Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint (original) (raw)
Abstract
Mad2 is a key component of the spindle checkpoint, a device that controls the fidelity of chromosome segregation in mitosis. The ability of Mad2 to form oligomers in vitro has been correlated with its ability to block the cell cycle upon injection into Xenopus embryos. Here we show that Mad2 forms incompatible complexes with Mad1 and Cdc20, neither of which requires Mad2 oligomerization. A monomeric point mutant of Mad2 can sustain a cell cycle arrest of comparable strength to that of the wild-type protein. We show that the interaction of Mad2 with Mad1 is crucial for the localization of Mad2 to kinetochores, where Mad2 interacts with Cdc20. We propose a model that features the kinetochore as a ‘folding factory’ for the formation of a Mad2–Cdc20 complex endowed with inhibitory activity on the anaphase promoting complex.
Keywords: anaphase promoting complex/Cdc20/Mad1/Mad2/mitotic spindle checkpoint
Introduction
The spindle checkpoint ensures fidelity of chromosome segregation by halting sister chromatid separation in cells with a defective mitotic spindle (reviewed in Hardwick, 1998; Amon, 1999; Shah and Cleveland, 2000). Mitotic-arrest deficient (MAD) and budding uninhibited by benzimidazole (BUB) mutants impair the ability of Saccharomyces cerevisiae to arrest cell cycle progression as a consequence of spindle damage (Hoyt et al., 1991; Li and Murray, 1991). Homologous proteins in higher eukaryotes are involved in functionally equivalent pathways. Vertebrate Mad1, Mad2, Bub1, Bub1R and Bub3 proteins associate with kinetochores in prometaphase, from which they detach in metaphase and anaphase, consistent with a role in processes that monitor attachment of sister chromatids to the spindle (Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998).
Genetic and biochemical evidence suggests that the spindle checkpoint requires the interaction of Mad2 with the anaphase promoting complex (APC). Among other substrates, this multisubunit ubiquitin ligase targets the anaphase inhibitor securin and B-type cyclins for degradation, triggering chromosome segregation at the metaphase–anaphase transition (reviewed in Zachariae and Nasmyth, 1999). APC inhibition during checkpoint activation is believed to occur through an interaction of Mad2 with Cdc20 (Li et al., 1997; Fang et al., 1998b; Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Wassmann and Benezra, 1998), a seven WD40-repeat-positive regulator of the APC. Yeast Cdc20 mutants that lose the ability to complex with Mad2 are unable to arrest cells in metaphase upon Mad2 or Mps1 overexpression (Hwang et al., 1998; Kim et al., 1998). The Cdc20–Mad2 interaction is direct, and mediated by a short fragment of Cdc20 located immediately upstream of the WD40 repeats (Hwang et al., 1998; Kim et al., 1998; Luo et al., 2000; Zhang and Lees, 2001). The formation of a Mad2–Cdc20 complex is not limited to cells with an active spindle checkpoint, as the complex can be precipitated from HeLa cells progressing through mitosis, starting during prometaphase, and until proper chromosome attachment to the mitotic spindle has been achieved (Wassmann and Benezra, 1998). Thus, the Mad2–Cdc20 complex may be required for the control of progression through mitosis in the normal cell. Consistent with this hypothesis, it was shown that Mad2 is an essential gene in mammals (Dobles et al., 2000). A correlation between ablation of spindle checkpoint function and the generation of chromosome instability (CIN), a property of many cancer types, has been proposed (Lengauer et al., 1998). In agreement with this proposal, disruption of one Mad2 allele causes high rates of chromosome loss, impaired checkpoint function, and tumorigenesis (Michel et al., 2001).
Human Mad2 is almost entirely spanned by the Horma domain (Aravind and Koonin, 1998). Its structure consists of a central helical layer flanked on one side by a large β-sheet, and on the other by a long and irregular β-hairpin. Several conserved Mad2 residues map to the solvent-exposed face of the large β-sheet, and possibly identify the location of a binding site for interacting proteins (Luo et al., 2000). Recombinant Mad2 exists with different quaternary structures (Fang et al., 1998a). When injected into Xenopus blastomeres, an oligomeric form of Mad2 (possibly corresponding to a tetramer, and thus defined as Mad2t) inhibits the destruction of histone H1 activity, revealing the induction of a metaphase block (Fang et al., 1998a). When added to frog mitotic extracts, Mad2t blocks the activity of the APC. A monomeric form of the protein (Mad2m), which co-exists with the tetramer in biochemical preparations and does not convert to Mad2t even at high protein concentrations, is unable to cause cell cycle arrest in Xenopus embryos (Fang et al., 1998a). Monomeric versions of Mad2 were also generated by 10 residue N- or C-terminal deletions (Mad2ΔN and Mad2ΔC). When incubated in frog extracts, these monomeric forms of Mad2 were not able to inhibit APC activity (Fang et al., 1998a).
Although a straightforward interpretation of these data is that the conversion of Mad2 to a monomer hampers Cdc20 binding, this does not appear to be the case. First, Mad2ΔN retains unaltered ability to interact with Cdc20 in vitro, suggesting that the oligomerization of Mad2 is not required for Cdc20 binding (Luo et al., 2000). Moreover, binding of Mad2t to Cdc20 elicits a dramatic conformational change in Mad2 that is associated with loss of Mad2 oligomerization, and results in the formation of a heterodimeric 1:1 complex devoid of further oligomerization (Luo et al., 2000). Also in this regard Mad2ΔN behaves indistinguishably from the wild-type protein, indicating that the oligomerization of Mad2 is also not important for the conformational change imposed by Cdc20 binding (Luo et al., 2000). Furthermore, when introduced into Xenopus mitotic extracts, both Mad2m and Mad2ΔC have a dominant-negative effect on the ability of Mad2t to cause APC inhibition, indicating that their Cdc20 binding functionality may be preserved at least in part (Fang et al., 1998a).
The role of oligomerization in Mad2 function is made less clear by the fact that self-assembling forms of this protein in vivo have not been described. In this study, we decided to investigate whether the oligomerization of Mad2 may be relevant for the interaction of this protein with other components of the spindle checkpoint. In particular, we concentrated our attention on Mad1, a 718 residue protein that is predicted to form a coiled coil along most of its length (Hardwick and Murray, 1995; Chen et al., 1998; Jin et al., 1998). Budding yeast Mad1 and Mad2 interact tightly in a high-molecular-weight complex, and immunodepletion of Xmad2 from Xenopus extract leads to co-depletion of Xmad1, suggesting the existence of a strong complex between these proteins (Chen et al., 1998, 1999). The relevance of Mad1 for the spindle checkpoint is emphasized by the finding that neither Mad2 nor Mad3 associates with Cdc20 in a budding yeast strain lacking Mad1, indicating that Mad1 is essential for the formation of APC inhibitory complexes (Hwang et al., 1998). It was also shown that the localization of Mad2 to kinetochores is impaired in Xenopus egg extracts depleted of Mad1, suggesting that Mad1 may play a central role in the localization of Mad2 (Chen et al., 1998). Here, we show that the binding of Mad2 to Mad1 and Cdc20, and not its oligomerization, is essential for spindle checkpoint function. We show that Mad2 forms distinct complexes with Mad1 and Cdc20, and that its oligomerization is dispensable for either interaction. Furthermore, we show that a monomeric point mutant of Mad2 that preserves its ability to interact with Mad1 and Cdc20 in vitro and in vivo supports cell cycle arrest upon co-expression of Mad1 to a level indistinguishable from that of the wild-type protein. The implications of our results are discussed.
