long-term culture in cells with compromised p53 or RB function corresponds to direct microscopic counts of centrosome numbers during interphase in the different cell lines (Fig. 1B). The small number of cells with more than two distinguishable centrosomes, as determined by γ-tubulin immunofluorescence in control REF-52 cells, is not enhanced by the absence of p53 or RB function. We also assayed centrosome number in a nontransformed human cell line, IMR-90, and a T-antigen transformed cell line derived from IMR-90, VA-13 (Fig. 1B). These cells also show no increase in number of centrosomes.
Increased numbers of centrosomes generate aneuploidy by creating more than two spindle poles during mitosis, forcing random anaphase distribution of chromosomes. An assay of the number of spindle poles during mitosis in the same cell lines (Fig. 1C) found that the frequency of multiple (more than two) spindle poles was equivalently negligible in all cell lines examined.
Duplication of the centrosomes occurs during S phase under the control of Cdk2 and cyclins A and E (22–25). A transformed Chinese hamster ovary cell line, CHO, amplifies centrosome numbers during prolonged S phase arrest in the presence of hydroxyurea (24, 25, 37), an inhibitor of DNA replication (38). However, this effect is not evident in other mammalian cells that have been assayed. To determine whether the absence of p53 function had any influence on centrosome duplication, we assayed for centrosome numbers in REF-52 and in derivative cells expressing p53DD or T-antigen during prolonged arrest with hydroxyurea. Results show little evident effect of compromised p53 or RB function on centrosome duplication during prolonged S phase arrest (Fig. 2).
Figure 2.
Absence of p53 and RB pocket protein family functions does not lead to centrosome amplification in S phase arrested cells. REF-52, p53DD REF-52, and TAG cells were exposed to 2 mM hydroxyurea for varying times and analyzed for centrosome numbers by using anti-γ tubulin antibodies. Cells were counted as having normal number of centrosomes (1 or 2 γ-tubulin spots) or excessive numbers of centrosomes (>2 spots) at each time. Values are means of three counts of at least 200 cells each, ±SD.
Amplification of Centrosome Numbers Occurs in p53DD Cells After Cleavage Failure.
We have previously shown that cleavage failure leads to G1 arrest in p53-competent tetraploid cells, but that the arrest was abrogated by suppression of p53 function in p53DD-expressing cells (19). The implication was that tetraploid status would generate aneuploidy in p53-null cells by their failure to arrest in G1, as cells in the next mitosis would contain supernumerary centrosomes and multiple spindle poles. However, it remained possible that a subpopulation of cells that escaped tetraploidy arrest would go through mitosis with two spindle poles, and thus would inherit abnormal centrosome numbers rather than become aneuploid. Previous studies have shown that multiple centrosomes could indeed cluster at spindle poles (39, 40).
We assayed for this possibility by treating randomly cycling REF-52 p53DD cells with dihydrocytochalasin B (DCB), a drug that interferes with actin assembly and causes cleavage failure (41, 42). At 30 h after release from DCB, nearly a third of the population of interphase cells had more than two centrosomes (Fig. 3A), and the absolute number of centrosomes continued to rise with time, so that by 72 h a subpopulation with four or more centrosomes was evident.
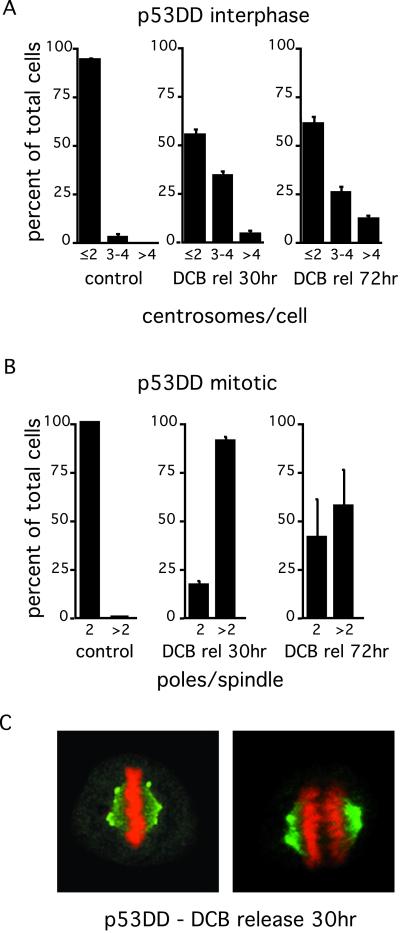
Figure 3.
