Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores (original) (raw)
Abstract
The kinetochore checkpoint pathway, involving the Mad1, Mad2, Mad3, Bub1, Bub3 and Mps1 proteins, prevents anaphase entry and mitotic exit by inhibiting the anaphase promoting complex activator Cdc20 in response to monopolar attachment of sister kinetochores to spindle fibres. We show here that Cdc20, which had previously been shown to interact physically with Mad2 and Mad3, associates also with Bub3 and association is up-regulated upon checkpoint activation. Moreover, co-fractionation experiments suggest that Mad2, Mad3 and Bub3 may be concomitantly present in protein complexes with Cdc20. Formation of the Bub3–Cdc20 complex requires all kinetochore checkpoint proteins but, surprisingly, not intact kinetochores. Conversely, point mutations altering the conserved WD40 motifs of Bub3, which might be involved in the formation of a β-propeller fold devoted to protein–protein interactions, disrupt its association with Mad2, Mad3 and Cdc20, as well as proper checkpoint response. We suggest that Bub3 could serve as a platform for interactions between kinetochore checkpoint proteins, and its association with Mad2, Mad3 and Cdc20 might be instrumental for checkpoint activation.
Keywords: Bub3/Cdc20/kinetochore/mitotic checkpoint/WD40
Introduction
In order to preserve genome stability, eukaryotic cells have evolved checkpoint mechanisms that respond to errors in the structure or segregation of chromosomes and delay cell cycle progression until errors have been corrected. Genetic instability involves gain or loss of genetic information and is thought to be one of the major causes of cancer development (Lengauer et al., 1998). Mitotic checkpoints delay progression through and exit from mitosis in case of mistakes in the attachment of chromosomes to the mitotic spindle and in their segregation. These controls are also thought to play a crucial role during alignment of chromosomes on the metaphase plate in normal mitoses. A mitotic checkpoint pathway, also called ‘kinetochore checkpoint’, monitors the lack of bipolar attachment of kinetochores to microtubules and prevents sister chromatid separation and exit from mitosis by inhibiting the anaphase promoting complex (APC), which in turn promotes degradation of the anaphase inhibitor securin and inactivation of cyclin B-dependent kinases at the end of mitosis (reviewed in Zachariae and Nasmyth, 1999). In budding yeast this checkpoint pathway involves the proteins Mad1, Mad2, Mad3, Bub1, Bub3 and Mps1, which are conserved throughout evolution. Verte brate homologues of Mad1, Mad2, Mad3, Bub1 and Bub3 localize at unattached kinetochores during prophase and prometaphase (reviewed in Wassmann and Benezra, 2001) and prevent APC activation by inhibiting its accessory factor Cdc20 (Fang et al., 1998; Hwang et al., 1998; Kim et al., 1998). Another branch of the yeast mitotic checkpoint involves a two-component GTPase-activating protein, formed by Bub2 and Bfa1, which inhibits the G protein Tem1 in response to errors in spindle positioning. Tem1 is in turn involved in activating the mitotic exit network and thereby mitotic exit and cytokinesis (for reviews see Hoyt, 2000; Schuyler and Pellman, 2001).
Thanks to the efforts of a number of laboratories, several details about the relationships between different Mad and Bub proteins involved in the kinetochore checkpoint have been elucidated. Mad1 has been shown to interact constitutively with Mad2 (Chen et al., 1998, 1999) and to be phosphorylated in yeast cells concomitantly to checkpoint activation. Its phosphorylation depends on Mad2, Bub1, Bub3 (Hardwick and Murray, 1995) and on Mps1 (Hardwick et al., 1996), a protein kinase required for both duplication of the microtubule-organizing centre (spindle pole body, SPB) and checkpoint activation (Weiss and Winey, 1996). Mad3 binds to Mad2 mostly in response to microtubule depolymerization and to Bub3 throughout the cell cycle; the former interaction depends on all the other proteins involved in the same checkpoint pathway, whereas the latter does not require any of them (Hardwick et al., 2000). Bub3 binds constitutively to Bub1 (Roberts et al., 1994; Brady and Hardwick, 2000), whereas the Bub1–Bub3 complex interacts with Mad1 only during S phase, when presumably the newly replicated centromeres have yet to form stable kinetochore–microtubule attachment, and during checkpoint activation by nocodazole treatment (Brady and Hardwick, 2000). Mad2 and Mad3 also bind budding yeast Cdc20 (Hwang et al., 1998; Hardwick et al., 2000). Furthermore, Mad2 associates with the Cdc20 homologues in vertebrate and Schizosaccharomyces pombe cells (Fang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Wassmann and Benezra, 1998) and inhibits their activity in vitro (Fang et al., 1998). While interaction between Mad2 and Cdc20 strictly depends only upon Mps1 and Mad1 function (Hwang et al., 1998; Hardwick et al., 2000), Mad3–Cdc20 complex formation requires Mad1, Mad2, and, to a lesser extent, also Bub1 and Bub3 (Hardwick et al., 2000). Putting together all the data concerning the physical interactions between different Mad and Bub proteins and Cdc20 it is possible to hypothesize the existence of at least three different constitutive subcomplexes in yeast cells, one including Mad1 and Mad2 and the others formed by Bub1–Bub3 and Mad3–Bub3, respectively. Due to the cell cycle-regulated interaction between Mad1 and Bub1–Bub3 (Brady and Hardwick, 2000), as well as between Mad2 and Mad3 (Hardwick et al., 2000), the three complexes might associate with each other only during a narrow window of the cell cycle and upon nocodazole treatment, thereby leading to interaction with and inhibition of Cdc20. However, the nature of the protein complex(es) with inhibitory activity towards Cdc20–APC has so far remained elusive. Recently, a complex including Bub3, BubR1, the putative human homologue of Mad3, and probably Mad2, has been purified from HeLa cells and shown to inhibit Cdc20–APC specifically in mitosis (Sudakin et al., 2001; Tang et al., 2001). The Bub3 protein is a good candidate to mediate assembly of protein complexes in response to kinetochore checkpoint activation because it contains WD40 motifs, which have been implicated in protein–protein interactions (Smith et al., 1999). Another interesting question concerns how structural kinetochore components might regulate assembly and function of checkpoint protein complexes. In fact, an involvement of the kinetochore in these control mechanisms has been hypothesized based on its central role in mitotic checkpoint activation (Rieder et al., 1995). In this paper we show that Saccharomyces cerevisiae Bub3 interacts physically with Cdc20, as well as with Mad1 and Mad2. Bub3–Cdc20 complex formation is upregulated upon checkpoint activation and depends on all the other kinetochore checkpoint proteins including Mps1, but does not require intact kinetochores. Moreover, co-fractionation experiments provide evidence that Mad2, Mad3 and Bub3 might concomitantly be present in a complex with Cdc20. Finally, association of Bub3 with Mad2, Mad3 and Cdc20 depends on Bub3 WD40 repeats and its loss correlates with defective checkpoint response.
