Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination (original) (raw)
Abstract
Escherichia coli ribosomal RNA (rRNA) operons contain antitermination motifs necessary for forming terminator-resistant transcription complexes. In preliminary work, we isolated ‘antiterminating’ transcription complexes and identified four new proteins potentially involved in rRNA transcription antitermination: ribosomal (r-) proteins S4, L3, L4 and L13. We show here that these r-proteins and Nus factors lead to an 11-fold increase in terminator read-through in in vitro transcription reactions. A significant portion of the effect was a result of r-protein S4. We show that S4 acted as a general antitermination factor, with properties very similar to NusA. It retarded termination and increased read-through at Rho-dependent terminators, even in the absence of the rRNA antiterminator motif. High concentrations of NusG showed reduced antitermination by S4. Like rrn antitermination, S4 selectively antiterminated at Rho-dependent terminators. Lastly, S4 tightly bound RNA polymerase in vivo. Our results suggest that, like NusA, S4 is a general transcription antitermination factor that associates with RNA polymerase during normal transcription and is also involved in rRNA operon antitermination. A model for key r-proteins playing a regulatory role in rRNA synthesis is presented.
Keywords: antitermination/r-protein S4/rrn transcription
Introduction
In Escherichia coli, premature arrest of translation and release of ribosomes from mRNA often causes inhibition of downstream transcription, a phenomenon known as polarity. It is thought that the denuding of the mRNA of ribosomes permits the transcription termination factor Rho to gain access to the message and inhibit RNA polymerase (for review see Adhya and Gottesman, 1978). Despite the fact that they are not translated, and have been shown to contain Rho-dependent termination sites, the ribosomal RNA (rRNA) operons (rrn) in E.coli are not subject to polarity (Morgan, 1980; Brewster and Morgan, 1981; Aksoy et al., 1984; Li et al., 1984; Berg et al., 1989). This is obviously vital in ensuring stoichiometric production of the co-transcribed 16S, 23S and 5S rRNAs. The absence of polarity in the rrn operons can be attributed to an antitermination mechanism invoked just as RNA polymerase is about to begin transcribing the 16S rRNA and, again, just as it is about to enter the 23S rRNA coding sequence (Heinrich et al., 1995; Pfeiffer and Hartmann, 1997).
The rRNA antitermination mechanism (reviewed in Condon et al., 1995) has much in common with the well studied N/nut antitermination system of the bacteriophage lambda. In the lambda system, RNA polymerase is modified by the lambda N protein and several E.coli host proteins, NusA, NusB, NusE [ribosomal (r)-protein S10] and NusG, to form a termination-resistant elongation complex capable of proceeding for many kilobases and through many transcription terminators without stopping (DeVito and Das, 1994). Assembly of the elongation complex requires a promoter-proximal sequence in the RNA known as the nut site, which consists of three conserved elements known as boxA, boxB and boxC. The rrn operons also contain A, B and C box motifs, just downstream of the P2 promoter, although the order of the boxA and boxB elements is reversed. boxA and boxC are conserved sequences, while boxB is a stem–loop structure with no obvious sequence conservation. The boxA sequence is found in the leader and spacer regions of all rRNA operons in E.coli, and is a prominent feature of rRNA operons in many other bacteria (Berg et al., 1989). It is necessary and sufficient to promote read-through of Rho-dependent terminators both in vivo and in vitro (Li et al., 1984; Berg et al., 1989; Albrechtsen et al., 1990; Squires et al., 1993). Several studies have shown that boxA mutations in the leader or spacer regions of a plasmid-borne rrn operon result in significant rRNA synthesis defects, testament to the importance of the rrn antitermination mechanism in vivo (Gourse et al., 1983; Heinrich et al., 1995; Pfeiffer and Hartmann, 1997).
Previous experiments suggested that the rRNA antitermination system shares most, if not all of the Nus factors required by the lambda system. A NusB mutant allele was shown to cause premature transcription termination within rrn operons in vivo (Sharrock et al., 1985). NusA has been shown to be responsible for the increased transcription elongation rate of RNA polymerase on rrn operons relative to mRNA and to be necessary for rrn transcription antitermination in a _boxA_-dependent manner in vivo (Vogel and Jensen, 1995; Vogel and Jensen, 1997). NusB and NusG have been identified in isolated rrn antiterminated complexes (Li et al., 1992) and are also required for the _boxA_-dependent increase in transcription elongation rate (Zellars and Squires, 1999). Mason et al. (1992) and Nodwell and Greenblatt (1993) have shown that NusB and NusE dimerize and bind the rrn boxA sequence in vitro, while NusA, NusE and NusG bind core RNA polymerase directly (Greenblatt and Li, 1981; Mason and Greenblatt, 1991; Li et al., 1992). These observations prompted us to attempt to reconstitute the ribosomal antitermination system in vitro using purified Nus factors and RNA polymerase (Squires et al., 1993). We showed that the addition of all four Nus factors and unidentified cellular factor(s), provided in an S100 extract, were necessary for optimal read-through of the _trp t_′ Rho-dependent terminator in a _boxA_-dependent manner. In the work presented here, we have examined the influence of four ribosomal proteins (in addition to NusE) on rRNA transcription. The details of the isolation of these r-proteins in antiterminated transcription complexes (C.Condon, M.Torres and C.L.Squires, unpublished data) will be presented elsewhere. One of the identified r-proteins, S4, was studied here in detail and was shown to bind directly to RNA polymerase and to display clear antitermination activity in vitro. This prompts us to propose a model in which key r-proteins, such as S4, play a dual role in the regulation of both r-protein and rRNA synthesis.
Results
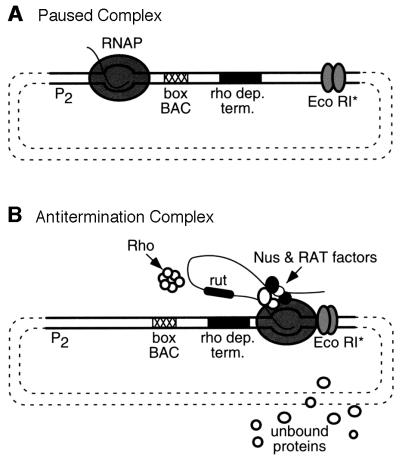
Using a template containing the boxBAC antitermination signal (AT) from rrnG and a Rho-dependent terminator, we showed in previous experiments that reconstitution of the rrn antitermination mechanism was possible in vitro (Squires et al., 1993). In the presence of Rho, addition of purified Nus factors and an S100 extract was necessary for optimal terminator read-through by RNA polymerase. We devised a strategy to isolate stable ‘antiterminating’ transcription complexes for the identification of essential antitermination factor(s) present in the S100 extract. This strategy exploited a mutant of the restriction endonuclease _Eco_RI, _Eco_RI*, which binds tightly to its restriction site, but does not cleave the DNA. Pavco and Steege (1990) have shown that _Eco_RI* serves as a roadblock for RNA polymerase without causing its dissociation from the template. We predicted that, using _Eco_RI*, we could isolate antitermination complexes stalled on the DNA template by virtue of their size. We used this system, shown in Figure 1, to isolate and identify five proteins by N-terminal sequencing that may be involved in rrn antitermination: the r-proteins S4, L3, L4 and L13, and the DNA binding protein, H-NS (M.Condon, C.Torres, and C.Squires, unpublished data). We assumed H-NS was present because it binds to DNA and did not analyze its role further.