Centrosome amplification correlates with induction of cleavage failure by DCB in p53-incompetent cells (p53DD REF-52). At the indicated times after release from DCB and after induction of cleavage failure, centrosomes were labeled with anti-γ-tubulin antibodies, and cells were counterstained for DNA with PI. Interphase cells (A) were counted as having 2 or less, 3 or 4, or >4 centrosomes (γ-tubulin spots), and mitotic cells (B) were counted as having 2 or >2 spindle poles. Control refers to random cycling cells. Histograms in A and B represent three counts of 100–200 cells each; bars represent SD. (C) Representative metaphase (Left) and mid-anaphase (Right) cells 30 h after release from DCB, demonstrating bipolar spindles with centrosome clustering at the spindle poles.
These results suggested that centrosome clustering at single poles was indeed occurring during mitosis to create multiple centrosomes in daughter cells. This possibility was assayed in two ways: (i) by determining the percent of mitotic cells with only two spindle poles in the tetraploid mitosis that followed cleavage failure, and (ii) by immunofluorescence demonstration of centrosome clustering at spindle poles in these cells. Thirty hours after release from DCB, approximately 20% of p53DD cells with tetraploid status formed two pole spindles (Fig. 3B), and this population approached 40% by 72 h. By immunofluorescence analysis, clustering of multiple centrosomes at a single spindle pole was common in mitotic cells forming bipolar spindles (Fig. 3C). The presence of multiple centrosomes neither interfered with formation of a normal metaphase plate nor with normal anaphase separation of two chromatid sets (Fig. 3C).
p53−/− and TKO MEF Cells Increase in Centrosome Number by Bypassing Natural Tetraploidy Arrest.
Contrary to the evidence presented above, it has been demonstrated that p53−/− MEFs normally contain augmented centrosome numbers in the absence of experimental manipulation or the introduction of other mutations (26). Thus, we examined MEFs to determine what might explain this exceptional behavior. p53-competent MEFs are unusual in that they stop proliferating after approximately eight passages in vitro (34, 43, 44, and data not shown). In contrast, p53−/− MEFs (32) and TKO MEFs with triple deletion for the RB pocket proteins, RB, p107, and p130 (34, 44), continue to proliferate indefinitely.
FACscan analysis of MEFs at successive passages after introduction into culture showed, to our surprise, that the cells become increasingly tetraploid at each passage (Fig. 4A), such that on progressing from passage 5 to 8, the ratio of 4N to 2N cells rose from 37 to 63%. Thus, there appear to be two causes for cessation of cycling in wild-type MEF. A portion of the 2N population ceases cycling even at early passages, as is evident from failure of a subpopulation to progress from 2N after exposure to DCB and release into nocodazole at passage 3 (Fig. 4B), but another subpopulation increasingly stops cycling with tetraploid status.
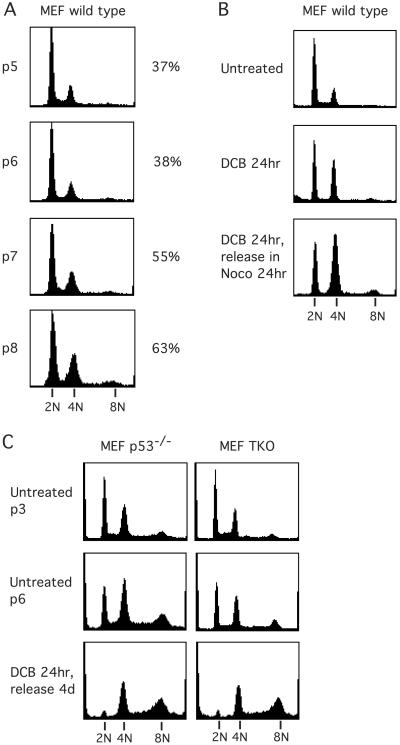
Figure 4.