Results
Bub3 interacts physically with Mad1, Mad2 and Cdc20
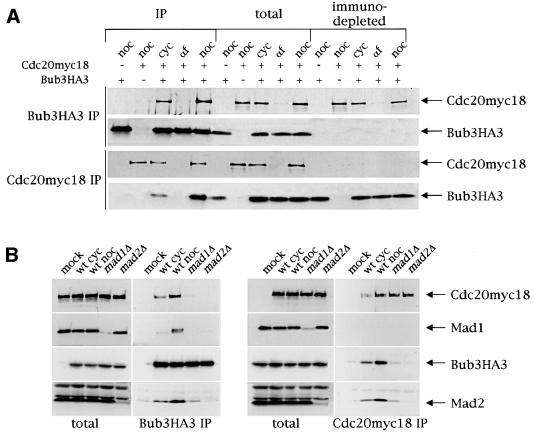
All Mad and Bub proteins except Bub2 were shown by genetic means to be involved in the same mitotic checkpoint pathway that monitors kinetochore–microtubule attachment and delays entry into anaphase and exit from mitosis by inhibiting the APC regulator Cdc20 (Hardwick and Murray, 1995; Wang and Burke, 1995; Hardwick et al., 1996; Pangilinan and Spencer, 1996; Alexandru et al., 1999). In addition, all these proteins display a similar localization pattern at unattached kinetochores in vertebrate cells (Li and Benezra, 1996; Chen et al., 1996, 1998; Taylor and McKeon, 1997; Gorbsky et al., 1998; Taylor et al., 1998). Although it has been shown that budding yeast Bub3 interacts physically with Mad3, which in turn can be co-immunoprecipitated with Cdc20 (Hardwick et al., 2000), no evidence that it can form a complex with Cdc20 had been provided so far. We therefore tested by co-immunoprecipitation experiments whether Bub3 and Cdc20 could interact physically by using protein extracts from strains expressing fully functional Bub3 tagged with three hemagglutinin (HA) epitopes (Bub3HA3, see Materials and methods), or Cdc20 tagged with 18 copies of the myc epitope (Cdc20myc18) (Shirayama et al., 1998), or both. As shown in Figure 1A, Bub3HA3 immunoprecipitates from extracts of the latter strain contained Cdc20myc18 and vice versa, suggesting that the two proteins physically interact in vivo. The amount of Bub3–Cdc20 complex was negligible in α-factor-arrested cells, where Cdc20myc18 was barely detectable in total extracts, whereas it was apparent in cycling cells and was even further increased when the checkpoint was activated by nocodazole treatment (Figures 1 and 2). It is worth noting that immunodepletion of Bub3HA3 did not deplete the extracts of Cdc20myc18 (Figure 1), indicating that there might exist a pool of Cdc20 free from Bub3 even during checkpoint activation. This is not due to limiting amounts of Bub3 versus Cdc20, because a significant fraction of Bub3 remained in the extracts after Cdc20 immunodepletion. We also found that, in addition to Cdc20myc18, both Mad1 and Mad2 associated with Bub3HA3 and inter action increased upon nocodazole treatment (Figure 1B). Conversely, Cdc20myc18 immunoprecipitates contained Bub3HA3 and Mad2, but not Mad1, suggesting that either Mad1 is not part of the Cdc20–Bub3 complex, or the levels of Mad1 bound to Cdc20 are below the detection limit. Interestingly, deletion of either MAD1 or MAD2 dramatically impaired formation of the Bub3–Cdc20 complex, as well as interaction of Mad2 with both Cdc20 and Bub3 and of Mad1 with Bub3 (Figure 1B). In particular, Mad2 was indispensable for the association of Bub3 with both Cdc20 and Mad1, whereas some residual Bub3–Cdc20–Mad2 interactions could still be detected in the absence of Mad1 (Figure 1B).
Fig. 1. Bub3 interacts physically with Cdc20, Mad1 and Mad2. (A) A cycling culture (cyc) of strain ySP1444, expressing both myc-tagged Cdc20 (Cdc20myc18) and HA-tagged Bub3 (Bub3HA3), was arrested either in G1 with α-factor (αf) or in G2 with nocodazole (noc). As negative controls, nocodazole-arrested cells expressing either Cdc20myc18 (ySP1413) or Bub3HA3 (ySP1346) were used. The presence (+) or absence (–) of the corresponding tagged proteins in the strains is indicated in the top part of the panel. Bub3 was immunoprecipitated from the extracts with anti-HA antibodies, whereas Cdc20 was immunoprecipitated with anti-myc antibodies. Immunoprecipitates (IP) with the antibodies indicated on the left side of the figure, along with the same amounts of total and immunodepleted extracts, were then run on SDS–PAGE and immunoblotted with both anti-HA (top) and anti-myc antibodies (bottom). (B) Cycling cultures (cyc) of wild-type (wt, ySP1444), _mad1_Δ (ySP1506) and _mad2_Δ (ySP1507) cells, all expressing both Bub3HA3 and Cdc20myc18, as well as nocodazole-arrested ySP1444 cells (wt noc), were harvested and protein extracts used for immunoprecipitations with anti-HA (left panel) or anti-myc (right panel) antibodies and analysed as in (A). Polyclonal antibodies were used to detect Mad1 and Mad2. Negative controls (mock) are represented by extracts from logarithmically growing strains expressing only either Cdc20myc18 (ySP1414) or Bub3HA3 (ySP1346) (left and right panel, respectively).
The interaction between Cdc20 and Bub3 is cell cycle regulated and is stimulated upon checkpoint activation
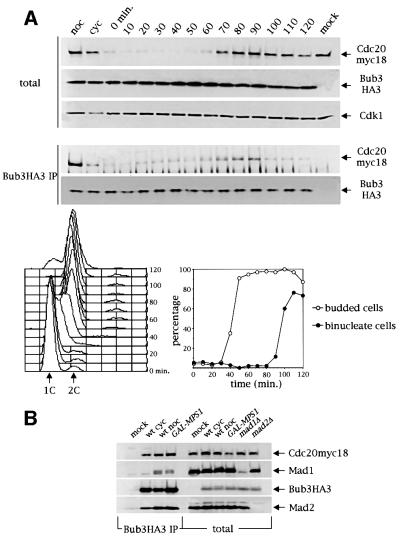
In order to gain insights into the physiological role of the Bub3–Cdc20 complex, we first analysed its appearance during an unperturbed cell cycle. For this purpose, cells expressing Bub3HA3 and Cdc20myc18 were synchronized in G1 by α-factor and the amount of Cdc20myc18 bound to immunoprecipitated Bub3HA3 was analysed at different time points after release into fresh medium (Figure 2A). In agreement with previous findings (Brady and Hardwick, 2000), Bub3 protein levels were constant throughout the cell cycle, whereas Cdc20 accumulated in mitosis and disappeared before cytokinesis (Figure 2A). These kinetics are similar to those of the mitotic cyclin Clb2 (Shirayama et al., 1998). As shown in Figure 2A, the amount of the Bub3–Cdc20 complex varied during the cell cycle and did not reflect exactly the periodicity of Cdc20 protein levels. Association increased at the G1–S transition, when Cdc20 protein levels were almost undetectable in the total extracts (see also Figure 3B). Interestingly, at this stage of the cell cycle Bub1 and Bub3 were found to interact with Mad1 (Brady and Hardwick, 2000). Subsequently, the amount of the Bub3–Cdc20 complex increased, reflecting the accumulation of Cdc20, and dropped again at the onset of anaphase, which took place at ∼90–100 min, as shown by the appearance of binucleate cells (Figure 2A, bottom right). If nocodazole was added after the release from α-factor the amount of Bub3–Cdc20 remained constantly high (data not shown).