Fig. 1. Antitermination complexes. (A) Paused complex. Step 1 of in vitro transcription reaction. A paused complex is formed by ‘walking’ RNA polymerase out to +6 by addition of the first two template encoded nucleotides. The plasmid template contains the rrnG P2 promoter (P2), the rrn antiterminator motif (boxBAC), the _trp t_′ Rho-dependent terminator (rho dep. term.) and a downstream _Eco_RI site. The coordinates of the fragment containing the rrnG P2 promoter are –88 to +3, relative to the initiating nucleotide. The boxBAC sequence corresponds to coordinates +4 to +64 of the transcript originating from rrnG P2. The mature 16S rRNA starts at position +174. The thick lines represent the PCR template used in all subsequent in vitro transcription reactions. RNA polymerase (RNAP), the short initial transcript and the _Eco_RI* mutant dimer are shown. (B) Antitermination complex. Step 2 of in vitro transcription reaction. The remaining two nucleotides, Nus factors and crude extract are added to the reaction to allow the formation of stalled ternary (RNA, DNA, protein) complexes at the _Eco_RI site. rut is the Rho utilization site for Rho binding. RAT factors represent the additional unknown factors required for rRNA antitermination. Proteins from the S100 extract that do not bind the complex are presented as the ‘unbound proteins’.
Assay of r-proteins in in vitro transcription reactions
We obtained purified samples of the r-proteins identified in the antitermination complex and assayed them individually to determine their influence on terminator read-through. Linear DNA template fragments containing the rrnG P2 promoter and boxBAC element and the _trp t_′ terminator were used. In vitro transcription reactions in the presence of Rho showed that, individually, S4, L3 and L13 all decreased Rho-dependent transcription termination, while L4 had no activity on its own (data not shown). In this study, we analyze the influence of r-protein S4 on transcription termination. The effect of r-proteins L3 and L13 will be reported elsewhere.
The addition of S4 to in vitro transcription reactions resulted in an increased amount of full-length transcript, a decreased amount of Rho-dependent termination and a shift in the Rho-dependent termination pattern towards higher molecular weight transcripts (Figure 2). Titration with S4 resulted in increased terminator read-through as the concentration of S4 increased. At 230 nM S4, there was a 6-fold (12 versus 2%) increase in read-through; 470 nM S4 caused a 15-fold (31 versus 2%) increase (Figure 2, compare lanes 1 and 6). These observations suggest that r-protein S4 functions as an antitermination factor.
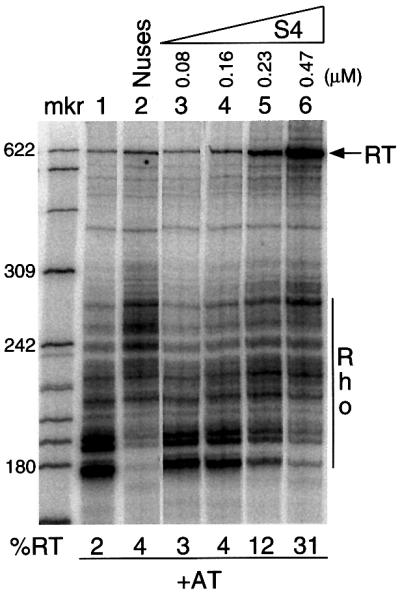
Fig. 2. Titration of S4 in the in vitro transcription system. Direction of increasing S4 concentration is represented by right-angled triangle. The template used contained the boxBAC motif (+AT). All reactions contained Rho. ‘RT’ refers to the read-through band. The percentage read-through (%RT) was calculated from the ratio of the intensity of the RT band to the total intensity, i.e. RT and termination bands. ‘Rho’ refers to the multiple Rho-dependent termination bands. Nuses used contained 219 nM NusA, 848 nM NusB, 1.02 µM NusE and 570 nM NusG. mkr is an end-labeled _Hpa_II digest of pBR322 DNA.
Combination of r-proteins and Nus factors increase Rho-dependent terminator read-through
We have previously shown that addition of four Nus factors and an S100 extract result in 100% read-through of the _trp t_′ terminator in in vitro transcription assays (Squires et al., 1993). To determine whether the r-proteins S4, L3, L4 and L13 represent all of the S100 factors necessary for complete terminator read-through, we analyzed the four newly identified r-proteins together with the four Nus factors (Figure 3). In the absence of Rho and added S100 factors, ∼78% of RNA polymerase molecules read-through the terminator (lane 1). This value was reduced to 5% upon addition of Rho (lane 2). Addition of a roughly equimolar mixture of the r-proteins in the presence of Rho increased terminator read-through 8-fold, to 39% (lane 4), and this value could be increased further to 54% (11-fold) upon addition of an equimolar mixture of the four Nus factors (lane 7). This value still falls below the maximal amount of read-through possible, 78%, indicating that while both the Nus factors and the r-proteins contribute significantly, the system is not yet optimized. It is possible that we have not yet identified all of the necessary antitermination factors; there were bands on the gel of the antitermination complex that did not give a homogenous N-terminal sequence. It is also possible that larger proteins were not transferred to the membrane or that we have not yet identified the optimal factor stoichiometry or reaction conditions.
Fig. 3. Influence of four r-proteins and four Nus factors on Rho-dependent terminator read-through. The 1× mixture of r-proteins contained 300 nM S4, 217 nM L4, 538 nM L3 and 270 nM L13. The preparation of L3 contained ∼50% of r-protein L6; therefore, the actual amount of L3 present was ∼270 nM. The 1× Nus factor mix (Nuses) contained 292 nM NusA, 464 nM NusE, 380 nM NusG and 566 nM NusB. All reactions contained Rho. The template used in the reactions in lanes 1–7 contained the antiterminator (+AT) and the template in lanes 8–14 lacked the antiterminator motif (–AT). ‘Rho’ refers to the multiple Rho-dependent termination bands. The percent read-through is given at the bottom of the gel and was calculated as described for Figure 2.