Results
Isolation of recombinant Mad1–Mad2 complexes
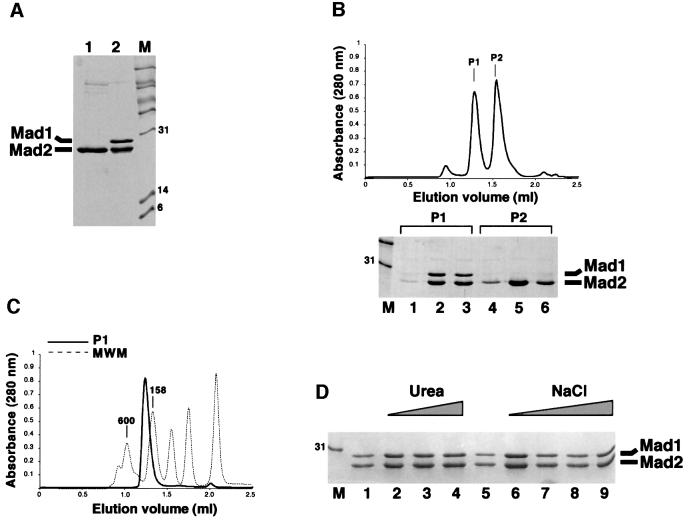
Residues 485–718 of human Mad1 (Mad1485–718) are sufficient for Mad2 binding (Jin et al., 1998). Recombinant Mad1–Mad2 was generated by bacterial co-expression of Mad1486–719 with His-tagged Mad2 (Figure 1A). For simplicity, this complex will be referred to as Mad1–Mad2. The complex was purified from bacterial lysates by immobilized metal-affinity chromatography (IMAC). N-terminal sequencing confirmed that the 27 kDa protein product co-purifying with Mad2 is Mad1. Further purification of the complex after imidazole elution was carried out by size-exclusion chromatography (SEC). This revealed the presence of two peaks (P1 and P2, Figure 1C). P1 contained the Mad1–Mad2 complex, while P2 contained only oligomeric Mad2. This excess of Mad2 originates from higher expression levels of this protein relative to Mad1, and not from disruption of the complex. In fact, Mad1 eluted exclusively in P2, indicating that disruption of the Mad1–Mad2 complex does not occur during SEC. Furthermore, when P1 was concentrated to 4 mg/ml and reloaded onto the size-exclusion column, a single peak was observed, suggesting that there is no significant disruption of the complex upon gel filtration. The elution volume of Mad1–Mad2 was compatible with a molecular mass of 250 kDa (Figure 1C).
Fig. 1. (A) Purification of Mad2 and Mad1485–718–Mad2 (lanes 1 and 2, respectively) by IMAC. (B) SEC profile of the material eluted after IMAC. Mad1–Mad2 is contaminated with free Mad2, resulting from higher expression levels of this protein relative to Mad1. By SDS–PAGE, peak 1 (P1) contains only Mad1–Mad2. The excess of Mad2 is contained in peak 2 (P2). (C) The P1 peak shown in (B) was concentrated and analysed by SEC, and a single peak appeared (thick line). The elution volume can be compared with that of known protein standards (thin dashed lines). (D) Mad1–Mad2 was incubated with urea concentrations of 1, 2 or 4 M (lanes 2–4), and NaCl concentrations of 1, 2, 3 or 4 M (lanes 6–9), and metal-affinity beads were added. Mad1 still co-purified with Mad2 after this treatment, indicating that the interaction between these proteins is very strong.
Budding yeast Mad1–Mad2 is resistant to high concentrations of salt and chaotropic agents (Chen et al., 1999). To test whether this was also a characteristic of the purified human complex, we incubated Mad1–Mad2 with different concentrations of urea or NaCl. Metal-affinity beads were then added to purify Mad2 and any Mad2-bound Mad1. SDS–PAGE of this material showed that concentrations of urea up to 4.0 M and of NaCl up to 2.0 M failed to produce significant disruption of the Mad1–Mad2 complex (Figure 1D). No association of Mad1 with Mad2 was observed in non-reducing gels, indicating that the two molecules are not covalently bound (not shown). Thus, Mad1485–718 is sufficient to establish a direct, high-affinity interaction with Mad2.
Effects of short N- and C-terminal deletions of Mad2 upon Mad1 binding
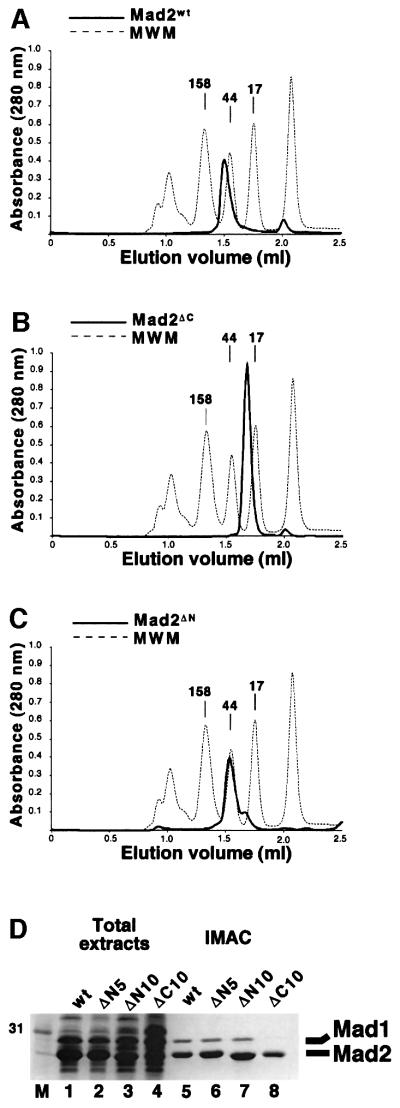
We expressed human Mad2 (Mad2wt) together with 10 residue N- and C-terminal deletion mutants (Mad2ΔN and Mad2ΔC, respectively) in bacteria as fusions to an N-terminal His tag. As it has been reported that Mad2ΔN and Mad2ΔC have altered oligomerization properties relative to Mad2wt (Fang et al., 1998a), we analysed Mad2 and its deletion mutants by SEC. The proteins were purified by IMAC, eluted, and loaded onto a Superdex 200 column at concentrations as low as 1 mg/ml. Mad2wt eluted as an oligomer of ∼80 kDa (Figure 2A). In contrast, Mad2ΔC eluted as a monomeric species (Figure 2B). Although this confirms that the C-terminal region of Mad2 is important for oligomerization, a synthetic peptide containing the Mad2 C-terminal region was unable to reverse the oligomerization of Mad2wt (data not shown). Mad2ΔN showed a prevalent peak corresponding to the size of a dimer, followed by a partly overlapping peak corresponding to a monomer (Figure 2C). In our hands, the effects of this deletion on Mad2 oligomerization are somewhat milder than previously reported (Fang et al., 1998a).
Fig. 2. (A) His-Mad2wt (thick line) was purified by IMAC and analysed by SEC. The elution profile was compared with that of known standards (MWM, thin dashed line). (B and C) SEC analysis of Mad2ΔC and Mad2ΔN. (D) The ability of Mad2wt (wt), 5- and 10-residue N-terminal deletions of Mad2 (ΔN5 and ΔN10, respectively) and Mad2ΔC (ΔC) to interact with Mad1 was evaluated using the co-expression system described in the text, followed by IMAC purification. Note that Mad2ΔC is unable to interact with Mad1.