Spontaneous induction of tetraploidy in MEF cells. (A) DNA content of randomly cycling wild-type MEFs at different passages in culture (as determined by flow cytometry) show that those cells become increasingly tetraploid between passages 5 and 8. Passage number is indicated by “p5-p8” (at left), and the percent indicates the ratio of 4N to 2N cells at that passage (at right). (B) Exposure of wild-type MEFs to DCB at passage 3 leads to accumulation of a subpopulation with 4N DNA. Release from DCB into nocodazole shows no further progression to >4N DNA content, indicating that wild-type MEF cells have a functioning tetraploidy checkpoint. Even at passage 3, a substantial subpopulation remains 2N after DCB and nocodazole exposure and, thus, is not cycling. (C) p53−/− and TKO MEFs also become increasingly tetraploid in culture. These passage 6 cells do not arrest when made tetraploid after induction of cleavage failure by DCB. They continue dividing (data not shown) and progress to hyperdiploid status.
The simplest interpretation of these findings is that a subpopulation of wild-type MEFs become tetraploid through an unknown mechanism, and arrest, probably because of the tetraploidy checkpoint. As we have shown previously for several cell types (19), MEFs treated with DCB for 24 h accumulate a 4N G1 subpopulation, creating a durable block that is retained after release into nocodazole for 24 h, indicating that wild-type MEFs have a tetraploidy checkpoint (Fig. 4B). In contrast, p53−/− and TKO MEFs do not exhibit a tetraploidy checkpoint but progress to hyperdiploid status on release from DCB (Fig. 4C).
As shown above for other cell lines, passage beyond tetraploidy arrest in p53−/− and TKO MEFs generates a substantial subpopulation of cells with amplified centrosome numbers (Fig. 5A). Comparison of the results with p53−/− and TKO MEFs reveals no distinction in centrosome number, regardless of whether p53 or the RB proteins have been suppressed. p53 status, per se, does not directly determine centrosome number in MEFs, because an indistinguishable result is obtained in TKO cells.
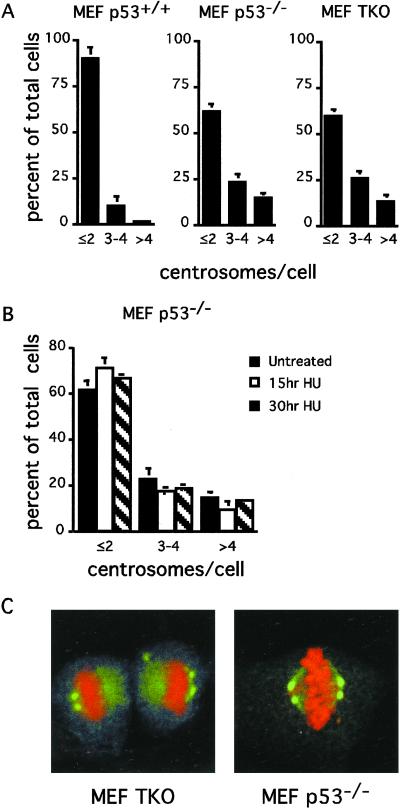
Figure 5.
Neither p53 nor RB pocket protein status per se generates centrosome amplification in MEF cells. (A) Random cycling wild-type, p53−/−, and TKO MEFs were analyzed by microscopy for centrosome number at passage 6. (B) Alternatively, p53−/− MEFs were exposed to 2 mM hydroxyurea (HU) for the indicated times, followed by centrosome number counts. Cells were counted as having a normal number of centrosomes: 1 or 2, 3 or 4, or >4 centrosomes (γ-tubulin spots). Values represent the average of three counts of 200 cells each ± SD. (C) Representative random cycling TKO and p53−/− MEFs that have formed bipolar spindles with centrosome clustering at spindle poles.
In accord with these results, uncontrolled amplification of centrosomes in p53−/− MEFs does not occur during a prolonged S phase block (Fig. 5B). Even though centrosome numbers are amplified in p53−/− cells in the absence of hydroxyurea treatment, S phase arrest does not change the number of centrosomes present. We conclude that the change in centrosome number in MEFs is not caused by deregulation of centrosome duplication controls during S phase.