A template lacking the AT motif (–AT) also gave an increased amount of full-length transcript with the addition of the r-proteins and the combination of the r-proteins with the Nus factors (Figure 3, compare lane 9 with lanes 11, 13 and 14). The increase in read-through, although substantial, was not altered by the addition of higher concentrations of the proteins (compare lanes 13 and 14). We interpreted these results as showing that some factor(s) in the mixture had a general (AT motif-independent) antitermination effect on Rho-dependent termination (see below).
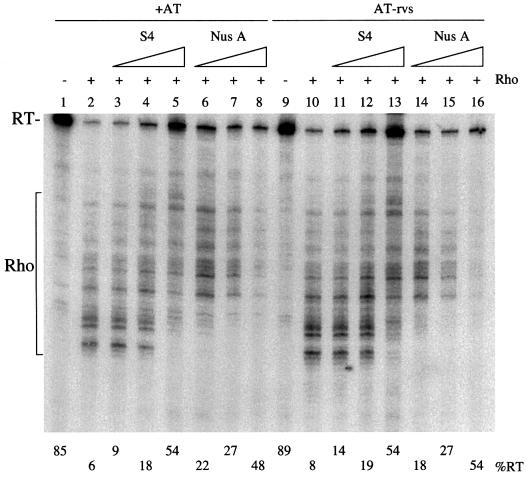
S4 and NusA have similar, antagonistic effects on Rho-dependent termination
As noted above, in addition to causing increased terminator read-through, S4 caused a shift in the Rho-dependent termination pattern towards higher molecular weight bands (Figure 2). This was reminiscent of what we and others (Squires et al., 1993; Burns et al., 1998) had previously observed upon addition of NusA to in vitro transcription reactions. We therefore performed in vitro transcription reactions where we directly compared the effect of S4 and NusA on Rho-dependent termination (Figure 4). These reactions were carried out using templates containing the AT motif in either the normal or reverse orientation (AT-rvs). Although NusA had a slightly inhibitory effect on transcription at higher concentrations, S4 and NusA had similar effects on Rho-dependent termination, increasing the size of the Rho-dependent termination bands and increasing terminator read-through (compare lanes 3–5 with lanes 6–8). Because we do not know the concentration of ‘active’ protein in the samples, this is only a qualitative assay and does not necessarily reflect the relative strengths of NusA and S4 as antitermination factors. Antitermination was independent of the AT motif, as very similar increases in read-through values were obtained with the AT-rvs template (lanes 11–16) and the –AT template (data not shown). These results suggest that, although S4 was isolated from an rRNA antitermination complex, it acts as a general transcription antitermination factor, like NusA.
Fig. 4. Comparison of the effect S4 and NusA in in vitro transcription reactions. Reactions containing Rho are indicated with a + sign. The percentage read-through is given at the bottom of the autoradiogram. The templates used contained the AT motif either in the correct (+AT) or reverse (AT-rvs) orientation. The read-through and Rho-dependent termination bands are shown to the side of the gel. S4 and NusA were added at 84, 253 and 760 nM, with the increasing concentrations depicted by the right-angled triangle.
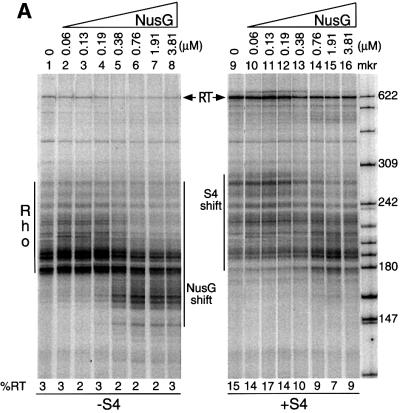
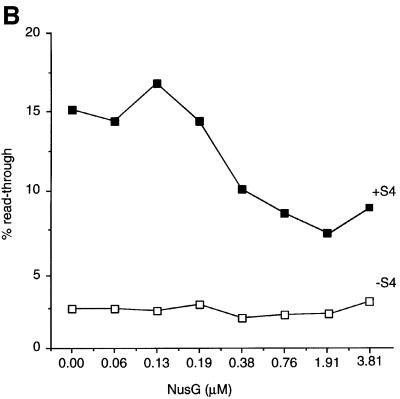
NusG decreases S4-mediated antitermination
NusG was revealed as a necessary component of the lambda N/nut antitermination system in early in vitro studies (Li et al., 1992), and we have evidence that NusG is required for rrn antitermination in vivo (our unpublished results). Paradoxically, however, NusG has been shown to increase the efficiency of some Rho-dependent terminators and cause early termination, as seen by a decrease in the average size of the termination bands, in vitro (Nehrke et al., 1993; Burns et al., 1998; Figure 5A, compare lanes 1 and 5). Thus, NusG plays a role in both termination and antitermination. Because of this dual function of NusG, we examined the consequence of addition of NusG to in vitro transcription reactions containing S4. The addition of increasing concentrations of NusG to in vitro transcription reactions caused a concomitant decrease in S4-mediated terminator read-through (Figure 5A, lanes 9–16). In the absence of S4, increasing concentrations of NusG had no effect on terminator read-through (Figure 5A, lanes 1–8), although the base read-through level (∼2.5%) may be too low to see such effects. Thus, while we believe that NusG directly antagonizes the antitermination activity of S4, we can not exclude the possibility that NusG has a direct effect on RNA polymerase in these experiments. Although the effect of NusG is small, at most 2-fold, for a 4-fold excess of NusG (Figure 5B), this result highlights the importance of the stoichiometry of the different factors in the antitermination reaction.
Fig. 5. Titration of NusG in the presence and absence of S4. The template used contained the antiterminator. All reactions contained Rho. The direction of increasing NusG concentration is represented by right-angled triangles. The increase or decrease in the average molecular weight of the termination bands is indicated as ‘S4 shift’ or ‘NusG shift’, respectively. ‘Rho’ refers to the multiple Rho-dependent termination bands. ‘RT’ refers to the read-through band. (A) (left) The titration of NusG alone; (right) the titration of NusG in the presence of 468 nM S4. (B) Quantitation of gels in (A) represented as line graphs.