Next, we tested the ability of the Mad2 mutants to interact with Mad1. Mad2ΔN and Mad2ΔC were co-expressed with Mad1486–719. The lysates were incubated with metal-affinity beads, and bound proteins were analysed by SDS–PAGE. Deletion of 5 or 10 residues from the N-terminus of Mad2 did not affect its ability to interact with Mad1 (Figure 2D, lanes 5–7), a result that was confirmed by SEC analysis of the eluates (not shown). On the other hand, binding of the C-terminal deletion mutant to Mad1 was completely impaired (Figure 2D, lane 8), despite the presence of normal levels of Mad1 in the bacterial lysates.
Monomeric Mad2 can bind Mad1 and Cdc20 in vitro
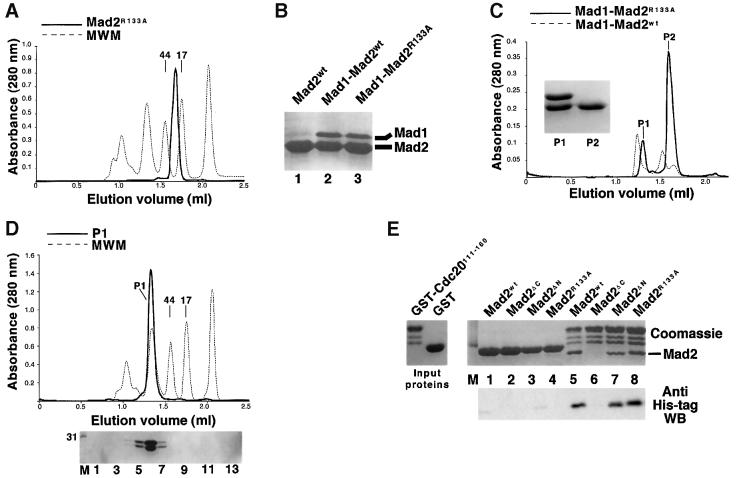
These data suggest a possible correlation between Mad2 oligomerization and Mad1 binding, in a way that is reminiscent of the Mad2–Cdc20 interaction. It should be noted, however, that our Mad1 binding assay does not allow us to establish whether the monomers and dimers that co-exist in our preparation of Mad2ΔN can both bind to Mad1 or not. An equally plausible interpretation is that the C-terminal region of Mad2 is required both for Mad1 and Cdc20 binding, and for oligomerization, so that its removal affects all three functions. On the other hand, the N-terminal deletion may partially affect oligomerization without seriously affecting Mad1 or Cdc20 binding. To verify this hypothesis, we searched for Mad2 point mutants with an enhanced monomerizing effect relative to the N-terminal deletion mutant, but maintaining the ability of the wild-type protein to interact with Cdc20 and Mad1. To isolate such a mutant, we introduced individual alanine point mutations into eight positions of Mad2 (listed in Materials and methods). On the assumption that oligomerization may require a solvent-exposed residue of Mad2, we selected surface residues of Mad2 using its three-dimensional structure as a guide (Luo et al., 2000). We used SEC analysis to evaluate the propensity of the mutants for self-association. Mad1 binding was evaluated using the co-expression strategy described above, while Cdc20 binding was studied using a semi-quantitative solid-phase binding assay described below. SEC analysis showed that the Arg133 to Ala mutant (Mad2R133A) behaved as a monomer, with no evidence of oligomeric species (Figure 3A). However, the mutant retained unaltered ability to complex with Mad1 (Figure 3B). The Mad1–Mad2R133A complex could be purified to homogeneity using a strategy similar to that followed with the complex containing Mad2wt. A SEC run was sufficient to separate Mad1–Mad2R133A from the excess of Mad1-free Mad2R133A purified from bacteria (Figure 3C). When the complex was separated again using SEC, we did not observe free Mad1 or Mad2 in high- or low-molecular-weight fractions, suggesting that there was no disruption of the Mad1–Mad2R133A complex, and that its stability is similar to that of the wild-type complex (Figure 3D). Interestingly, the mutant complex eluted with an apparent mol. wt of 160 kDa, showing a shift in elution volume relative to Mad1–Mad2wt (250 kDa).
Fig. 3. (A) Mad2R133A (thick line) is monomeric by gel filtration. (B) The ability of Mad2R133A to interact with Mad1 was evaluated using the co-expression system described in the text, followed by IMAC purification, and separation of the bound proteins by SDS–PAGE. Similarly to Mad2wt, the Mad2R133A mutant co-purifies with Mad2. (C) Elution profile of Mad1–Mad2R133A (thick line) compared with the elution profile of Mad1–Mad2wt (thin dashed line). (D) The P1 peak shown in (C) was concentrated and analysed by SEC. A single peak appeared (thick line). There is no disruption of the Mad1–Mad2R133A complex upon SEC, as indicated by the complete absence of free Mad1 or Mad2R133A upon SDS–PAGE separation (bottom panel). Thirteen 100 µl fractions (1–13) were collected and analysed, starting at an elution volume of 0.8 ml. (E) The forms of Mad2 indicated were incubated either with GST (lanes 1–4) or GST–Cdc20111–160 on GSH–agarose beads. After washing, bound proteins were resolved by SDS–PAGE. Mad2ΔC (lane 6) was unable to bind to Cdc20, whereas the N-terminal deletion mutant (lane 7) and Mad2R133A (lane 8) bound like the wild-type protein (lane 5). The results were confirmed by western blotting using an anti-His tag antibody.
Next, we tested the ability of Mad2R133A to interact with Cdc20. A glutathione _S_-transferase (GST) fusion protein containing residues 111–160 of Cdc20 (GST–Cdc20111–160) was adsorbed to glutathione–agarose beads, and the beads were diluted to obtain a final protein concentration of 0.4 µM. Mad2wt, Mad2ΔC, Mad2ΔN and Mad2R133A were added at a concentration of 4 µM (see Materials and methods for details). After incubation, the beads were washed, and all bound proteins were boiled in sample buffer and separated by SDS–PAGE. Mad2wt, Mad2ΔN and Mad2R133A bound with apparently similar affinity to GST–Cdc20111–160 (Figure 3E, lanes 5, 7 and 8), while Mad2ΔC did not bind. We also carried out a surface plasmon resonance analysis using the same combination of immobilized and liquid-phase proteins using a Biosensor instrument. An accurate evaluation of the dissociation constant (_K_D) of the Mad2–Cdc20 interaction was hampered by the slow kinetics of Mad2 binding to Cdc20, probably a consequence of the conformational change imposed on Mad2 by the interaction with Cdc20 (Luo et al., 2000). The Biosensor analysis, however, confirmed that Mad2R133A binds Cdc20 as effectively as the wild-type protein (not shown). We estimate that the _K_D of the Mad2–Cdc20 interaction might be in the nanomolar range, as even at the low protein concentrations used in our binding assay we often observed saturation of immobilized Cdc20 by Mad2, even after extensive washing.