As shown above, an increase of centrosome number that occurs after passage of cells beyond G1 tetraploidy also involves clustering of centrosomes to form bipolar spindles with multiple centrosomes in tetraploid cells that proceed to mitosis. As expected, multiple centrosomes that are clustered at a single spindle pole in cells with bipolar spindles are common in both p53−/− and TKO populations (Fig. 5C). Remarkably, despite the dispersed nature of centrosome clustering at spindle poles, such unfocused poles are still competent to form functioning bipolar spindles.
Discussion
We have demonstrated that suppression of p53 or of the RB pocket protein family does not directly cause altered centrosome numbers in any of several mammalian primary cell lines. Instead, amplification of centrosome number occurs principally through failure to arrest at a G1 tetraploidy checkpoint after failure to segregate the genome in mitosis. Cells that are tetraploid in G1 contain two centriole pairs and will contain four pairs on entry into the next mitosis.
The second critical element in the generation of cells with amplified numbers of centrosomes is the clustering of multiple centrosomes at a single spindle pole. We demonstrate that centrosome clustering commonly happens in p53−/−- and RB pocket protein-deficient cells with multiple centrosomes. Depending on the distribution of the centrosomes in mitosis, a cell that continues to cycle after failure to arrest in tetraploid G1 can form a two-pole, three-pole, or four-pole spindle in the next mitosis. With a four-pole spindle, a normal number of centrosomes will be restored to highly aneuploid progeny. In contrast, if centrosomes cluster to form a two-pole spindle, the tetraploid status will be maintained in a cell with double the normal complement of centrosomes. All intermediates between these scenarios also can occur, eventually leading to a subpopulation of aneuploid cells with multiple centrosomes.
Equivalent Loss of G1 Tetraploidy Control with Either p53 or RB Suppression.
p53 and the RB pocket proteins are important to the control of G1 progression. p53 is activated in response to DNA damage (45, 46) or after induction of tetraploidy in G1 (16, 17, 19) and operates upstream of the RB pocket proteins. One of its important functions is transactivation of the cyclin-dependent kinase inhibitor p21, which in turn is critical to G1 arrest. As p21 acts largely to suppress RB phosphorylation and the release of E2F, inactivation of the RB pocket protein family abrogates p53/p21 control (for a review, see ref. 29). As a result of the linkage between p53 and RB control, the consequences of evasion of G1 tetraploidy arrest could be, in principle, equivalent in p53 or RB suppressed cells.
An important conclusion from our results is that suppression of p53 or of RB pocket proteins yields an equivalent outcome with respect to induction of aneuploidy and loss of centrosome control in cells that escape G1 tetraploidy control. These results are in accord with our recent demonstration that tetraploidy arrest in G1 after DNA damage depends on p21 activity (47) and, thus, should involve the pathway linking p53 and RB function. Therefore, we conclude that the G1 tetraploidy checkpoint is suppressed in cells with either compromised p53 or RB pocket protein function. Because RB pocket protein function is suppressed in the great majority of tumors, the role of RB pocket proteins in tetraploidy control creates the potential for aneuploidy and centrosome number augmentation in virtually all tumor cells.
Both p53 and RB Status Alter Centrosome Number as a Result of Tetraploidy Checkpoint Override and Mitotic Clustering of Centrosomes.
We have found that neither p53 nor RB pocket protein suppression leads to augmented centrosome numbers during prolonged S phase arrest in the presence of hydroxyurea.
On the basis of work in MEFs, it has been proposed that p53 deficiency directly causes augmentation in centrosome numbers and that this amplification in turn causes aneuploidy (26). Several lines of evidence argue against this possibility. First, p53 knockout mice are euploid and viable (32) and second, CIN in tumors strongly correlates with mutant bub1, bubR1 (48), and APC (49) but does not appear to correlate with p53 status (47). Further, as we show here, with the exception of MEF cells, cells deficient in p53 remain indistinguishable from nontransformed parental cells with respect to DNA profile and centrosome number during long-term culture.