The S4 antitermination activity is specific for Rho-dependent terminators
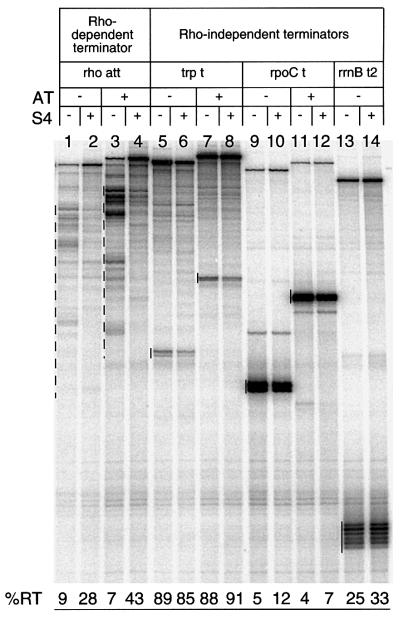
While the lambda antitermination mechanism promotes read-through of both Rho-dependent and Rho-independent terminators, we have previously shown that the rrn antitermination mechanism is most efficient with terminators requiring Rho action (Albrechtsen et al., 1990). We thus wished to see whether the terminator read-through activity of S4 was also specific to Rho-dependent terminators. To this end, we compared the effect of S4 on another Rho-dependent terminator (the rho attenuator; Figure 6, lanes 1–4) and three Rho-independent terminators (trp t, rpoC t and rrnB t2; Figure 6, lanes 5–14) in the presence or absence of boxBAC in vitro. The addition of S4 significantly increased terminator read-through of the rho attenuator, and further enhanced it if boxBAC was present on the template (Figure 6, lanes 3 and 4). At the same time, only minor effects were seen with the three Rho-independent terminators tested, suggesting that, like the rrn antitermination mechanism, S4 had a preference for Rho-dependent terminators.
Fig. 6. Effect of S4 on Rho-dependent and Rho-independent terminators in the presence or absence of antiterminator motif. Templates were created by PCR using the following plasmids: pHBA41 (–AT) and pHBA42 (+AT) to create the rho att templates; pHBA14 (–AT) and pHBA15 (+AT) to create the trp t templates; pHBA24 (–AT) and pHBA25 (+AT) to create the rpoC t templates; and pSL206 (–AT) to create the rrnB t2 template. All plasmids are from Albrechtsen et al. (1990). The primers were the same as those used to create the _trp t_′ template described in Materials and methods. The S4 concentration was 1.5 µM. Termination bands are marked with dashed (Rho-dependent) or solid (Rho-independent) lines.
S4 is associated with RNA polymerase in vivo and binds to it directly in vitro
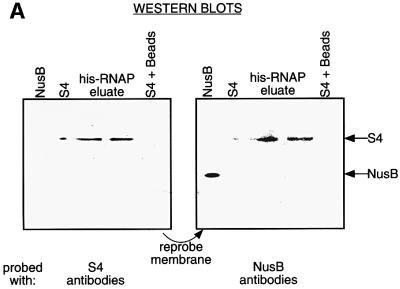
The similarity between the modes of action of S4 and NusA at Rho-dependent terminators suggested that S4, like NusA might be capable of binding RNA polymerase (Greenblatt and Li, 1981). To examine this possibility, we prepared an extract of a strain over-expressing His-tagged RNA polymerase, and incubated it with nickel beads. After repeated washes, bound proteins were eluted from the column, run on an SDS–PAGE gel and examined for the presence of S4 by western blot analysis (Figure 7A). S4 was detected in the final elution samples, while a Nus factor not known to bind RNA polymerase, NusB, was not. S4 does not bind to His-tagged yeast FIP protein over-expressed in E.coli, showing that S4 does not simply bind avidly to any His-tagged protein (Figure 7B). This result suggested that S4 bound tightly and specifically to RNA polymerase, although it did not permit us to determine whether this association was direct or indirect, via another protein or RNA.
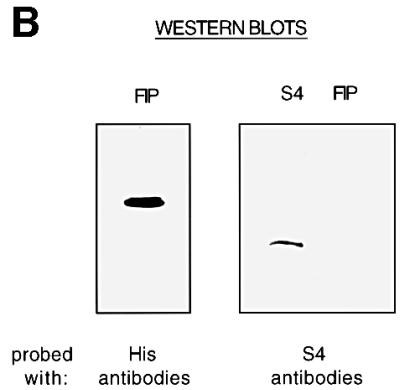
Fig. 7. S4 binds RNA polymerase in vivo. (A) A western blot of proteins eluted from nickel beads incubated with a whole-cell extract of a strain producing His-tagged RNA polymerase. Reference lanes of NusB and S4 are included. The membrane was first probed with S4 antibodies (left), and then re-probed with NusB antibodies (right). (B) A western blot of proteins eluted from nickel beads incubated with a whole-cell extract of a strain producing His-tagged FIP. Identical membranes were probed with anti-His antibodies (left) or anti-S4 antibodies (right).
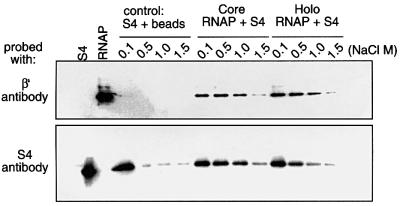
To determine whether S4 could bind RNA polymerase in vitro in the absence of additional proteins or RNA, His-tagged holo and core RNA polymerases were first reconstituted from purified components. We then mixed S4 with these polymerase preparations and nickel agarose beads, and analyzed the eluate after the different salt washes for the presence of S4 by western blot analysis. This experiment showed that at 1 M NaCl, S4 remained bound to both core and holo polymerases (Figure 8), and indicated that S4 binds to RNA polymerase without the aid of other factors. This result argues for a direct interaction of S4 with RNA polymerase, rather than with RNA potentially co-purifying with the enzyme in vivo. The experiment also showed that S4 binds tenaciously to both core and holo forms of the enzyme. This is in contrast to NusA, which only binds to RNA polymerase after sigma-factor has cycled off the complex (Li et al., 1992). Thus, although both NusA and S4 interact directly with RNA polymerase and have very similar effects on Rho-dependent termination, their modes of action may be different.
Fig. 8. S4 binds reconstituted RNA polymerase in vitro. His-tagged core or holo RNA polymerase was mixed with purified S4 and the mixture was absorbed onto nickel agarose beads. The beads were washed with the concentrations of NaCl shown, samples eluted with EDTA and run on a 12% SDS–PAGE gel. The gel was then blotted onto a PVDF membrane. The membrane was cut in half and the upper half was probed with mAbs against the β′ subunit of RNA polymerase, and the lower half was probed with polyclonal antibodies against S4. The control was a mix of nickel beads + S4 alone, treated similarly.
Discussion
The addition of a combination of four r-proteins, and the four Nus factors to in vitro transcription reactions, using templates containing the rrnG boxBAC antitermination motif upstream of the _trp t_′ Rho-dependent terminator, led to an 11-fold increase in terminator read-through. In this work, close examination of the individual contribution of each of the r-proteins suggested that a large portion of the antiterminator effect could be attributed to the presence of S4. We further characterized the role of S4 and showed that it acts as a general antitermination factor, with properties very similar to NusA, in that it binds tightly to RNA polymerase and provokes read-through of Rho-dependent terminators in a _boxBAC_-independent fashion. These results suggest that S4 and possibly other r-proteins, in addition to S10 (NusE), are important components of the rrn antitermination system.