Fig. 2. Bub3–Cdc20 interaction is cell cycle regulated and is stimulated upon checkpoint activation. (A) A cycling culture (cyc) of strain ySP1444 expressing both Cdc20myc18 and Bub3HA3, was either blocked with nocodazole (noc) or arrested in G1 with α-factor (t = 0 min) and then released. At the indicated time points after α-factor release cell samples were collected to produce protein extracts (upper panel), FACS analysis of DNA contents (bottom left) and kinetics of budding and nuclear division (bottom right). Protein extracts were analysed by immunoblotting either directly to detect at each time point the total amounts of Cdc20myc18 and Bub3HA3 (total) or after immunoprecipitation of Bub3 with anti-HA antibodies (Bub3HA3 IP). The negative control (mock) is represented by strain ySP1414, expressing Cdc20myc18 and untagged Bub3. (B) Protein extracts from the following strains were used for immunoprecipitations of Bub3HA3 as described for Figure 1: wild type (wt, ySP1444), either cycling (cyc) or arrested for 3 h in nocodazole (noc), a strain carrying the galactose-inducible GAL-MPS1 construct (ySP2025) arrested by 3 h of incubation in galactose, and a negative control (mock, ySP1414). Both total extracts and immunoprecipitates (Bub3HA3 IP) were analysed by western blot. As controls, total extracts from _mad1_Δ (ySP1506) and _mad2_Δ (ySP1507) cells were similarly analysed.
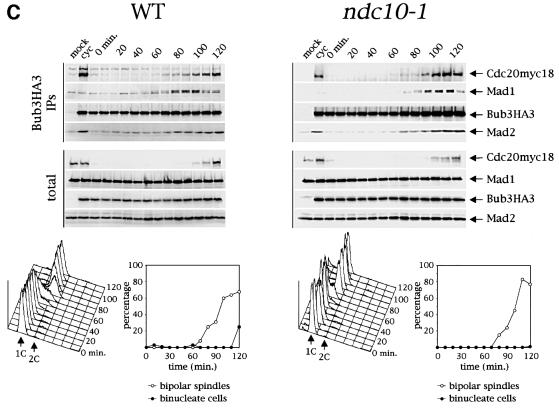
Fig. 3. Bub3–Cdc20 association does not require intact kinetochores. (A) Cell cultures of either a wild-type strain (wt) or the indicated checkpoint mutants, all expressing both Bub3HA3 and Cdc20myc18 were arrested in the same stage of the cell cycle by HU. Whereas wt (ySP1444), _mad1_Δ (ySP1506), _mad2_Δ (ySP1507), mad3 (ySP1512), bub1-1 (ySP1508) and _bub2_Δ (ySP1513) cell cultures were arrested by HU treatment (150 mM) for 3 h at 25°C, ndc10-1 (ySP1510) and mps1-1 (ySP1553) cells were first synchronized in G1 by α-factor at 25°C and then released in the presence of HU (100 mM) at 37°C for 4 h, to obtain a homogeneous arrest. Bub3 was immunoprecipitated with anti-HA antibodies and both total extracts and immunoprecipitates were analysed by western blot. As a negative control (mock), a strain expressing only Cdc20myc18 (ySP1414) was used. (B) Cycling cultures (cyc) of strains ySP1444 (wt), ySP2171 (ndc10-1), ySP2162 (ndc80-1), ySP2164 (spc25-1), ySP2166 (spc24-1), ySP1507 (_mad2_Δ) and ySP1413 (mock) were arrested in G1 by α-factor at 25°C and released from α-factor at 37°C for 4 h in the presence of HU. Cell extracts (total) and Bub3HA3 immunoprecipitates (Bub3HA3 IP) were analysed by western blot with anti-myc (Cdc20myc18) and anti-Mad1, -Mad2 and -Mad3 antibodies. (C) Wild-type (ySP1444) and ndc10-1 (ySP1510) cycling cells (cyc) were arrested in G1 by α-factor at 25°C (t = 0 min) and then released from α-factor at 37°C. At the indicated times cell samples were collected to produce protein extracts (upper panel), FACS analysis of DNA contents and kinetics of bipolar spindle formation and nuclear division (bottom panel). Western blot analysis with anti-myc (Cdc20myc18), anti-HA (Bub3HA3), anti-Mad1 and anti-Mad2 antibodies was performed on protein extracts (total) and Bub3HA3 immunoprecipitates (Bub3HA3 IP). The negative control (mock) is represented by strain ySP1414, expressing Cdc20myc18 and untagged Bub3.
To verify whether formation of specific Bub3 complexes is up-regulated in response to checkpoint activation, we investigated whether association of Mad1, Mad2 and Cdc20 to Bub3 could be induced by high levels of Mps1 (Figure 2B), which activates the kinetochore checkpoint in the absence of spindle damage (Hardwick et al., 1996). Overexpression of MPS1 from the galactose-inducible GAL1-10 promoter (GAL–MPS1 fusion) caused Mad1, Mad2 and Cdc20myc18 to interact with Bub3HA3 to levels similar to those obtained by nocodazole treatment (Figure 2B). Therefore, Bub3 association with Mad1, Mad2 and Cdc20 is up-regulated during mitotic checkpoint activation, although the most striking differences concern the Mad1–Bub3 interaction.
The Bub3–Cdc20 interaction requires other checkpoint proteins but not an intact kinetochore
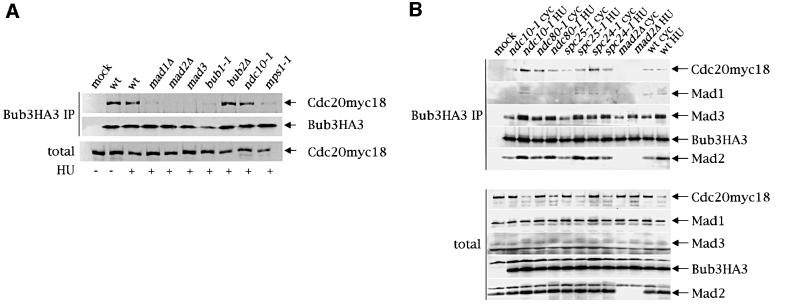
As shown in Figure 1B, Bub3–Cdc20 interaction requires Mad1 and Mad2. To examine whether other mitotic checkpoint proteins are important for this interac tion, we performed Bub3–Cdc20 co-immunoprecipitation experiments using extracts from strains bearing mutations in different checkpoint genes. In order to analyse all strains at the same cell cycle stage, the co-immunoprecipitation experiments were carried out from cells arrested in S phase with hydroxyurea (HU), due to the fact that nocodazole would not arrest the checkpoint mutants. As shown in Figure 3A, interaction between Bub3 and Cdc20 was severely impaired, if not absent, in mad1, mad2, mad3, bub1 and mps1 mutants, consistent with the notion that they all affect the same kinetochore checkpoint signal transduction cascade which also involves Bub3 (Hardwick and Murray, 1995; Wang and Burke, 1995; Hardwick et al., 1996; Pangilinan and Spencer, 1996; Alexandru et al., 1999). Conversely, interaction was unaltered, as expected, in extracts from _bub2_Δ cells, which are unable to activate a different pathway detecting errors in spindle orientation (Bardin et al., 2000; Bloecher et al., 2000; Pereira et al., 2000). Surprisingly, we found that the ndc10-1 mutation, which disrupts kinetochore function (Goh and Kilmartin, 1993) as well as checkpoint response (Ciosk et al., 1998; Tavormina and Burke, 1998; Fraschini et al., 2001), did not affect the formation of the Bub3–Cdc20 complex (Figure 3A), suggesting that the association between these two proteins does not require intact kinetochores. We therefore repeated this analysis on other temperature-sensitive mutants affecting either both kinetochore function and mitotic checkpoint response, such as spc24-1 and spc25-1 (Janke et al., 2001; Wigge and Kilmartin, 2001; our unpublished observations), or only kinetochore function, like ndc80-1 (Wigge et al., 1998). The Spc24, Spc25 and Ndc80 proteins have recently been shown to be part, together with Nuf2, of a protein complex residing on kinetochores and SPBs, which is essential for chromosome segregation (Wigge et al., 1998; Janke et al., 2001; Wigge and Kilmartin, 2001). We synchronized in α-factor at 25°C wild type, ndc10-1, _mad2_Δ, ndc80-1, spc24-1 and spc25-1 strains expressing both Cdc20myc18 and Bub3HA3, followed by release from α-factor in the presence of HU at the restrictive temperature for 4 h. Cell extracts were then prepared, and the presence of Cdc20myc18, Mad1, Mad2 and Mad3 in Bub3HA3 immunoprecipitates was analysed. Similar amounts of Cdc20myc18 could be co-immunoprecipitated with Bub3HA3 from both cycling and HU-blocked wild-type cells, despite the lower levels of total Cdc20myc18 in the latter conditions, thus confirming that the interaction between these proteins increases during S phase (Figure 3B). Cdc20myc18, Mad2 and Mad3 co-immunoprecipitated with Bub3HA3 in all kinetochore mutants, whereas, as expected, Bub3HA3–Cdc20myc18 interaction was abolished by deletion of MAD2, which did not affect association of Bub3HA3 to Mad3, as previously shown (Hardwick et al., 2000). The extremely low levels of Mad1 pulled down with Bub3HA3 even in wild-type cells under these conditions did not allow us to establish unequivocally whether the ndc80-1, spc24-1 and _mad2_Δ mutations affected the Mad1–Bub3HA3 interaction, which, however, did not seem to require Ndc10 and Spc25 (Figure 3B).