An alternative possibility to account for augmentation of centrosome number in p53-compromised cells is through failure to arrest in G1 after tetraploidization. Frequently, tetraploidy override rapidly results in aneuploidy (15, 19, 47). As we have proposed (15), progression of tumors to a highly aneuploid condition may occur in at least two discrete steps. First, a primary control in mitosis or cell cleavage fails, leading to tetraploid daughter cells. Second, failure of a p53, or of RB-dependent surveillance mechanisms that normally arrest such tetraploid cells in G1, leads to aberrant cell cycles. As we show here, progression to aneuploidy is conditional on the clustering of centrosomes at the mitotic spindle poles in tetraploid cells. When centrosomes cluster, a two-pole spindle forms and tetraploidy is maintained but centrosome number augments in progeny cells. When centrosomes fail to cluster, a normal centrosome number can be retained in highly aneuploid progeny.
What causes the immediate mitotic or cleavage failure that sets the stage for generation of aneuploidy and change in centrosome number in p53- or RB-compromised cells? A reasonable possibility is that mutations in bub1, bubR1, APC, BRCA1, or other proteins controlling mitotic checkpoint function and accuracy of mitotic segregation could create the conditions for p53- or RB-compromised cells to continue cycling past a G1 tetraploid status, with evident consequences for tumorigenesis.
The only exception to the requirement for mitotic or cleavage failure to set the stage for G1 tetraploidy checkpoint override and consequent centrosome number abnormality are MEF cells that have served as the model system to support the claim that p53 directly controls centrosome number (26). As we show here, MEFs are indeed exceptional. They spontaneously cease cycling in both euploid G1 and in tetraploid G1 after approximately eight passages in vitro. The cause of euploid G1 arrest remains unknown, as does the cause of mitotic or cleavage failure that leads to tetraploid G1 arrest. The outcome, however, is clear. Both aneuploidy and abnormal centrosome numbers arise spontaneously in both p53−/− and in TKO MEFs, most likely by override of the G1 tetraploidy checkpoint. Thus, the generation of abnormal centrosome numbers in MEFs arises in the same manner as in other mammalian cells.
Consequences for Tumor Chemotherapy.
Mitotic inhibitors such as vinblastine, estramustine, and taxol, that cause mitotic failure and entry into tetraploid G1, are used successfully in chemotherapy for a variety of tumors (50). Although nontransformed cells arrest in tetraploid G1, p53-compromised cells continue to cycle, a property, as we show here, shared by RB pocket protein-compromised cells. It is possible that the capacity of these drugs to kill tumor cells is linked to their progression to a largely nonviable aneuploid status. We show that clustering of centrosomes is a major phenomenon in response to augmentation in centrosome number, and clustering can maintain cells by repeatedly generating two-pole spindles and, thus, retaining tetraploid status. Success with mitotic inhibitors in tumor therapy thus may be linked to the relative success of tetraploid cells in clustering centrosomes, which may vary among cell types. Clustering would lead to progeny cells with multiple centrosomes but no aneuploidy, whereas lack of clustering would generate normal centrosome numbers but gross aneuploidy.
Acknowledgments
We thank Julien Sage and Tyler Jacks for providing TKO MEFs, and Moshe Oren for providing p53DD retrovirus producing cells. This work was supported by grants to R.L.M. from La Ligue Nationale contre le Cancer (Laboratoire Labelisé) and l'Association pour la Recherche sur le Cancer (ARC). F.B. was supported by AssociationEspoir Isère contre le Cancer and Agence Nationale de Recherche sur le Sida. O.D.L. was supported by le Commissariat à l'Energie Atomique and ARC.