The presence of ribosomal subunit proteins in the rrn transcription antitermination complex is perhaps not surprising in hindsight. The participation of r-protein S10 (NusE) in lambda N/nut antitermination is well documented (Das et al., 1985; Horowitz et al., 1987). It has also been shown to bind directly to RNA polymerase (Mason and Greenblatt, 1991). Its role in rrn antitermination has not been characterized to the same extent, but S10 has been shown to form heterodimers with NusB in vitro that bind to the rrn boxA sequence. The r-protein S1, can also bind boxA, interfering with NusB–E binding (Mason and Greenblatt, 1992; Nodwell and Greenblatt, 1993; Mogridge and Greenblatt, 1998), and this S1 interference raises the intriguing possibility of a negative regulatory role for S1 in rrn antitermination in vivo. In this context, the discovery that other r-proteins may be involved in rrn antitermination is not so unusual.
Three of the four r-proteins identified in the antitermination complex are known to bind directly to rRNA (L3, L4 and S4), and two of them, S4 and L4, regulate the expression of their own operons (Noller and Nomura, 1996). S4 is one of the first proteins to interact with nascent 16S rRNA and initiate the proper folding and assembly of the 30S ribosomal subunit (Noller and Nomura, 1996). It interacts with a 460 nucleotide fragment of 16S rRNA [nucleotides 39–500 of the mature sequence, none of which is present on the RNAs produced in our _in vitro_ assay system (Vartikar and Draper, 1989)]. S4 is the regulatory protein for translational control of the α-operon, a complex operon containing genes for four r-proteins and the α-subunit of RNA polymerase (Keener and Nomura, 1996). S4 also has the unusual property of binding to RNA pseudoknot structures, making its interactions with RNA somewhat special (Tang and Draper, 1989). Recent crystal structure work on the S4 protein from Bacillus stearothermophilus showed that it contains an ETS structural domain, a DNA binding motif normally found in regulatory transcription factors in eukaryotes (Davies et al., 1998). L4 is also a regulatory r-protein. In addition to its normal role in binding 23S rRNA, it is involved in a complex set of interactions necessary for both NusA-dependent transcription termination (Sha et al., 1995) and translational control of the S10 (NusE) operon, an operon that contains 11 r-protein genes (Zengel et al., 1980; Zengel and Lindahl, 1993). L3, another primary rRNA binding protein, is also located within this same operon. Indeed, the first three genes of this operon are those of S10, L3 and L4, in that order (Lindahl et al., 1990). L3 can be cross-linked to L13 on the ribosome, suggesting a close physical association between these two proteins (Walleczek et al., 1989), which may, or may not, be relevant to antitermination complex formation. L13 is located in a short operon, containing only itself and S9, and regulation of this operon has not been studied in detail.
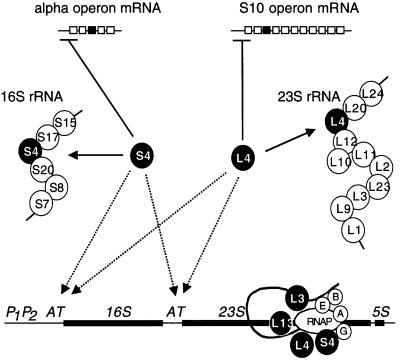
The participation of at least two regulatory r-proteins in rrn transcription antitermination is of great significance to current models on the coordinated production of rRNA and r-proteins. r-protein operons are regulated by a feedback repression mechanism, where one of the encoded r-proteins, which binds to rRNA, also binds its own mRNA to inhibit its translation under conditions of excess. Up to now, it has generally been believed that because of this phenomenon, the production of r-proteins will be determined automatically by the synthesis of rRNA in the cell, and that this suffices to explain how imbalances between r-protein and rRNA synthesis are redressed. Our data suggest that there may also be a flow of information in the opposite direction, i.e. r-protein to rRNA (Figure 9). Excess r-proteins such as S4, and possibly L4, in addition to shutting off expression of their own operons, may simultaneously increase the synthesis of functional rRNA through transcription antitermination, permitting a more rapid return to homeostasis. Overproduction of S4 has indeed been shown to stimulate rRNA synthesis (Takabe et al., 1985). Although this effect is probably primarily a result of feedback de-repression of rRNA synthesis caused by inhibition of ribosome assembly, resulting from the decrease in α-operon expression, a contribution from a direct stimulatory effect of S4 on rrn operon transcription would also be consistent with the increase in rRNA synthesis observed. We thus propose the addition of a new and previously unsuspected regulatory loop to current models on the regulation of ribosome synthesis. Its role is presumably to fine-tune the coordination of the production of r-proteins and rRNA.
Fig. 9. Model of how regulatory r-proteins present in the anti termination complex could act both positively and negatively to restore r-protein and rRNA imbalances. Regulatory r-proteins, S4 and L4, that are in excess over rRNA will simultaneously decrease expression of their own operons by translational feedback control (solid lines), and by increased synthesis of rRNA caused by stimulated assembly of antitermination complexes at the leader and spacer AT motifs. The dashed arrows represent this proposed new control loop. Schematic structures of the alpha and S10 r-protein operons are shown, with the black boxes representing the respective coding sequences for S4 and L4, while the horizontal under-bars represent the binding sites for S4 and L4 repression. The 16S and 23S rRNAs are also shown associated with the known primary binding proteins (Noller and Nomura, 1996). The scheme shows the proposed complex, with A, B, E and G referring to the four Nus factors. While this study has shown that S4, like S10 (S10 = NusE), binds directly to RNAP, it remains unknown how r-proteins L4, L3 and L13 are associated with the antitermination complex.
Several studies have suggested a role for the rrn AT sequence in ribosome assembly, more so than in antitermination (Theißen et al., 1990; Pardon and Wagner, 1995; Balzer and Wagner, 1998). The identification of several r-proteins with rRNA binding capabilities in the antitermination complex suggests a way in which these two seemingly unrelated phenomena might be reconciled. It is not inconceivable that the antitermination complex, in addition to ensuring unimpeded transcription to the end of the 16S and 23S genes, serves to ‘deliver’ these r-proteins to their binding sites on the nascent transcript. Such a mechanism might be predicted to considerably aid ribosome assembly by increasing local concentrations of r-proteins and by ‘locking-in’ certain structures as soon as they are transcribed.
By adjusting the concentration of proteins in our in vitro transcription reactions, we achieved up to 70% of maximal possible terminator read-through using a combination of r-proteins S4, L3, L4 and L13, and Nus factors A, B, E and G. These may, in fact, be all of the factors that are required for rrn antitermination, and further optimization of the reaction conditions will result in 100% read-through. However, it is quite likely that additional factors may be necessary to achieve the same terminator read-through that is possible with a fresh S100 extract (Squires et al., 1993). When we examined the contribution of each of the r-proteins individually, it became clear that the bulk of the antitermination effect could be accounted for by the presence of S4. Other primary rRNA binding proteins that have a great avidity for RNA, such as S15 or L4, had no significant effect on terminator read-through on their own (data not shown), suggesting that S4 has antitermination activity in the strict sense, rather than simply by ‘coating’ the RNA and thereby blocking Rho access.