Mad2 forms distinct complexes with Mad1 and Cdc20 in vitro
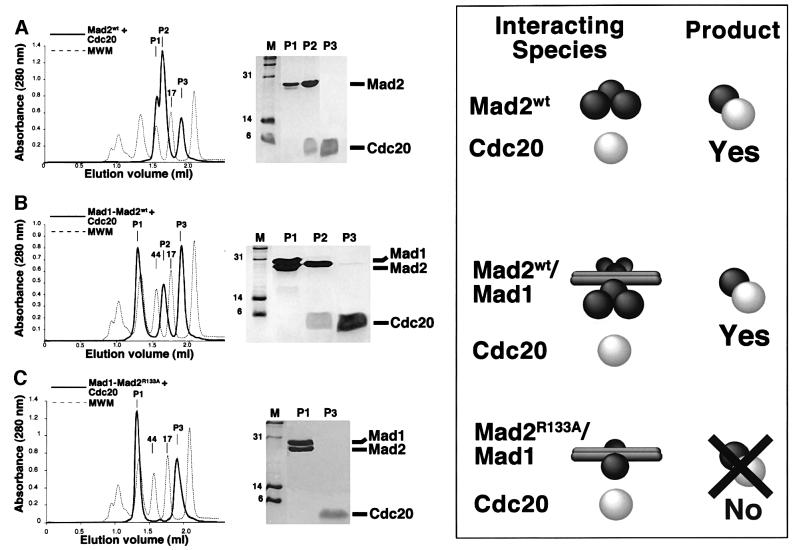
In summary, Mad2R133A is a monomeric point mutant retaining the ability to interact with Mad1 and Cdc20. We next asked whether Mad2 was able to form a ternary complex with Cdc20 and Mad1, or rather, whether these interactions were mutually incompatible. For this we generated a synthetic peptide corresponding to residues 111–154 of human Cdc20 (Cdc20111–154). This region of Cdc20 is sufficient for a high-affinity interaction with Mad2 (Luo et al., 2000; Zhang and Lees, 2001). Purified Mad2 was mixed with this peptide at an approximate molar ratio of 1:5, and the resulting mixture separated by SEC. While Mad2wt ran as a single peak under these conditions (Figure 2A), three peaks (P1–3) appeared after incubation with Cdc20. SDS–PAGE analysis revealed that the three peaks corresponded to unliganded oligomeric Mad2 (P1), a Mad2–Cdc20111–154 complex (P2) and free Cdc20111–154 (P3), respectively (Figure 4A). Mad2– Cdc20111–154 eluted at a volume compatible with a mol. wt of ∼30 kDa, indicating the existence of a 1:1 complex devoid of further oligomerization, confirming that Cdc20 reverses the oligomerization of Mad2 in vitro (Luo et al., 2000).
Fig. 4. (A) Mad2wt was incubated with a synthetic peptide containing residues 111–154 of Cdc20, and the mix separated by SEC (left panel, thick line). The content of the peaks was visualized by SDS–PAGE (central panel). The right-hand site of the figure is a cartoon representing our interpretation of the results. (B) When the same experiment was carried out with purified Mad1–Mad2wt complex (running as a single peak, Figure 1C), two new peaks appeared. P2 contains Mad2 and the Cdc20 peptide, while P3 contains an excess of peptide. (C) The same experiment was carried out with the purified Mad1–Mad2R133A complex (running as a single peak, Figure 3D). Relative to the wild-type complex shown in (B), we observe the absence of P2. Thus, the appearance of P2 in (B) is probably due to Mad2 oligomerization.
Next, we incubated purified Mad1–Mad2wt (which runs as a single peak, as shown in Figure 1C) with Cdc20111–154 and analysed the resulting mixture by SEC (Figure 4B). Peak 1 (P1) contained Mad1–Mad2. P2 contained Mad2– Cdc20111–154, and its elution volume was identical to that of P2 obtained from the incubation of Mad2wt with Cdc20111–154. Finally, P3 contained an excess of Cdc20111–154. There was no trace of Cdc20111–154 in P1, suggesting that a ternary complex, Mad1–Mad2–Cdc20, does not form in vitro, and that binding of Mad2 to Mad1 or Cdc20 is exclusive. The appearance of Mad2– Cdc20111–154, however, suggests that Mad1–Mad2 contains a source of Mad2 available for Cdc20 binding. The absence of free Mad1, even in the presence of a very large excess of the Cdc20 peptide (not shown), suggested that the Mad2–Cdc20 complex is not generated as an effect of competition by Cdc20111–154 on the Mad1–Mad2 interaction.
We imagined that Mad2wt preserves an oligomeric state when bound to Mad1. If the Mad1–Mad2 complex consisted of a core complex, with accessory Mad2 subunits linked via Mad2 oligomerization, there would be a pool of Mad2 available for the interaction with Cdc20 without the need to invoke a disruption of the core complex. Mad1–Mad2R133A was a good reagent to test this hypothesis, because Mad2R133A is unable to form oligomers and its complex with Mad1 should only consist of the high-affinity core complex without an extra pool of oligomeric Mad2. Thus, we incubated Mad1–Mad2R133A (previously purified and running as a single peak, as shown in Figure 3D) with Cdc20111–154 and separated the resulting species by SEC. Only two peaks formed. Peak 1 (P1) contained intact Mad1–Mad2R133A, and P2 contained Cdc20111–154. No Mad2R133A–Cdc20111–154 complex formed, confirming our supposition that the source of Mad2wt available for Cdc20111–154 is linked to Mad1–Mad2wt via Mad2 self-association. Our data suggest that Mad2 self-association, albeit unnecessary, is compatible with Mad1 binding in vitro. A pictorial interpretation of the results is shown on the right-hand side of Figure 4. Mad1 is likely to form a coiled coil, and interacts with itself in the two-hybrid assay (Jin et al., 1998). This prompted us to depict Mad1 as an elongated dimeric molecule in Figure 4. In summary, we propose that the Mad1–Mad2wt complex consists of a core Mad1–Mad2 complex, with accessory Mad2 subunits linked through the oligomerization of Mad2. Cdc20111–154 is unable to disrupt the high-affinity core complex between Mad2 and Mad1 in vitro, but can interact with the loosely associated Mad2 subunits in this complex.
Oligomerization of Mad2 is not required for kinetochore binding in vivo
Our biochemical investigation is consistent with previous reports that Mad2 forms oligomers in vitro (Fang et al., 1998a). We show that the oligomerization of Mad2 is not required for its interaction with Cdc20 (which indeed reverses the oligomerization of Mad2) and with Mad1. However, we show that Mad2 is oligomeric when bound to Mad1 in vitro, and that this oligomerization is required for the interaction of Mad2 with Cdc20 (Figure 4), as shown by the fact that the monomeric mutant Mad2R133A cannot bind Cdc20 when engaged in a complex with Mad1. Because the interaction of Mad2 with Mad1 might be important for the kinetochore localization of Mad2, this result suggests a mechanism through which the oligomerization of Mad2 may play a role in the spindle checkpoint. In particular, the ‘core’ interaction of Mad2 with Mad1 might be required for the kinetochore localization of Mad2, whereas the ‘loose’ interactions based on Mad2 oligomerization might be required to generate a pool of Mad2 available for Cdc20 binding at the kinetochore. The proposal that Mad2 oligomerization is essential for the spindle checkpoint was based very heavily on data obtained using the Mad2ΔC mutant (Fang et al., 1998a). We show that this deletion mutant is defective not only in oligomerization, but also in binding to Mad1 and Cdc20, making it unsuitable to study the role of Mad2 oligomerization in the spindle checkpoint. The generation of Mad2R133A, a mutant that retains the ability to bind Cdc20 and Mad1 but is fully monomeric, allowed us to test rigorously the influence of oligomerization on Mad2 function in vivo.