Abbreviations
MEF
mouse embryo fibroblasts
REF
rat embryo fibroblasts
TAG
simian virus–40 T antigen-transformed REF-52 cells
DCB
dihydrocytochalasin B
TKO
RB pocket protein triple knockout
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 2.Sandberg A A. Cancer Res. 1977;37:2950–2956. [PubMed] [Google Scholar]
- 3.Rabinovitch P S, Reid B J, Haggitt R C, Norwood T H, Rubin C E. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- 4.Giaretti W. Lab Invest. 1994;71:904–910. [PubMed] [Google Scholar]
- 5.Schutte B, Reynders M M, Wiggers T, Arends J W, Volovics L, Bosman F T, Blijham G H. Cancer Res. 1987;47:5494–5496. [PubMed] [Google Scholar]
- 6.Stephenson R A, James B C, Gay H, Fair W R, Whitmore W F, Jr, Melamed M R. Cancer Res. 1987;47:2504–2507. [PubMed] [Google Scholar]
- 7.Fallenius A G, Auer G U, Carstensen J M. Cancer. 1988;62:331–341. doi: 10.1002/1097-0142(19880715)62:2<331::aid-cncr2820620218>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. Proc Natl Acad Sci USA. 2000;97:3236–3241. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 10.Shackney S E, Smith C A, Miller B W, Burholt D R, Murtha K, Giles H R, Ketterer D M, Pollice A A. Cancer Res. 1989;49:3344–3354. [PubMed] [Google Scholar]
- 11.Reid B J, Haggitt R C, Rubin C E, Rabinovitch P S. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- 12.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ornitz D M, Hammer R E, Messing A, Palmiter R D, Brinster R L. Science. 1987;238:188–193. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- 14.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 15.Andreassen P R, Martineau S N, Margolis R L. Mutat Res. 1996;372:181–194. doi: 10.1016/s0027-5107(96)00138-8. [DOI] [PubMed] [Google Scholar]
- 16.Minn A J, Boise L H, Thompson C B. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 17.Lanni J S, Jacks T. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart Z A, Leach S D, Pietenpol J A. Mol Cell Biol. 1999;19:205–215. doi: 10.1128/mcb.19.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreassen P R, Lohez O D, Lacroix F B, Margolis R L. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuriyama R, Dasgupta S, Borisy G G. Cell Motil Cytoskeleton. 1986;6:355–362. doi: 10.1002/cm.970060402. [DOI] [PubMed] [Google Scholar]
- 21.Piel M, Nordberg J, Euteneuer U, Bornens M. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 22.Hinchcliffe E H, Li C, Thompson E A, Maller J L, Sluder G. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 23.Lacey K R, Jackson P K, Stearns T. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Hayashi K, Nishida E. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 25.Meraldi P, Lukas J, Fry A M, Bartek J, Nigg E A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 26.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, Kuang J, Zhong L, Kuo W L, Gray J W, Sahin A, Brinkley B R, Sen S. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 28.Meraldi P, Honda R, Nigg E A. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 30.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C J. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 32.Harvey M, Sands A T, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 33.Perry M E, Commane M, Stark G R. Proc Natl Acad Sci USA. 1992;89:8112–8116. doi: 10.1073/pnas.89.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sage J, Mulligan G J, Attardi L D, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trielli M O, Andreassen P R, Lacroix F B, Margolis R L. J Cell Biol. 1996;135:689–700. doi: 10.1083/jcb.135.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalvide J, DeCaprio J A. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balczon R, Bao L, Zimmer W E, Brown K, Zinkowski R P, Brinkley B R. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timson J. Mutat Res. 1975;32:115–132. doi: 10.1016/0165-1110(75)90002-0. [DOI] [PubMed] [Google Scholar]
- 39.Ring D, Hubble R, Kirschner M. J Cell Biol. 1982;94:549–556. doi: 10.1083/jcb.94.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingle W L, Salisbury J L. Am J Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubin J E, Osborn M, Weber K. Exp Cell Res. 1981;136:63–79. doi: 10.1016/0014-4827(81)90038-0. [DOI] [PubMed] [Google Scholar]
- 42.Martineau S N, Andreassen P R, Margolis R L. J Cell Biol. 1995;131:191–205. doi: 10.1083/jcb.131.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 44.Dannenberg J H, van Rossum A, Schuijff L, te Riele H. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 46.Cox L S, Lane D P. BioEssays. 1995;17:501–508. doi: 10.1002/bies.950170606. [DOI] [PubMed] [Google Scholar]
- 47.Andreassen P R, Lacroix F B, Lohez O D, Margolis R L. Cancer Res. 2001;61:7660–7668. [PubMed] [Google Scholar]
- 48.Cahill D P, Kinzler K W, Vogelstein B, Lengauer C. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 49.Tighe A, Johnson V L, Albertella M, Taylor S S. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan M A, Wilson L. Curr Opin Cell Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]