The terminator suppression activity of S4 closely resembles that of NusA in several regards. First, both S4 and NusA altered the Rho-dependent termination pattern identically, increasing the average size of the termination bands. Secondly, both S4 and NusA exhibited antitermination activity with control templates, suggesting that both are general antitermination factors. We suspected that, in order for S4 to exercise its antitermination activity, it would have to interact with either Rho or RNA polymerase. Given the similarity between the modes of action of S4 and NusA at Rho-dependent terminators it was not surprising that, like NusA, S4 turned out to bind RNA polymerase. S4 was found bound to His-tagged RNA polymerase in whole-cell extracts, suggesting that this factor is associated with RNA polymerase during normal transcription, consistent with the idea that it serves as an antitermination factor in the absence of boxBAC sequences. Stringent washes were required to remove S4, suggesting that the binding of this protein to RNA polymerase is much stronger than the binding of NusE or NusG, other Nus factors known to associate with RNA polymerase (Mason and Greenblatt, 1991; Li et al., 1992)
The identification of four ribosomal subunit proteins, in addition to NusE, in the rrn antitermination complex adds significantly to the list of r-proteins with extra-ribosomal functions, in both prokaryotes and eukaryotes (for reviews see Wool, 1996; Squires and Zaporojets, 2000). Escherichia coli r-protein S1 has been shown to be involved in a host of cellular and phage processes including poly(A)-RNA binding (Kalapos et al., 1997), Qβ phage replication (Miranda et al., 1997) and stimulation of the T4 _Reg_B endonuclease activity (Jayasena et al., 1996), and, as mentioned above, it also binds the rrn boxA motif, inhibiting NusB and NusE binding (Mogridge and Greenblatt, 1998). L14 stimulates the Rep helicase in bacteriophage replication (Yancey and Matson, 1991), S12 is involved in phage T4 intron splicing in vitro (Coetzee et al., 1994) and S9 participates with UmuC in the SOS DNA repair process (Woodgate et al., 1989). These observations support the interesting argument that the r-proteins were co-opted from a set of proteins that had a role in different cellular processes at least one time in their evolutionary past.
Materials and methods
In vitro transcription reactions
Two primers (upstream primer 5′-TGAAAATCTCGTCGAAGCTCGG GGCGG-3′; downstream primer 5′-CGGATTTGAACGTTGCGAAGC AACG-3′) were used to synthesize templates by PCR from pHBA17 (Albrechtsen et al., 1990), pRATT-1 (Squires et al., 1993) and pCAB (this work). The first plasmid contains the rrnG P2 promoter (–88 to +3, relative to transcription start) and a 250 nucleotide fragment containing the _trp t_′ Rho-dependent terminator. The second plasmid is identical to pHBA17 except it has a _Cla_I–_Bam_HI fragment containing the rrnG antiterminator boxBAC (+4 to +64, relative to transcription start) cloned between the _Cla_I–_Bam_HI site. pCAB is identical to pRATT-1 except that it has the 61 nucleotide boxBAC sequence inserted in the reverse orientation downstream of rrnG P2. Step 1: the in vitro transcription reactions contained 50–60 nM PCR fragment, 41.8 nM RNA polymerase (Epicentre) or 20 nM RNA polymerase (Sigma), 62 nM _Eco_RI*, 100 µM CpC (Sigma), 2 µM CTP, GTP and ATP (Pharmacia), 12 U of RNasin (Promega) and 2 µCi of [α-32P]GTP (NEN). The reaction volume was brought up to 12.5 µl with Buffer A [20 mM Tris–glutamate pH 8.0, 5 mM magnesium glutamate, 100 mM potassium glutamate, 5% glycerol and 1 mM dithiothreitol (DTT)] (DeVito and Das, 1994). Reactions were incubated for 6 min at 30°C. Step 2: a mix of UTP, GTP and CTP ribonucleotides was added to a final concentration of 100 µM, ATP was added to a final concentration of 4 mM and rifampicin was added to a final concentration of 10 ng/µl. In the reactions that required Rho, a final concentration of 22.4 nM of hexamers was added. The amount of Nus factors or r-proteins added to the reactions is given in the figure legends. The volume was then brought up to 25 µl with Buffer A. The reactions were incubated at 30°C for 5 min and stopped by adding 100 µl of a mix containing 0.1 M sodium acetate pH 5.2, 0.4% sodium dodecylsulfate and 1.3 mg of carrier yeast tRNA per ml. The samples were extracted with phenol–chloroform, ethanol-precipitated and resuspended in 4 µl of formamide plus dyes. The samples were boiled for 3 min, then loaded on 6% polyacrylamide, 7 M urea gels and electrophoresed. The gels were then dried, scanned by PhosphoImager (Storm; Molecular Dynamics) and the desired bands quantified by the ImageQuant program.
Isolation of His-tagged RNA polymerase from cell extracts
We prepared an extract of XL1 Blue strain containing plasmid pMKA201(Kashlev et al., 1993). This plasmid carries a His-tagged rpoC gene under the control of a lactose promoter. The strain also has pACYC184 bearing _lacI_q. Cells were grown at 37°C in LB + 100 µg/ml of ampicillin to an OD600 of 0.4 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2.5 h. The cells were spun at 13 000 g for 10 min and washed with cold phosphate buffered saline (PBS). The pellet was resuspended in half the culture volume of TSE buffer (100 mM Tris–acetate pH 8.2, 0.5 M sucrose, 5 mM EDTA). An equal volume of lysozyme (0.16 mg/ml) in cold magnesium sulfate (3.6 mM) was added to the suspension followed by 5 min incubation on ice. Spheroplasts were pelleted by centrifuging for 10 min at 13 000 g. The pellets were resuspended in a half volume of KI buffer (50 mM Tris–chloride pH 8.0, 150 mM sodium chloride, 2% Triton X-100, 1 mM EDTA), phenylmethylsulfonyl fluoride to 40 µg/ml, and aprotinin and leupeptin (Sigma) to 0.5 µg/ml each. Undisrupted cells were removed by centrifugation. The supernatant was dialyzed overnight against Buffer B (50 mM sodium phosphate pH 8.0, 300 mM NaCl). Five milliliters of lysate were incubated with 80 µl of equilibrated nickel agarose beads (Ni-NTA resin from Qiagen) with rotation overnight at 4°C. The beads were spun down, washed four times with 500 µl of Buffer B, washed four times with Buffer C (Buffer B at pH 6.0) and eluted with Buffer D (Buffer C plus 0.5 M imidazole). The samples were concentrated with Centricon-3 (Amicon), prepared for electrophoresis on a 12% SDS–PAGE gel, transferred to a nitrocellulose membrane and probed with antibodies against S4, NusB, NusE and NusG. Polyclonal antiserum against S4 was a gift from M.Nomura and NusG polyclonal antiserum was a gift from B.Stitt. Polyclonal antiserum to NusB and NusE were obtained from Biodesign International.