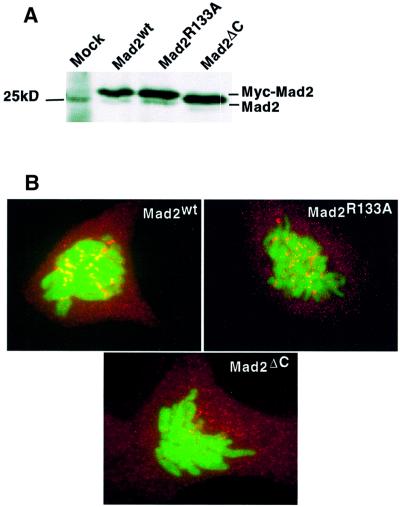
To this end, we generated Mad2wt, Mad2ΔC and Mad2R133A plasmids expressing the three proteins with an N-terminal Myc epitope tag. The three proteins were expressed at similar levels in a number of mammalian cell lines (Figure 5A and data not shown). In agreement with the localization of endogenous Mad2 and green fluorescent protein (GFP)-tagged Mad2 (Chen et al., 1996; Fang et al., 1998a; Gorbsky et al., 1998; Waters et al., 1998; Howell et al., 2000), overexpression of Mad2wt and of the two mutants led to a diffuse cytoplasmic localization of the proteins (data not shown). In agreement with these previously published results, Mad2wt showed both cytoplasmic and kinetochore staining in prometaphase (data not shown), and in cells undergoing a nocodazole-mediated metaphase arrest (Figure 5B). An identical localization was observed for the R133A mutant, demonstrating that oligomerization is not required for correct localization of Mad2 to the kinetochores. In contrast, Mad2ΔC was not enriched on the kinetochores either in prophase or metaphase (data not shown), or when the spindle checkpoint was induced (Figure 5B), which correlated with the fact that this mutant is unable to interact with Mad1 in vitro (Figure 2A). These results show that Mad1 binding, and not oligomerization, is required for kinetochore binding by Mad2 in vivo. This is consistent with previous findings in Xenopus extracts that Mad1 binding is required for Mad2 localization to unattached kinetochores (Chen et al., 1998).
Fig. 5. (A) The expression levels of Mad2R133A and Mad2ΔC are similar to that of Mad2wt. HeLa cells were transfected with Myc-tagged Mad2 expression plasmids; 24 h after transfection the cell lysates were prepared and analysed on a 12% polyacrylamide gel, followed by western blotting using Mad2 antibody (Santa Cruz). (B) Mad2R133A localizes to kinetochores in nocodazole-arrested cells, while the C-terminal deletion mutant does not. PTK1 cells were co-transfected with H2B–GFP and Myc-tagged Mad2 expression plasmids; 24 h after transfection the cells were treated with nocodazole for another 24 h, then fixed and stained with anti-Myc antibody (9E10; in red). Chromosomes were visualized by H2B–GFP fluorescence (in green).
Mad2 binding to Mad1 and Cdc20, but not its oligomerization, is required for the spindle checkpoint
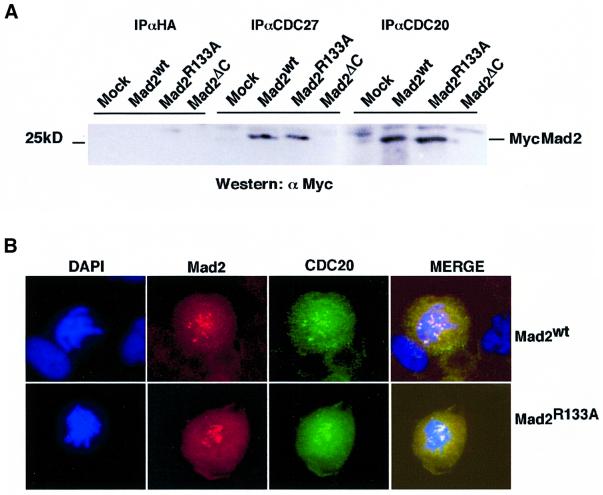
The association between Mad2 and Cdc20 is thought to occur early during mitosis, and the resulting temporary inactivation of the APC may be instrumental in granting all kinetochores enough time to attach to the spindle fibres (Fang et al., 1998a). To test whether Mad2 oligomerization is required for the in vivo interaction between Mad2 and Cdc20, HeLa cells were transfected with the Mad2 expression plasmids and treated with nocodazole (Figure 6A). Mad2wt and Mad2R133A bound to Cdc20 and to Cdc27, one of the subunits of the APC, apparently with identical efficiency (Figure 6A). Consistent with the results in vitro, the Mad2ΔC mutant was unable to interact with Cdc20 or Cdc27 in vivo.
Fig. 6. (A) Mad2R133A associates with Cdc20 and CDC27 in nocodazole-arrested cells. HeLa cells were transfected with Mad2 expression vectors and 24 h later treated with nocodazole for 16 h.The cells were lysed and Mad2 was immunoprecipitated with the antibodies indicated. The immunoprecipitates were analysed by western blotting using αMyc antibody (9E10). (B) Mad2R133A co-localizes with Cdc20 at kinetochores in nocodazole-arrested cells. Mad2 expression plasmids were transfected into PTK1 cells and the cells were treated with nocodazole for 24 h. Myc-tagged Mad2 was visualized with Cy3-conjugated antibody (in red). Cdc20 was visualized using anti-Cdc20 mouse monoclonal antibody.
We also tested whether Mad2wt and Mad2R133A co-localized with Cdc20 in nocodazole-treated cells. PTK1 cells were transfected with Myc-tagged Mad2 expression plasmids, and then treated with nocodazole to induce a mitotic block. Under these conditions, Cdc20 localized to the cytosol and to kinetochores (Figure 6B). The pattern of localization of Mad2wt was very similar (Figures 5B and 6B), and merging of the Cdc20 and Mad2wt immunostainings showed that these proteins appear to co-localize at kinetochores. Precisely the same result was obtained when Mad2R133A was used in this experiment (Figure 6B). The co-localization pattern suggests that Cdc20 and Mad2 are likely to associate at the kinetochores. In summary, these results show that the oligomerization of Mad2 is dispensable for Cdc20–APC binding in vivo, and for its correct subcellular localization.
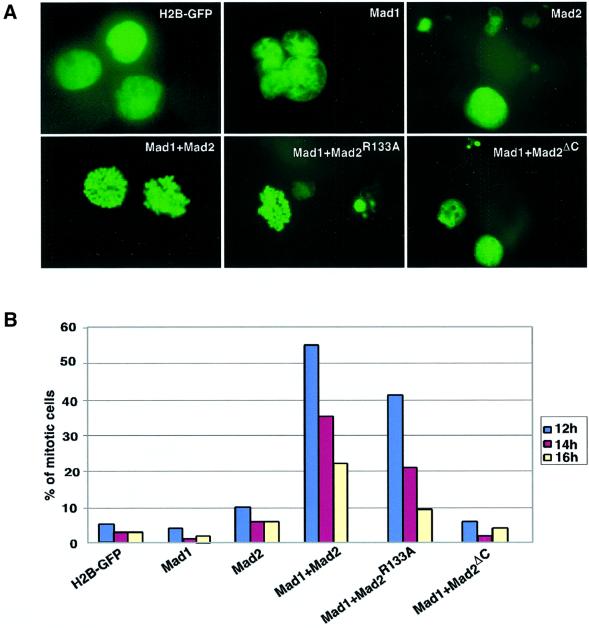
Overexpression of Mad2 in fission yeast arrests cells in mitosis (He et al., 1997; Kim et al., 1998). Similarly, high levels of recombinant Mad2 induce mitotic arrest in Xenopus extracts and in tissue culture cells (Chen et al., 1998; Fang et al., 1998a; Howell et al., 2000). However, the induction of mitotic arrest by high levels of Mad2 is independent of Mad1 and localization to the kinetochores (Chen et al., 1998). Since the results from Fang et al. (1998a) suggested that Mad2 oligomerization is an essential mechanism for the regulation of the mitotic spindle checkpoint, we tested the ability of Mad2wt, Mad2ΔC and Mad2R133A to induce a spindle checkpoint. Initially, we performed the experiments using purified Mad2 proteins. However, none of the Mad2 proteins induced a significant increase in cells arrested in metaphase (data not shown). We speculated that the lack of checkpoint response could be due to a rapid degradation of the recombinant Mad2 proteins, and we tested whether high levels of sustained Mad2 overexpression from expression plasmids were sufficient to induce the mitotic spindle checkpoint. As shown in Figure 7, overexpression of wild-type Mad2 in HeLa cells is not sufficient to induce the spindle checkpoint. However, when Mad2 is co-expressed with Mad1, the spindle checkpoint is induced (Figure 7). This result is in agreement with previous data showing that the mitotic spindle checkpoint is dependent on both Mad1 and Mad2, and that Mad2 is in excess over Mad1 (Chen et al., 1998). The R133A mutant was fully capable of inducing mitotic arrest, showing that oligomerization is not required for the biological function of Mad2. Mad2ΔC, which cannot bind to Mad1 or Cdc20 and does not associate with kinetochores, was unable to induce mitotic arrest. Interestingly, co-expression of Mad1 and Mad2 induces apoptosis in a substantial fraction of the transfected cells, an effect that was increased when Mad2R133A was used. Thus, although oligomerization is not important for the Mad2-induced mitotic checkpoint, it may reduce the apoptotic effects of Mad2 in some unknown manner.