Since the above results did not rule out the possibility that the checkpoint defect of kinetochore mutants could be related to defects in the timing of association of Bub3 with the other checkpoint proteins, we also determined the kinetics of complex formation between Bub3 and Mad1, Mad2 and Cdc20 in synchronized wild-type and ndc10-1 cells released from a G1 arrest at 37°C. Association of Bub3HA3 with Mad1, Mad2 and Cdc20myc18 took place at the onset of S phase and increased in mitosis in both strains (Figure 3C). Therefore, intact kinetochores do not appear to be required for the cell cycle regulation of these interactions.
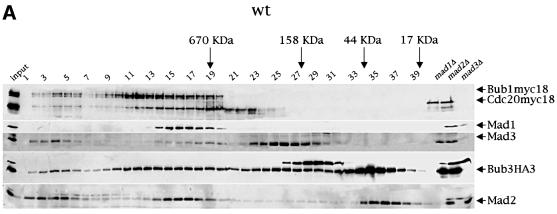
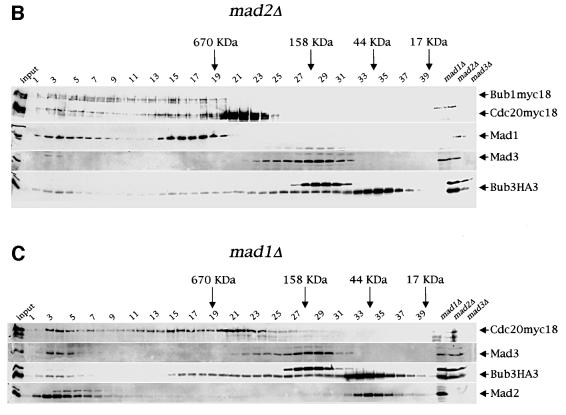
Co-fractionation of Mad2, Mad3, Bub3 and Cdc20 upon gel filtration depends on Mad1 and Mad2
The finding that Bub3 interacts not only with Mad1, Mad2, Mad3 and Bub1, but also with Cdc20, and that some of these interactions are up-regulated upon mitotic checkpoint activation, raises the possibility that all these proteins might assemble in the same complex, leading to inhibition of Cdc20–APC. Alternatively, Bub3 may associate with the above proteins to form different subcomplexes. To distinguish between these two possibilities, we fractionated by gel filtration protein extracts from cycling cells expressing simultaneously Bub3HA3, Cdc20myc18 and Bub1myc18. Cell lysates were loaded on a Superose 6 column and the collected fractions were analysed by western blotting. As shown in Figure 4A, we found that Mad2, Mad3, Bub1myc18, Bub3HA3 and Cdc20myc18 co-fractionated in two peaks (corresponding to fractions 2–6 and 14–19, respectively), which might identify two distinct complexes, both eluting with apparent molecular weights >670 kDa. Mad1 was present in the fractions corresponding to the hypothetical complex of lower molecular weight (Figure 4A), but in some experiments we could detect it in low amounts also in the other (Figure 4B and data not shown). No free Mad1, Mad3, Bub1myc18 and Cdc20myc18 proteins could be detected upon gel filtration, whereas quite large amounts of both Mad2 (22 kDa) and Bub3HA3 (45 kDa) were present as free pools (fractions 34–38). Interestingly, in addition to fractions 14–19, fractions 9–13 contained both Bub1myc18 and Bub3HA3, but not the other checkpoint proteins, whereas fractions 23–30 contained only Mad3 and Bub3, suggesting that constitutive interaction between Bub3 and either Bub1 (Roberts et al., 1994; Brady and Hardwick, 2000) or Mad3 (Hardwick et al., 2000) can take place within different subcomplexes.
Fig. 4. Co-fractionation of Mad2, Mad3, Bub3 and Cdc20 in gel filtration chromatography. (A) A protein extract from a cycling culture of a strain (ySP2156) expressing Bub1myc18, Cdc20myc18 and Bub3HA3 was fractionated by gel filtration (see Materials and methods) and fractions 1–40 were analysed by western blot with anti-HA (Bub3HA3), anti-myc (Bub1myc18 and Cdc20myc18), anti-Mad1, -Mad2 and -Mad3 antibodies. Total protein extracts from _mad1_Δ (ySP1506), _mad2_Δ (ySP1507) and _mad3_Δ (ySP1577) cell cultures were loaded on the same gel to identify aspecific immunoreactive bands. (B and C) Protein extracts from logarithmically growing _mad2_Δ (ySP2317) or _mad1_Δ (ySP1506) cells were fractionated and analysed as in (A).
To investigate whether the two putative protein complexes identified by all the co-fractionating proteins might have a physiological relevance in kinetochore checkpoint activation, we fractionated and analysed with the same method protein extracts from _mad2_Δ and _mad1_Δ cells (Figure 4B and C). Interestingly, the lack of either Mad2 or Mad1 led to the disappearance of Mad3 and to a decrease of Bub3 protein levels in the lower molecular weight complex (fractions 14–19). In addition, Mad2 was also missing from this complex in the extract from _mad1_Δ cells. Therefore, Mad2, Mad3 and Bub3 might be part of a large protein complex together with Cdc20, whose formation requires Mad1. The residual Bub3 protein found in fractions 14–19 of _mad2_Δ and _mad1_Δ extracts might derive from other Bub3 complexes, such as, for instance, Bub1–Bub3. Since the putative complex at very high molecular weight (fractions 2–6) remained unaffected in the absence of two key checkpoint proteins, we conclude that it is unlikely to play an important role in checkpoint activation. It is worth noting that deletion of MAD2 altered the elution profile of Cdc20, whose protein levels increased in the fractions at lower molecular weight (20–24), but not those of either Bub1 or Mad1, suggesting that Bub1 and Mad1 are part of different protein complex(es) from the one including Mad2, Mad3, Bub3 and Cdc20. The apparent molecular weight of the hypothetical checkpoint complex was higher than expected on the basis of association of single copies of the polypeptides under analysis (i.e. 218 kDa), suggesting that either additional proteins are associated or some or all of the above proteins are present in multiple copies within the complex. We found no major differences in the distribution of the above proteins upon gel filtration of protein extracts from nocodazole-treated wild-type cells (data not shown), in agreement with our co-immunoprecipitation results that indicate that association between Bub3 and Mad2 and Cdc20, although stimulated by checkpoint activation, takes place in all conditions.