In vitro interaction between His-tagged RNA polymerase and S4
E.Nudler kindly supplied us with holo and core-reconstituted His-tagged RNA polymerase. We incubated 7 µM His-tagged RNA polymerase with 38 µM S4 (provided by D.Draper) in a 10 µl volume at 30°C for 30 min. The volume was then brought up to 200 µl with Buffer B and 5 µl of washed nickel beads was added. The mix was incubated overnight with rotation at 4°C, spun down and washed three times with 1 ml of Buffer E (50 mM sodium phosphate pH 6.0, 1% glycerol, NaCl) containing either 0.1, 0.5, 1.0 or 1.5 M NaCl. The beads were transferred to another siliconized tube during the fourth wash, washed twice more with the same buffer and eluted with Buffer B plus 100 mM EDTA pH 8.0 plus 100 mM DTT. Loading dye was added and the samples were boiled and loaded on a 12% SDS–PAGE gel. The proteins were then blotted onto a PVDF membrane. The membrane was cut horizontally to allow individual probing of the top portion with β′ mAb, and the lower portion with S4 polyclonal antiserum. The β′ mAb was a generous gift from R.Burgess.
Western blot analysis
The protocol was from Bollag and Edelstein (1991). The samples were electrophoresed on SDS–PAGE gels for 45 min at 200 V. The proteins were then transferred onto a PVDF membrane according to manufacturer’s instructions (Bio-Rad). The membrane was blocked using 5% non-fat dry milk prepared in 1× Tris-buffered saline (TBS; 30 mM Tris–chloride pH 7.4, 150 mM NaCl) for 1 h at room temperature. A dilution of the rabbit serum or mAb in blocking solution was then added to the membrane. The membrane was incubated for 1 h at room temperature and then washed with 1× TBSTT (same as 1× TBS, but with 0.05% Tween-20 and 0.2% Triton X-100) three times for 10–15 min. After washing, a 1:10 000 dilution of the secondary antibody, goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) from Promega in 1× TBS, was added to the membrane. Incubation for 1 h at room temperature followed. The membrane was washed as before, and then rinsed in 1× TBS. The membrane was then incubated for 1 min in chemiluminescence reagent (ECL-Amersham Life Science) and exposed to film.
Acknowledgments
Acknowledgements
We thank M.Kashlev for plasmid pMKA201 containing a His-tagged rpoC gene, E.Nudler for purified His-tagged core and holo RNA polymerase, B.Stitt for antibodies to NusG and NusG protein and Rho protein, M.Nomura for r-proteins and antibodies to them, D.Draper for r-protein, S4, L.Lindahl for r-protein, L4, S.Helmling for the His-tagged FIP protein, D.Bear for a strain containing a mutated _Eco_RI*, J.Singh for assistance with the preparation of the _Eco_RI*, M.Gottesman for NusA, R.Zimmerman for NusE, R.Burgess for mAbs to RNA polymerase, B.Whalen for AD7333 and M.Springer for helpful comments on the manuscript. National Institutes of Health grant GM24751 to C.L.S supported this work.
References
- Adhya S. and Gottesman,M. (1978) Control of transcription termination. Annu. Rev. Biochem., 47, 967–996. [DOI] [PubMed] [Google Scholar]
- Aksoy S., Squires,C.L. and Squires,C. (1984) Evidence for antitermination in Escherichia coli rRNA transcription. J. Bacteriol., 159, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen B., Squires,C.L., Li,S. and Squires,C. (1990) Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J. Mol. Biol., 213, 123–134. [DOI] [PubMed] [Google Scholar]
- Balzer M. and Wagner,R. (1998) Mutations in the leader region of ribosomal RNA operons cause structurally defective 30S ribosomes as revealed by in vivo structural probing. J. Mol. Biol., 276, 547–557. [DOI] [PubMed] [Google Scholar]
- Berg K., Squires,C. and Squires,C.L. (1989) Ribosomal RNA operon antitermination: Function of leader and spacer region boxB_–_boxA sequences and their conservation in diverse micro-organisms. J. Mol. Biol., 209, 345–358. [DOI] [PubMed] [Google Scholar]
- Brewster J.M. and Morgan,E.A. (1981) _Tn_9 and _IS_1 inserts in a ribosomal ribonucleic acid operon of Escherichia coli are incompletely polar. J. Bacteriol., 148, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag D.M. and Edelstein,S.J. (1991) Immunoblotting. In Protein Methods. Wiley-Liss Inc., New York, NY, pp. 181–211.
- Burns C.M., Richardson,L.V. and Richardson,J.P. (1998) Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in _Escherichia coli._J. Mol. Biol., 278, 307–316. [DOI] [PubMed] [Google Scholar]
- Coetzee T., Herschlag,D. and Belfort,M. (1994) Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev., 8, 1575–1588. [DOI] [PubMed] [Google Scholar]
- Condon C., Squires,C. and Squires,C.L. (1995) Control of rRNA transcription in _Escherichia coli._Microbiol. Rev., 59, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Ghosh,B., Barik,S. and Wolska,K. (1985) Evidence that ribosomal protein S10 itself is a cellular component necessary for transcription antitermination by phage λ N protein. Proc. Natl Acad. Sci. USA, 82, 4070–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C., Gerstner,R.B., Draper,D.E., Ramakrishnan,V. and White,S.W. (1998) The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J., 17, 4545–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito J. and Das,A. (1994) Control of transcription processivity in phage λ: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl Acad. Sci. USA, 91, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R.L., Stark,M.J.R., and Dahlberg,A.L. (1983) Regions of DNA involved in the stringent control of plasmid encoded rRNA in vivo. Cell, 32, 1347–1354. [DOI] [PubMed] [Google Scholar]
- Greenblatt J. and Li,J. (1981) Interaction of the sigma factor and the nusA gene protein of E.coli with RNA polymerase in the initiation-termination cycle of transcription. Cell, 24, 421–428. [DOI] [PubMed] [Google Scholar]
- Heinrich T., Condon,C. Pfeiffer,T. and Hartmann,R.K. (1995) Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J. Bacteriol., 177, 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz R.J., Li,J. and Greenblatt,J. (1987) An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell, 51, 631–641. [DOI] [PubMed] [Google Scholar]
- Jayasena V.K., Brown,D., Shtatland,T. and Gold,L. (1996) In vitro selection of RNA specifically cleaved by bacteriophage T4 _Reg_B endonuclease. Biochemistry, 35, 2349–2356. [DOI] [PubMed] [Google Scholar]
- Kalapos M.P., Paulus,H. and Sarkar,N. (1997) Identification of ribosomal protein S1 as a poly(A) binding protein in Escherichia coli. Biochimie, 79, 493–502. [DOI] [PubMed] [Google Scholar]
- Kashlev M., Martin,E., Polyakov,A. Severinov,K., Nikiforov,V. and Goldfarb,A. (1993) Histidine-tagged RNA polymerase: dissection of the transcription cycle using immobilized enzyme. Gene, 130, 9–14. [DOI] [PubMed] [Google Scholar]
- Keener J. and Nomura,N. (1996) Regulation of ribosome synthesis. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1417–1428.