Fig. 7. Mad2R133A cooperates with Mad1 to arrest the cells in mitosis. HeLa cells were synchronized by double thymidine block, co-transfected with Mad1, Mad2 and H2B–GFP expression plasmids during release from first thymidine block. The nuclear morphology (A) and mitotic indexes (B) were analysed by H2B–GFP fluorescence 12, 14 and 16 h after release from double thymidine block.
Discussion
It is now clearly established that Mad2 oligomerization is not required for Cdc20 binding (Luo et al., 2000; this study). Here, we show that Mad2 oligomerization is not important for its function in the spindle checkpoint. Co-expression with Mad2 was instrumental in reversing the insolubility of Mad1485–718 in bacteria. Full-length Mad1 was insoluble in bacteria or insect cells, and the yields of the complex with Mad2 were unsuitable for detailed biochemical analyses. Similarly to the full-length complexes from budding yeast and frog, however, the complex containing Mad1485–718 is extremely stable, in the absence of other proteins or post-translational modifications. Thus, Mad1 phosphorylation is unlikely to play a role in the formation of Mad1–Mad2, contradicting previous suggestions (Waters et al., 1999). However, a function for phosphorylation in the checkpoint is not ruled out.
Using a monomeric point mutant (Mad2R133A), we showed that oligomerization of Mad2 is not required for binding Mad1 or Cdc20 (Figure 3). Despite this, Mad2 maintains an oligomeric state in vitro when bound to Mad1. Mad1–Mad2WT eluted from a gel filtration column at higher molecular weights than Mad1–Mad2R133A (Figure 3C), suggesting a different stoichiometry, as expected if Mad2WT, but not MadR133A, retained an oligomeric state when bound to Mad1. Moreover, incubation of Mad1–Mad2WT with a Cdc20 peptide resulted in a 1:1 Mad2–Cdc20 complex (Figure 4B), which was not observed when the experiment was carried out with purified Mad1–Mad2R133A (Figure 4C). Thus, the Cdc20 peptide reverses the oligomerization of Mad2WT even when this is bound to Mad1 in vitro. This is consistent with the ability of the same peptide to reverse the oligomerization of Mad2WT in solution (Luo et al., 2000; this study). Mad2 forms distinct and incompatible complexes with Mad1 and Cdc20 in vitro (Figure 4), as we never observed a ternary complex containing Mad1–Mad2–Cdc20. Cdc20 is unable to disrupt the core Mad1–Mad2 complex, presumably because this interaction is of much higher affinity than that between Mad2 and Cdc20. Free Mad1 was not observed even when the Mad2–Mad1 complex was incubated with a large excess of Cdc20 peptide. The Cdc20 peptide was unable to compete the interaction of Mad2R133A with Mad1 (Figure 4C), even if the isolated Mad2R133A and Mad2WT bind Cdc20 with an apparently identical affinity (Figure 3E). Budding yeast Mad1 binds Cdc20 in a two-hybrid assay (Hwang et al., 1998). We were unable to test this interaction directly due to the unavailability of Mad2-free Mad1, but we show for the human proteins that Cdc20 is unable to bind Mad1 in the presence of Mad2. It is possible that Mad1 and Cdc20 interact in the absence of Mad2.
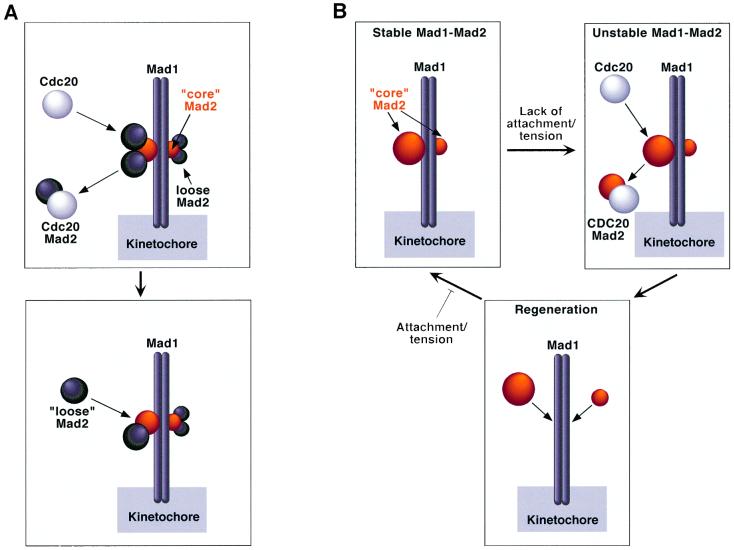
In vitro, Cdc20 interacts with Mad2 subunits associated with the Mad1–Mad2 complex via Mad2 oligomerization. A working model based on this finding proposes that the tight Mad1–Mad2 core complex may be required for the localization of Mad2 to kinetochores, while the subunits bound via oligomerization act as a store of Mad2 for Cdc20 (Figure 8A). We tested the relevance of this model in vivo starting from the observation that the interaction of Mad2R133A with Mad1 in vitro prevents Cdc20 binding (Figure 4C). Thus, binding of Mad2R133A to Mad1 should saturate the binding site for Mad2WT, preventing the localization of Mad2 oligomers at the kinetochore. We found that Mad2R133A localized at kinetochores like Mad2wt, while Mad2ΔC was unable to do so (Figure 5). Mad2R133A also behaved indistinguishably from Mad2wt with respect to Cdc20 and APC binding (Figure 6), and checkpoint activation upon co-expression with Mad1 (Figure 7). Thus, our findings contradict the model in Figure 8A and indicate that Mad2 oligomerization is not important for the spindle checkpoint. Consistently, we could not co-immunoprecipitate flag- and myc-tagged Mad2 (WT or R133A) in concomitantly transfected cells (data not shown). The ΔC mutant is unable to interact with Mad1 and Cdc20 in vitro. In vivo, this mutant did not localize at kinetochores, did not bind Cdc20 and the APC, and was unable to cause a cell cycle arrest upon co-expression with Mad1. Thus, impairment of Mad1 and Cdc20 binding, rather than of oligomerization, is the critical damaging event affecting Mad2ΔC. The reported dominant-negative effect of Mad2ΔC on the ability of Mad2wt to inhibit the APC in vitro (Fang et al., 1998a) may be due to the sequestration by Mad2ΔC of another protein required for effective inhibition of the APC. The oligomerization of recombinant Mad2 observed in vitro is plausibly a result of its harvesting in non-physiological conditions. Cross-linking experiments did not suggest a precise stoichiometry of the Mad2 oligomer (data not shown). Mad2 is polydisperse by dynamic light scattering (data not shown), and its early elution in SEC experiments may reflect unspecific aggregation. A closely related protein, Mad2L2, forms oligomers, but a monomeric form is fully functional in inhibiting Cdh1 (Chen and Fang, 2001; Pfleger et al., 2001). We have also found that budding yeast Mad2 is monomeric (data not shown).