Interactions between Bub3 and Mad2, Mad3 and Cdc20 require the Bub3 WD40 repeats
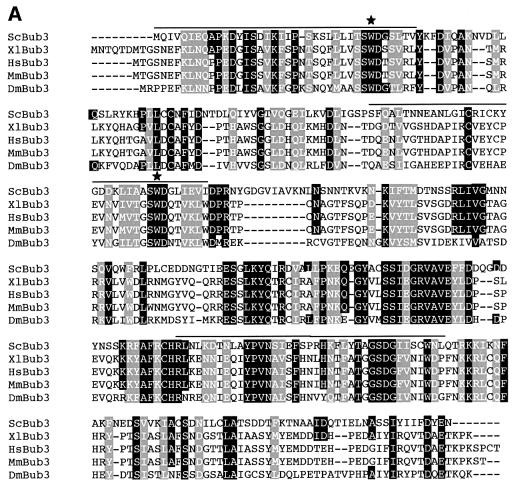
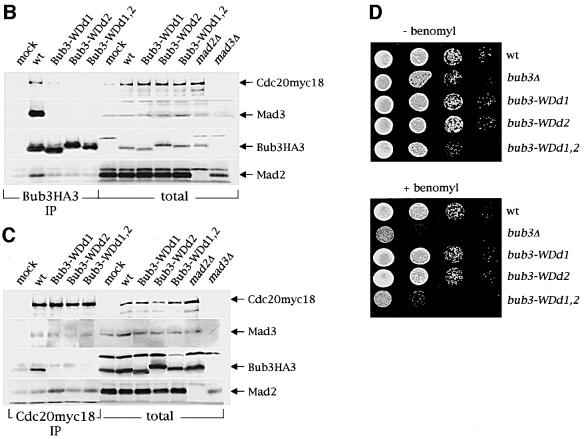
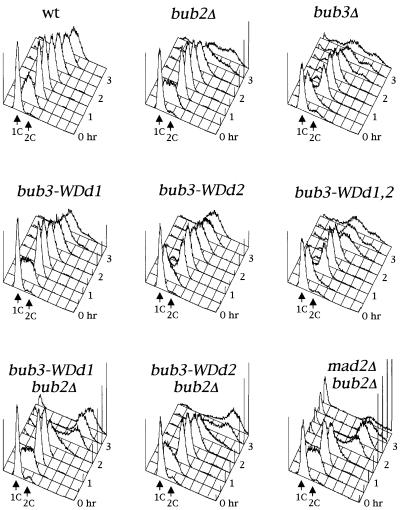
Three conserved regions in the budding yeast Bub3 protein sequence (overlined in Figure 5A) share significant homology with WD40 repeat domains, which contain a Trp-Asp motif. These motifs were first identified in β-subunits of trimeric G proteins (Neer et al., 1994) and have been proposed to adopt a β-propeller fold which mediates protein–protein interactions (Smith et al., 1999). This particular structure is highly symmetrical and consists of at least four structural repeats, or blades, of four β-strands each (Smith et al., 1999). In order to understand how the WD40 repeats of Bub3 might participate in its fold and function, we performed a threading analysis (Jones, 1999) using the PSIPRED server (http://insulin. brunel.ac.uk/psipred/). The top scores were characterized by the highest degree of statistical confidence, and all matched to β-propeller structures, including β-transducin, a well-characterized WD40-containing protein (Murzin, 1992). The Bub3 sequence aligned on the different structures with the same relative frame, further confirming the accuracy of the structural prediction and indicating that Bub3 is most likely to be a seven-bladed β-propeller. To address the physiological role of the Bub3 WD40 repeats, we then generated bub3 point mutations changing conserved solvent exposed amino acids. In particular, we selected residues W31 and W120 (Figure 5A) because, according to our structural alignment, these residues are predicted to be positioned in the β2–β3 loop of blades 1 and 3, respectively, and to be located on the top surface of the propeller. Residues in this position have been shown to be involved in the interaction of β-transducin with the α-subunit (Lambright et al., 1996), and their mutagenesis in Bub3 might be expected to reduce its functionality without affecting its stability. We generated three WD40-defective alleles (bub3-WDd) by introducing specific base-pair changes in the yeast BUB3 coding region, that caused the amino acid substitutions W31G (bub3-WDd1), W120G (bub3-WDd2) or both (bub3-WDd1,2) in the Bub3 protein (see Materials and methods), and analysed the in vivo effects of these alleles. As shown in Figure 5B, HA-tagged Bub3-WDd mutant proteins were no longer able to interact with Mad2, Mad3 and Cdc20 in co-immunoprecipitation experiments, although the mutant protein levels were comparable to those of wild-type Bub3 (Figure 5B). Conversely, binding of Mad2 and Mad3 to Cdc20 was unaffected by the presence of the bub3-WDd alleles (Figure 5C). Along with the defects in protein–protein interactions, bub3-WDd1,2 mutant cells displayed a slight slow growth phenotype and sensitivity to the microtubule depolymerizing drug benomyl similar to those of _bub3_Δ cells, whereas bub3-WDd1 and bub3-WDd2 mutants grew normally and were as sensitive to benomyl as wild-type cells (Figure 5D). To further investigate the possible effects of the bub3-WDd alleles on the mitotic checkpoint, the strains indicated in Figure 6 were synchronized in G1 by α-factor and then released from the block in the presence of nocodazole. As shown in Figure 6, nocodazole-treated bub3-WDd1,2 cells re-replicated their DNA, reaching DNA contents higher than 2C, and re-budded (not shown) with the same kinetics of _bub3_Δ cells under the same conditions, suggesting that point mutations affecting two WD40 repeats are sufficient to abolish completely Bub3 checkpoint function. Conversely, most bub3-WDd1 and bub3-WDd2 cells arrested cell cycle progression upon nocodazole treatment (Figure 6). However, both alleles turned out to cause a synthetic effect when combined with the deletion of BUB2, which acts in a different mitotic checkpoint pathway (Alexandru et al., 1999; Fesquet et al., 1999; Fraschini et al., 1999; Li, 1999). In fact, _bub3-WDd1 bub2_Δ and _bub3-WDd2 bub2_Δ cells re-replicated their DNA much faster and more efficiently than either _bub3_Δ or _bub2_Δ single mutants, although somewhat more slowly than a _mad2_Δ _bub2_Δ double mutant, which is totally defective in mitotic checkpoint signalling (Figure 6). Thus, the bub3-WDd1 and bub3-WDd2 alleles do indeed impair proper checkpoint response. Altogether, these results indicate that modification of a single WD40 motif might still allow Bub3 to interact in vivo with Mad2, Mad3 and Cdc20, but with reduced affinity, resulting in partial checkpoint defects and in disassembly of the complexes under the conditions used for the immunoprecipitation assays. Therefore, the interaction of Bub3 with Mad2, Mad3 and Cdc20 is likely to be mediated by its WD40 domains, which are also required for proper checkpoint response.