- Li J., Horwitz,R., McCracken,S. and Greenblatt,J. (1992) NusG, a new E.coli elongation factor required for processive antitermination of transcription by the N protein of phage λ. J. Biol. Chem., 267, 6012–6019. [PubMed] [Google Scholar]
- Li S., Squires,C.L. and Squires,C. (1984) Antitermination of E.coli rRNA transcription is caused by a control region segment containing lambda _nut_-like sequences. Cell, 38, 851–860. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Sor,F., Archer,R.H., Nomura,M. and Zengel,J.M. (1990) Transcriptional organization of the S10, spc and alpha operons of Escherichia coli. Biochim. Biophys. Acta., 1050, 337–342. [DOI] [PubMed] [Google Scholar]
- Mason S. and Greenblatt,J. (1991) Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev., 5, 1504–1512. [DOI] [PubMed] [Google Scholar]
- Mason S.W., Li,J. and Greenblatt,J. (1992) Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J. Mol. Biol., 223, 55–66. [DOI] [PubMed] [Google Scholar]
- Miranda G., Schuppli,D., Barrera,I., Hausherr,C., Sogo,J.M. and Weber,H. (1997) Recognition of bacteriophage Qbeta plus strand RNA as a template by Qβ replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol., 267, 1089–103. [DOI] [PubMed] [Google Scholar]
- Mogridge J. and Greenblatt,J. (1998) Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol., 180, 2248–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E.A. (1980) Insertions of _Tn_10 into an E.coli ribosomal RNA operon are incompletely polar. Cell, 21, 257–265. [DOI] [PubMed] [Google Scholar]
- Nehrke K.W., Zalatan,F. and Platt,T. (1993) NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr., 3, 119–133. [PMC free article] [PubMed] [Google Scholar]
- Nodwell J.R. and Greenblatt,J. (1993) Recognition of boxA anti terminator RNA by the E.coli antitermination factors NusB and ribosomal protein S10. Cell, 72, 261–268. [DOI] [PubMed] [Google Scholar]
- Noller H.F. and Nomura,M. (1996) Ribosomes. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 167–186.
- Pardon B. and Wagner,R. (1995) The Escherichia coli ribosomal RNA leader nut region interacts specifically with mature 16S RNA. Nucleic Acids Res., 23, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco P.A. and Steege,D.A. (1990) Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem., 265, 9960–9969. [PubMed] [Google Scholar]
- Pfeiffer T. and Hartmann,R.K. (1997) Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23S rRNA synthesis _in vivo._J. Mol. Biol., 265, 385–393. [DOI] [PubMed] [Google Scholar]
- Sha Y., Lindahl,L. and Zengel,J.M. (1995) Role of NusA in L4-mediated attenuation control of the S10 r-protein operon of Escherichia coli. J. Mol. Biol., 245, 474–485. [DOI] [PubMed] [Google Scholar]
- Sharrock R.A., Gourse,R.L. and Nomura,M. (1985) Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of _Escherichia coli._Proc. Natl Acad. Sci. USA, 82, 5275–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C.L. and Zaporojets,D. (2000) Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol., 54, 775–798. [DOI] [PubMed] [Google Scholar]
- Squires C.L., Greenblatt,J., Li,J. Condon,C. and Squires,C. (1993) Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl Acad. Sci. USA, 90, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabe Y., Miura,A., Bedwell,D., Tam,M. and Nomura,M. (1985) Increased expression of ribosomal genes during inhibition of ribosome assembly in Escherichia coli. J. Mol. Biol., 184, 23–30. [DOI] [PubMed] [Google Scholar]
- Tang C.K. and Draper,D.E. (1989) Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell, 57, 531–536. [DOI] [PubMed] [Google Scholar]
- Theißen G., Behrens,S.E. and Wagner,R. (1990) Functional importance of the Escherichia coli ribosomal RNA leader boxA sequence for post-transcriptional events. Mol. Microbiol., 4, 1667–1678. [DOI] [PubMed] [Google Scholar]
- Vartikar J.V. and Draper,D. (1989) S4–16S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J. Mol. Biol., 209, 221–234. [DOI] [PubMed] [Google Scholar]
- Vogel U. and Jensen,K.F. (1995) Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J. Biol. Chem., 270, 18335–18340. [DOI] [PubMed] [Google Scholar]
- Vogel U. and Jensen,K.F. (1997) NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem., 272, 12265–12271. [DOI] [PubMed] [Google Scholar]
- Walleczek J., Martin,T., Redl,B., Stoffler-Meilicke,M. and Stoffler,G. (1989) Comparative cross-linking study on the 50S ribosomal subunit from Escherichia coli. Biochemistry, 28, 4099–4105. [DOI] [PubMed] [Google Scholar]
- Woodgate R., Rajagopalan,M., Lu,C. and Echols,H. (1989) UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD’. Proc. Natl Acad. Sci. USA, 86, 7301–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool I. (1996) Extra-ribosomal functions of ribosomal proteins. Trends Biochem. Sci., 21, 164–165. [PubMed] [Google Scholar]
- Yancey J.E. and Matson,S.W. (1991) The DNA unwinding reaction catalyzed by Rep protein is facilitated by an RHSP-DNA interaction. Nucleic Acids Res., 19, 3943–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellars M. and Squires,C.L. (1999) Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol., 32, 1296–1304. [DOI] [PubMed] [Google Scholar]
- Zengel J. and Lindahl,L. (1993) Domain I of 23S rRNA competes with a paused transcription complex for ribosomal protein L4 of Escherichia coli. Nucleic Acids Res., 21, 2429–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J.M., Mueckl,D. and Lindahl,L. (1980) Protein L4 of the E.coli ribosome regulates an eleven gene r protein operon. Cell, 21, 523–535. [DOI] [PubMed] [Google Scholar]