Fig. 8. Models of spindle checkpoint function. (A) The Mad1–Mad2 complex contains ‘core’ Mad2 subunits (orange) forming a strong interaction with Mad1, and loosely associated subunits (dark grey) that are associated via Mad2 oligomerization. Cdc20 interacts with the loosely associated Mad2 subunits, whose stock is replenished by the addition of new subunits. (B) The Mad1–Mad2 complex is tight. Association of Cdc20 with Mad2 occurs upon destabilization of the Mad1–Mad2 complex. A new cycle starts when a new Mad2 molecule binds Mad1. The cycle is interrupted as a consequence of spindle attachment, and a stable Mad1–Mad2 complex is formed that does not permit further recruitment of Mad2. The precise regulatory mechanism impinging on the recruitment and release of Mad2 to/from Mad1 is not known at present.
Overexpression of Mad2 in fission yeast leads to a block in mitosis (He et al., 1997; Kim et al., 1998). A large excess of Mad2 is required to stabilize CDC2 activity in Xenopus eggs, and to cause mitotic arrest in Xenopus embryos (Chen et al., 1998; Fang et al., 1998a). These Mad1-independent effects may result from ‘systemic’ binding of Cdc20 by Mad2. We were unable to observe cell cycle arrest when overexpressing Mad2wt or Mad2R133A in mammalian cells, and co-expression of Mad2 with Mad1 was instrumental (Figure 7), confirming a recent report (Geley et al., 2001). Thus, Mad1-mediated kinetochore localization is critical for the formation of productive Mad2–Cdc20 complexes. Consistently, Cdc20 and Mad2 co-localize at kinetochores in nocodazole-treated cells (Figure 6). Thus, it is conceivable that individual Mad2 molecules are first captured by Mad1 and subsequently transferred to Cdc20 to mediate cell cycle arrest. In agreement with this idea, recent results show that Mad2 cycles very rapidly in and out of kinetochores (Howell et al., 2000). This leads to an alternative model for checkpoint function, which is depicted schematically in Figure 8B. How cycling of Mad2 is regulated remains unclear, but it probably involves an attachment/tension-sensitive modifying enzyme. In the absence of attachment/tension, this enzyme may promote the exchange of Mad2 from Mad1 to Cdc20. Extinction of activity upon attachment, possibly as a result of direct mechanical coupling, would result in the formation of a stable and unperturbed Mad1–Mad2 complex, preventing further translocation of Mad2 to Cdc20. A candidate modification is phosphorylation: Mad1 is phosphorylated during checkpoint activation (Hardwick and Murray, 1995; Hardwick et al., 1996; Jin et al., 1998; Chen et al., 1999; Waters et al., 1999), and the candidate kinases are Bub1 and Mps1, two critical components of the spindle checkpoint (Hardwick et al., 1996; Seeley et al., 1999). Whether modification is required to recruit Mad2 at kinetochores, or to cause its transfer to Cdc20, is currently unclear. In any case, the existence of a counteracting enzyme to re-initialize the system at every transfer cycle is implied. In summary, we propose that kinetochores act as regulated ‘folding factories’ for Mad2–Cdc20 complexes endowed with inhibitory function on the APC.
Materials and methods
Expression and purification of Mad2 constructs
PCR was used to amplify the coding sequence of human Mad2, using IMAGE clone 1047276 as a template. The PCR product was subcloned into the pQE30 vector (Qiagen) in-frame with the sequence encoding the polyhistidine tag. The same strategy was used to subclone Mad2ΔN. Mad2ΔC was obtained by introducing a stop codon after residue 196 in the Mad2wt sequence using the Quickchange kit (Stratagene). To co-express Mad1, we engineered the pQE30 vector so as to introduce downstream of Mad2 a new ribosome binding site followed by the necessary restriction sites. Fragment 485–718 of Mad1 was amplified from IMAGE clone 1860469 and subcloned into the dicistronic vector using the _Bgl_II and _Sal_I sites introduced after the first subcloning step. Point mutants were generated by substitution of the original codon using the Quickchange kit, and the wild-type Mad2 coding sequence in the pQE30 vector as a template. Residues Val85, Arg99, Arg133, Q134, L154, Y156 and H191 were all individually changed into alanine. All DNA constructs were confirmed by sequencing.
Very similar purification strategies were applied to the different constructs, based on the presence of the polyhistidine tag on Mad2. Bacterial expression was carried out in Escherichia coli strain XL1blue at room temperature. Induction of protein expression was carried out at an OD600 of 0.5–0.8 using 0.5 mM isopropyl-β-d-thiogalactopyranoside, and continued for ∼12 h. Cells were harvested by low-speed centrifugation, and the bacterial pellets were resuspended in buffer A [50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5% glycerol, 5 mM imidazole, and a tablet of cocktail inhibitors Complete, EDTA free (Roche)]. After sonication and ultracentrifugation, the bacterial lysates were incubated with Ni-NTA–agarose (Qiagen). The beads were washed with buffer A, and the bound protein was eluted using 150 mM imidazole in buffer A. The eluted proteins were purified further by SEC on a Superdex 200 equilibrated with buffer B [20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.5 mM EDTA] and concentrated by ultrafiltration to variable concentrations (between 2 and 15 mg/ml). Analytical chromatography was carried out on a SMART system (Amersham-Pharmacia Biotech) using a Superdex 200 column; 100 µl fractions were collected.
Binding assays
A cDNA fragment encoding residues 111–160 of human Cdc20 was amplified by PCR and subcloned in pGEX4T-2 (Amersham-Pharmacia Biotech) as a fusion to GST. GST–Cdc20111–160 was expressed in E.coli strain BL21(DE3). Expression, harvesting and lysis were carried out as described above. The bacterial lysates were resuspended in buffer C (10 mM HEPES–HCl pH 7.5, 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA) and purified using glutathione–agarose resin (Amersham-Pharmacia Biotech). Ten micrograms of bound GST–Cdc20111–160 fusion protein or GST control were incubated with 100 µg of Mad2, Mad2ΔN, Mad2ΔC or Mad2R133A, and the volume was brought to 1 ml using buffer C supplemented with 1% Triton X-100. Under these conditions, the concentration of GST–Cdc20111–160 in the binding assay was ∼0.4 µM, while that of Mad2 and its mutants was ∼4 µM. The binding reaction was incubated at 4°C for 1 h, after which beads and bound proteins were washed twice with buffer C (always containing 1% Triton X-100) and separated by SDS–PAGE.