Fig. 5. Bub3 WD40 repeats are required for interaction with Mad2, Mad3 and Cdc20. (A) Clustal W alignment of the Bub3 protein sequence from different organisms (Sc: Saccharomyces cerevisiae; Xl: Xenopus laevis; Hs: Homo sapiens; Mm: Mus musculus; Dm: Drosophila melanogaster). Lines above the ScBub3 sequence mark the three WD40 domains. Stars indicate the two Trp residues changed into Gly in the Bub3-WDd mutant protein. (B) Protein extracts from wild-type (wt, ySP2076), bub3-WDd1 (ySP2079), bub3-WDd2 (ySP2082) and bub3-WDd1,2 (ySP2085) mutant cells were used to analyse by western blot either the total levels of Mad2, Mad3, Bub3HA3 and Cdc20myc18 (total) or the amounts of the same proteins that could be co-immunoprecipitated with Bub3HA3 (Bub3HA3 IP). The negative control (mock) is represented by strain ySP1414, expressing Cdc20myc18 and untagged Bub3. Cell extracts from _mad2_Δ (ySP1507) and _mad3_Δ (ySP1577) cultures were loaded in parallel to identify aspecific bands recognized by anti-Mad2 and -Mad3 antibodies. (C) The same protein extracts as in (B) were used to also analyse the amounts of Mad3, Bub3HA3 and Mad2 in total extracts and in Cdc20myc18 immunoprecipitates (Cdc20myc18 IP). (D) Serial dilutions of exponentially growing cultures of wild-type (wt, W303), _bub3_Δ (ySP589), bub3-WDd1 (ySP2198), bub3-WDd2 (ySP2202) and bub3-WDd1,2 (ySP2211) strains were spotted on YEPD plates either lacking or containing benomyl and photographed after incubation at 30°C for 2 days.
Fig. 6. Mutations altering the WD40 repeats of Bub3 impair proper checkpoint response. Wild-type, _bub3_Δ (ySP589), _bub2_Δ (ySP1071), bub3-WDd1 (ySP2198), bub3-WDd2 (ySP2202), bub3-WDd1,2 (ySP2211), _bub3-WDd1 bub2_Δ (ySP2197) _bub3-WDd2 bub2_Δ (ySP2201) and _mad2_Δ _bub2_Δ (ySP1152) cells were arrested in G1 by α-factor and then released at 25°C in the presence of nocodazole. At the indicated times cell samples were collected for FACS analysis of the DNA contents.
Discussion
Several biochemical observations indicate that in yeast cells the Bub1–Bub3, Mad3–Bub3 and Mad1–Mad2 complexes are present throughout the cell cycle (Roberts et al., 1994; Hardwick et al., 2000). Mutations affecting any of these complexes disrupt the checkpoint response, although they do not interfere with formation of the other two, suggesting that anyone of these complexes is not sufficient for checkpoint activation. Mad2 interacts physically with Cdc20 and this association does not appear to be restricted to checkpoint activation (Hwang et al., 1998). Mad2–Cdc20 interaction depends on Mad1 and Mps1 (Hwang et al., 1998; Hardwick et al., 2000), the latter possibly regulating Mad2–Cdc20 interaction by Mad1 phosphorylation (Hardwick et al., 1996). Mad3 has also been shown to bind Cdc20 (Hwang et al., 1998; Hardwick et al., 2000), and interaction strictly depends on Mad1 and Mad2 and only partially on Bub1 and Bub3 (Hardwick et al., 2000). Finally, Mad1 has been found to bind to Bub1 and Bub3 and this interaction is not only stimulated by treatment with microtubule depolymerizing drugs, but is also cell cycle regulated, being maximal during the S phase of an unperturbed cell cycle, when presumably duplicated centromeres start capturing spindle microtubules via their kinetochores leading to checkpoint activation (Brady and Hardwick, 2000). In fact, not only in yeast cells centromeres are replicated very early (McCarroll and Fangman, 1988) and formation of a bipolar spindle occurs already during S phase, but separation of duplicated centromeric regions, which depends on kinetochore–microtubule binding (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000), seems to occur before the end of S phase (Goshima and Yanagida, 2000).
Altogether, the above observations suggest that some of the above proteins might be brought together preferentially during a cell cycle stage in which the checkpoint is activated due to kinetochore capturing of microtubules. So far, it was unclear whether Bub1 and Bub3 could be physically part of a complex together with Mad1, Mad2, Mad3 and Cdc20, leaving open the question of whether Bub1 and Bub3 might be upstream regulators of Mad1, Mad2 and Mad3. We have shown here that Bub3 does indeed co-immunoprecipitate with Cdc20, as well as with Mad1, Mad2 and Mad3, suggesting that all these proteins do interact in vivo. In addition, we found that Mad2, Mad3, Bub3 and Cdc20 also co-fractionate in gel filtration chromatography (Figure 4), suggesting the existence of a Mad2–Mad3–Bub3–Cdc20 complex, whose assembly depends on Mad1 (Figure 4C). Accordingly, the Bub3– Cdc20 interaction requires all proteins involved in the kinetochore checkpoint, and Bub3 no longer co-immunoprecipitates with Mad2 in the absence of Mad1 (Figure 1B). Our results are in agreement with the recent discovery in HeLa cells of a mitotic checkpoint complex (MCC) containing the human Mad3 homologue BubR1, Bub3, Cdc20, and presumably Mad2, which inhibits the activity of mitotic APC (Sudakin et al., 2001; Tang et al., 2001). Altogether, these data suggest that the mechanism by which the kinetochore checkpoint operates is conserved throughout evolution. In a scenario where the Mad2– Mad3–Bub3–Cdc20 complex directly inhibits APC activity, it is possible to speculate that Mad1, Bub1 and Mps1 play an early function in kinetochore checkpoint signalling and positively regulate the assembly of checkpoint complexes.
Bub3 contains WD40 domains, which probably participate, together with other protein regions, in the folding of a β-propeller three-dimensional structure, as indicated by structural prediction analysis. Substitution with glycines of two Bub3 conserved tryptophan residues, which should be exposed on the top surface of the propeller, abolishes the interaction of Bub3 with Cdc20, Mad2 and Mad3. The observation that these mutations do not seem to compromise Bub3 stability suggests that the Bub3 WD40 repeats might be specifically involved in protein–protein interactions. Indeed, WD-repeat propeller structures have been proposed to create a stable platform that can coordinate sequential and/or simultaneous interactions of several sets of proteins (Smith et al., 1999). In this view, Bub3 might mediate the interactions between different subcomplexes of kinetochore checkpoint proteins and Cdc20. Indeed, formation of the Mad2–Mad3–Bub3– Cdc20 complex seems to correlate with the metaphase arrest caused by checkpoint activation, since mutations altering the Bub3 WD40 repeats impair not only complex formation, but also the ability to properly arrest cell cycle progression in the presence of microtubule depolymerizing drugs.