For the experiments shown in Figure 4, purified Mad2 and the wild-type and mutant Mad2–Mad1 complex were incubated in buffer B at room temperature for 1 h with a 3- to 4-fold molar excess of synthetic peptide corresponding to residues 111–154 of Cdc20. The incubation mixes were separated on the SMART system using a Superdex 200 column equilibrated with buffer B, and the resulting fractions were analysed by SDS–PAGE.
Cell culture and transfections
HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM) (Euroclone) supplemented with 10% bovine calf serum (Hyclone) and 2 mM l-glutamine (Euroclone). To synchronize the cells at the G1/S border, 2.5 mM thymidine was added to the cells for 16 h, followed by release for 8 h and a second block for 16 h. During the release from the first thymidine block, the cells were transfected with Mad2 and Mad1 expression plasmids using lipofectamine (Gibco-BRL). Growing HeLa cells were transfected using a standard calcium phosphate precipitation procedure. PTK1 cells were cultured in DMEM (Euroclone) supplemented with 10% fetal bovine serum. To obtain metaphase-arrested PTK1 cells, 100 ng/ml nocodazole was added to the culture for 24 h. Transfection of PTK1 cells was performed using Lipofectamine-Plus reagent (Gibco-BRL) according to the manufacturer’s instructions.
Antibodies
Mouse monoclonal antibodies against hCdc20 and hCDC27 were raised against full-length GST fusion proteins. Briefly, Balb/C mice were immunized subcutaneously with GST–CDC27 and with urea-denatured His6 hCdc20 recombinant proteins. Monoclonal antibodies were generated by fusing the splenocytes to NS-2 mouse myeloma cells 3 days after the final boost. One cell line producing mouse monoclonal antibodies against hCdc20 (AR12) and two cell lines producing anti-hCDC27 antibodies (ZC51 and SO27) were generated by single-cell cloning. 9E10 mouse monoclonal antibody was used for Myc-tagged Mad2 proteins. Cy3-conjugated anti-Myc antibody (9E10) was purchased from Sigma. Goat polyclonal antibody for Mad2 was obtained from Santa Cruz.
Plasmids
Mad2 expression plasmids were generated by ligation of _Bam_HI–_Not_I PCR fragments containing the full-length Mad2 open reading frame into pCMV Neo-Bam expression vector, and expressed with an N-terminal Myc tag. The histone-2B (H2B)–GFP expression vector was a kind gift of G.Wahl.
Cell lysis and immunoprecipitations
The cells were lysed in NP-40 lysis buffer containing 100 mM Tris–HCl pH 7.4, 150 mM NaCl, 10% glycerol and 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml each aprotinin and leupeptin, 1 mM NaF, 0.1 mM sodium vanadate for 15 min on ice, followed by sonication for 30 s. After centrifugation at 14 000 r.p.m. in an Eppendorf microfuge for 10 min, the lysates were precleared with protein G–Sepharose beads for 1 h at 4°C, then incubated with anti-hCdc20 (AR12) and anti-hCDC27 (SO27) antibodies for 1 h, followed by addition of protein G–Sepharose beads for an additional 1 h. Immunoprecipitates were washed for three times in lysis buffer and analysed on a 12% polyacrylamide gel, blotted to nitrocellulose membranes and probed with the antibodies indicated.
Immunofluorescence and determination of the mitotic index
PTK1 cells were grown on glass cover slips, fixed with 4% paraformaldehyde for 10 min at room temperature, and permeabilized using 0.1% Triton X-100. To detect Myc-tagged Mad2, a mouse monoclonal antibody against the Myc tag was used. Cdc20 staining was performed using AR12 mouse monoclonal antibody. For determination of mitotic indexes, HeLa cells were collected by centrifugation, washed twice with phosphate-buffered saline (PBS) and fixed in 1% paraformaldehyde–PBS for 5 min at room temperature. The cell suspension was spun at 500 r.p.m. on glass cover slips using a Cytospin centrifuge, and the cover slips were dried for 30 min, washed twice with PBS and mounted. Nuclear morphology was determined by H2B–GFP fluorescence. The cover slips were analysed under a Provis microscope (Olympus), and images were acquired and analysed using Adobe Photoshop 3.0 software (Adobe Inc.).
Acknowledgments
Acknowledgements
We wish to thank Kevin Hardwick, Luca Jovine, Marina Mapelli and Simonetta Piatti for helpful discussions and critical reading of the manuscript. We also wish to thank Professor Kuan-Teh Jeang for the Mad1 expression vector. A.M. is a Scholar of the Italian Foundation for Cancer Research (FIRC) and an EMBO Young Investigator. This work was supported by grants from the Giovanni Armenise–Harvard Foundation for Advanced Scientific Research and the Italian Association for Cancer Research.
References
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin,E.V. (1998) The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci., 23, 284–286. [DOI] [PubMed] [Google Scholar]
- Chen J. and Fang,G. (2001) MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev., 15, 1765–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko,A., Mann,M. and Murray,A.W. (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol., 143, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady,D.M., Smith,D., Murray,A.W. and Hardwick,K.G. (1999) The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell, 10, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal,V., Scott,M.L., Benezra,R. and Sorger,P.K. (2000) Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell, 101, 635–645. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998a) The checkpoint protein MAD2 and the mitotic regulator Cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev., 12, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998b) Direct binding of Cdc20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Geley S., Kramer,E., Gieffers,C., Gannon,J., Peters,J.M. and Hunt,T. (2001) Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol., 153, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen,R.H. and Murray,A.W. (1998) Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol., 141, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G. (1998) The spindle checkpoint. Trends Genet., 14, 1–4. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G. and Murray,A.W. (1995) Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol., 131, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss,E., Luca,F.C., Winey,M. and Murray,A.M. (1996) Activation of the budding yeast spindle checkpoint without mitotic spindle disruption. Science, 273, 953–956. [DOI] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Hoffman,D.B., Fang,G., Murray,A.W. and Salmon,E.D. (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers and at spindle poles in living cells. J. Cell Biol., 150, 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis,L. and Roberts,B.T. (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Jin D.Y., Spencer,F. and Jeang,K.T. (1998) Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell, 93, 81–91. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein,J., Daum,J.R., Burke,D.J. and Gorbsky,G.J. (1998) Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol., 141, 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Li R. and Murray,A. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea,C., Mahaffey,D., Rechsteiner,M. and Benezra,R. (1997) MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl Acad. Sci. USA, 94, 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Fang,G., Coldiron,M., Lin,Y., Yu,H., Kirschner,M.W. and Wagner,G. (2000) Structure of the mad2 spindle assembly checkpoint protein and its interaction with cdc20. Nature Struct. Biol., 7, 224–229. [DOI] [PubMed] [Google Scholar]
- Michel L.S. et al. (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature, 409, 355–359. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Salic,A., Lee,E. and Kirschner,M.W. (2001) Inhibition of Cdh1–APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev., 15, 1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley T.W., Wang,L. and when, JY (1999) Phosphorylation of human Mad1 by the BUB1 kinase in vitro. Biochem. Biophys. Res. Commun., 257, 589–595. [DOI] [PubMed] [Google Scholar]
- Shah J.V. and Cleveland,D.W. (2000) Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell, 103, 997–1000. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (1998) Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc. Natl Acad. Sci. USA, 95, 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Chen,R.H., Murray,A.W. and Salmon,E.D. (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol., 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Chen,R.H., Murray,A.W., Gorbsky,G.J., Salmon,E.D. and Nicklas,R.B. (1999) Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr. Biol., 9, 649–652. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Lees,E. (2001) Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol., 21, 5190–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]