The Bub3–Cdc20 interaction, although constitutively present at a basal level, is cell cycle regulated: it increases as cells enter S phase, when both Mad1 (Brady and Hardwick, 2000) and Mad2 (Figure 3B and C) form a complex with Bub3, and peaks in mitosis, subsequently dropping at the onset of anaphase. Furthermore, formation of the Bub3–Cdc20 complex is stimulated upon checkpoint activation by both nocodazole treatment and Mps1 overproduction. Surprisingly, although there is an increase in the amount of Cdc20 bound to either Mad3 (Hardwick et al., 2000) or Bub3 (this paper) during nocodazole arrest compared with other cell cycle phases, we found that immunodepletion of Bub3 does not deplete the extracts of Cdc20, indicating that even in these conditions there might be a pool of free Cdc20. Why is this pool of Cdc20 unable to drive mitotic progression? One possibility is that this fraction of Cdc20 is compartmentalized elsewhere in the cell and cannot promote sister chromatid separation. Alternatively, the amount of free Cdc20 might go below a threshold level necessary to trigger anaphase. Although further experiments will be required to address this issue, we think that our observation is consistent with the notion that the action of the inhibitor(s) produced by unattached kinetochores is not freely diffusible but is restrained to the spindle containing the unattached kinetochore(s). In fact, in cells containing two independent spindles in the same cytoplasm, the anaphase onset of a mature spindle is not delayed by one or more unattached kinetochores in the adjacent spindle (Rieder et al., 1997), suggesting that in these conditions there exists in the cell a fraction of Cdc20 able to drive anaphase in one of the spindles but not in the other.
Another important issue concerns the role of the kinetochore in checkpoint activation. We found that binding of Bub3 to Mad1, Mad2, Mad3 and Cdc20 does not require the kinetochore proteins Ndc10, Spc24 and Spc25, although the mutations used for our analysis cause a mitotic checkpoint defect (Tavormina and Burke, 1998; Fraschini et al., 2001; our unpublished observations). In addition, interaction of Bub3 with Mad1, Mad2 and Cdc20 occurs in a similar cell cycle regulated manner in both wild-type and ndc10-1 mutant cells. If, as previously suggested (Brady and Hardwick, 2000), association between the above proteins coincides with checkpoint activation, then ndc10-1, spc24-1 and spc25-1 cells are proficient in transmitting the inhibitory signals. This result is particularly revealing for two reasons: first, the possible formation of a complex containing Mad2, Mad3, Bub3 and Cdc20 would be necessary but not sufficient for checkpoint activation. Secondly, formation of this complex would not require intact kinetochores, in spite of the fact that at least some of the described physical interactions are thought to take place at the kinetochore. Consistently, localization of Drosophila Bub3 at centromeres does not require the kinetochore proteins ZW10 and Rod (Basu et al., 1998) and the mitotic checkpoint complex from interphase HeLa cells has APC inhibitory activity despite the fact that mature kinetochores are not present at this cell cycle stage (Sudakin et al., 2001). If association between checkpoint proteins is the first step towards checkpoint activation, then part or all of the signal transduction cascade leading to checkpoint activation might still be functional in kinetochore mutants that are unable to arrest cell cycle progression in the presence of nocodazole.
Materials and methods
Strains, media and reagents
All yeast strains were derivatives of or were backcrossed at least three times to W303 (ade2-1, trp1-1, leu2-3 112, his3-11,15, ura3, ssd1). Strains used for this work are listed in Table I. Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) supplemented with 2% glucose (YEPD) or 2% raffinose (YEPR). Galactose was added to YEPR at 1% to make YEPRG. α-factor was used at 2 µg/ml, nocodazole at 15 µg/ml, benomyl at 10 µg/ml and HU at 100 mM, unless otherwise specified. All strains were normally grown at 25°C and subsequently shifted to 37°C to inactivate the function of temperature-sensitive alleles.
Table I. Yeast strain genotypes.
| Name | Relevant genotype |
|---|---|
| ySP589 | MATa, bub3::LEU2 |
| ySP1071 | MATa, bub2::HIS3 |
| ySP1152 | MATa, bub2::HIS3, mad2::TRP1 |
| ySP1346 | MATa, bub3::BUB3-HA3::KlURA3 |
| ySP1413 | MATa, cdc20::CDC20-myc18::TRP1 |
| ySP1414 | _MAT_α, cdc20::CDC20-myc18::TRP1 |
| ySP1444 | MATa, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1506 | MATa, mad1::LEU2, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1507 | MATa, mad2::TRP1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1508 | MATa, bub1-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1510 | MATa, ndc10-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1512 | MATa, mad3, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1513 | MATa, bub2::HIS3, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1553 | MATa, mps1-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP1577 | MATa, mad3::KlTRP1 |
| ySP2025 | MATa, ura3::URA3 GAL-MPS1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2061 | MATa, bub3::LEU2, cdc20::CDC20-myc18::TRP1 |
| ySP2076 | MATa, bub3::LEU2, ura3::URA3::BUB3-HA3, cdc20::CDC20-myc18::TRP1 |
| ySP2079 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd1-HA3, cdc20::CDC20-myc18::TRP1 |
| ySP2082 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd2-HA3, cdc20::CDC20-myc18::TRP1 |
| ySP2085 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd1,2-HA3, cdc20::CDC20-myc18::TRP1 |
| ySP2156 | MATa, bub1::BUB1-myc18::KlTRP1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2162 | MATa, ndc80-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2164 | MATa, spc25-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2166 | MATa, spc24-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2171 | MATa, ndc10-1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
| ySP2197 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd1-HA3, bub2::HIS3 |
| ySP2198 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd1-HA3 |
| ySP2201 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd2-HA3, bub2::HIS3 |
| ySP2202 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd2-HA3 |
| ySP2211 | MATa, bub3::LEU2, ura3::URA3::bub3-WDd1,2-HA3 |
| ySP2317 | MATa, mad2::kanMX4, bub1::BUB1-myc18::KlTRP1, bub3::BUB3-HA3::KlURA3, cdc20::CDC20-myc18::TRP1 |
Plasmid constructions and genetic manipulations
Standard genetic techniques were used to manipulate yeast strains (Sherman, 1991) and standard protocols were used for genetic manipulations (Maniatis et al., 1992). BUB1 and BUB3 were tagged immediately before the stop codon by one-step gene tagging (Knop et al., 1999). The bub3-WDd alleles were generated by PCR site-directed mutagenesis: the yeast integrative plasmid pSP135, carrying an HA3-tagged BUB3 gene was used as a template for two independent PCRs where either the oligos 5′-GTCACTGCTTTTGATTACGTCTGGGGATGGCTCTTTAACAGTCTAC-3′ (coding) and 5′-GTAGACTGTTAAAGAGCCATCCCCAGACGTAATCAAAAGCAGTGAC-3′ or the oligos 5′-CGATAAACTCATTGCCGCGTCAGGGGATGGCCTGATAGAGGTTATC-3′ (coding) and 5′-GATAACCTCTATCAGGCCATCCCCTGACGCGGCAATGAGTTTATCG-3′ were used as primers. The resulting plasmids pSP139 and pSP140 carry the bub3-WDd1 and bub3-WDd2 alleles, encoding a Bub3 mutant protein where the tryptophan residues at position 31 or 120 were substituted by glycines, respectively. Plasmid pSP139 was used as template for a second round of mutagenesis with the latter set of primers described, to generate plasmid pSP141, which carries the bub3-WDd1,2 allele, encoding a Bub3 mutant protein with both the W31G and W120G amino acid substitutions. The presence of the expected mutations in all the plasmids was confirmed by DNA sequencing. Plasmids pSP139, pSP140 pSP141 were then cut with _Nco_I to direct integration at the URA3 locus of a haploid strain carrying a bub3::LEU2 deletion and the CDC20myc18 allele (ySP2061) to generate strains ySP2079, ySP2082 and ySP2085, respectively. Correct integration of the plasmids was assessed by Southern analysis.
Immunoprecipitations, gel filtration and western blot analysis
For immunoprecipitations cells were lysed with glass beads in 50 mM HEPES pH 7.4, 150 mM NaCl, 20% glycerol, 1 mM orthovanadate, 60 mM β-glycerophosphate supplemented with a cocktail of protease inhibitors (Complete, Boehringer Mannheim). Cleared extracts (1–2 mg) were incubated for 2 h with either antibodies and protein A–Sepharose which had been pre-incubated for 1 h, or with antibodies directly crosslinked to protein A–Sepharose. The slurry was washed three times with lysis buffer and three times with phosphate-buffered saline before loading. For gel filtration chromatography, cell lysates were prepared as for immunoprecipitations; ∼1 mg of extracts was loaded on a Superose 6 column (Pharmacia-Amersham, final volume 3 ml) and fractionated at 4°C using a SMART system (Pharmacia Biotech). Thirty microlitre fractions were collected at a flow rate of 50 µl/min after the first 1 ml had passed and analysed by SDS–PAGE followed by western blotting. The column was calibrated with native protein standards (Bio-Rad) according to instructions provided by the supplier. For western blot analysis proteins were transferred to Protran membranes (Schleicher and Schuell). myc-tagged proteins were detected with 9E10 mAb, HA-tagged proteins with mAb 12CA5 whereas Mad1, Mad2, Mad3 and Cdk1 were detected with polyclonal antibodies (Hardwick and Murray, 1995; Hardwick et al., 2000). Secondary antibodies were purchased from Amersham and proteins were detected by an enhanced chemiluminescence system according to the manufacturer.
Other techniques
Flow cytometric DNA quantitation was determined according to Epstein and Cross (1992) on a Becton-Dickinson FACScan. In situ immunofluorescence was performed as described in Nasmyth et al. (1990).
Acknowledgments
Acknowledgements
We are grateful to Kevin Hardwick for antibodies against Mad1, Mad2 and Mad3, John Kilmartin and Kim Nasmyth for yeast strains, Maria Pia Longhese for critical reading of the manuscript. This work has been supported by grants from Associazione Italiana Ricerca sul Cancro and Training and Mobility of Researchers contract ERBFMRXCT98-0212 to S.P., Cofinanziamenti 1999 and 2000 MURST-Università di Milano-Bicocca to G.L. and CNR Target Project on Biotechnology Grant CT.97.01180.PF49(F).
References
- Alexandru G., Zachariae,W., Schleiffer,A. and Nasmyth,K. (1999) Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J., 18, 2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Visintin,R. and Amon,A. (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell, 102, 21–31. [DOI] [PubMed] [Google Scholar]
- Basu J., Logarinho,E., Herrmann,S., Bousbaa,H., Li,Z., Chan,G.K., Yen,T.J., Sunkel,C.E. and Goldberg,M.L. (1998) Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma, 107, 376–385. [DOI] [PubMed] [Google Scholar]
- Bloecher A., Venturi,G.M. and Tatchell,K. (2000) Anaphase spindle position is monitored by the BUB2 checkpoint. Nature Cell Biol., 2, 556–558. [DOI] [PubMed] [Google Scholar]
- Brady D.M. and Hardwick,K.G. (2000) Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol., 10, 675–678. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko,A., Mann,M. and Murray,A.W. (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol., 143, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady,D.M., Smith,D., Murray,A.W. and Hardwick,K.G. (1999) The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell, 10, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., Zachariae,W., Michaelis,C., Shevchenko,A., Mann,M. and Nasmyth,K. (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell, 93, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Epstein C.B. and Cross,F.R. (1992) CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev., 6, 1695–1706. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick,P.J., Johnson,A.L., Kramer,K.M., Toyn,J.H. and Johnston,L.H. (1999) A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J., 18, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti,E., Lucchini,G. and Piatti,S. (1999) Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol., 145, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta,A., Lucchini,G. and Piatti,S. (2001) Role of the kinetochore protein Ndc10 in mitotic checkpoint activation. Mol. Gen. Genom., 266, 115–125. [DOI] [PubMed] [Google Scholar]
- Goh P.Y. and Kilmartin,J.V. (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol., 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen,R.H. and Murray,A.W. (1998) Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol., 141, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G. and Yanagida,M. (2000) Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell, 100, 619–633. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G. and Murray,A.W. (1995) Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol., 131, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss,E., Luca,F.C., Winey,M. and Murray,A.W. (1996) Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science, 273, 953–956. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Johnston,R.C., Smith,D.L. and Murray,A.W. (2000) MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p and Mad2p. J. Cell Biol., 148, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana,S. and Sorger,P.K. (2000) Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell, 101, 763–775. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A. (2000) Exit from mitosis: spindle pole power. Cell, 102, 267–270. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint [published erratum appears in Science (1998) 280, 1331]. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Janke C., Ortiz,J., Lechner,J., Shevchenko,A., Magiera,M.M., Schramm,C. and Schiebel,E. (2001) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J., 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. (1999) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol., 287, 797–815. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein,J., Daum,J.R., Burke,D.J. and Gorbsky,G.J. (1998) Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol., 141, 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lambright D.G., Sondek,J., Bohm,A., Skiba,N.P., Hamm,H.E. and Sigler,P.B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature, 379, 311–319. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Li R. (1999) Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl Acad. Sci. USA, 96, 4989–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.F. and Sambrook,J. (1992) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- McCarroll R.M. and Fangman,W.L. (1988) Time of replication of yeast centromeres and telomeres. Cell, 54, 505–513. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1992) Structural principles for the propeller assembly of β-sheets: the preference for seven-fold symmetry. Proteins, 14, 191–201. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Adolf,G., Lydall,D. and Seddon,A. (1990) The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell, 62, 631–647. [DOI] [PubMed] [Google Scholar]
- Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature, 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Pangilinan F. and Spencer,F. (1996) Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell, 7, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Hofken,T., Grindlay,J., Manson,C. and Schiebel,E. (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell, 6, 1–10. [PubMed] [Google Scholar]
- Rieder C.L., Cole,R.W., Khodjakov,A. and Sluder,G. (1995) The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol., 130, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Khodjakov,A., Paliulis,L.V., Fortier,T.M., Cole,R.W. and Sluder,G. (1997) Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl Acad. Sci. USA, 94, 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Farr,K.A. and Hoyt,M.A. (1994) The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol., 14, 8282–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C. and Pellman,D. (2001) Search, capture and signal: games microtubules and centrosomes play. J. Cell Sci., 114, 247–255. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae,W., Ciosk,R. and Nasmyth,K. (1998) The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J., 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan,G.K.T. and Yen,T.J. (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J. Cell Biol., 154, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs,J., Loidl,J. and Nasmyth,K. (2000) Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol., 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj,R., Li,B. and Yu,H. (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell, 1, 227–237. [DOI] [PubMed] [Google Scholar]
- Tavormina P.A. and Burke,D.J. (1998) Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics, 148, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. and Burke,D.J. (1995) Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 6838–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (1998) Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc. Natl Acad. Sci. USA, 95, 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (2001) Mitotic checkpoints: from yeast to cancer. Curr. Biol., 11, 83–90. [DOI] [PubMed] [Google Scholar]
- Weiss E. and Winey,M. (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol., 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen,O.N., Holmes,S., Soues,S., Mann,M. and Kilmartin,J.V. (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol., 141, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A. and Kilmartin,J.V. (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol., 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]